Abstract
Structures of prM-containing dengue and yellow fever virus particles were determined to 16 and 25 Å resolution, respectively, by cryoelectron microscopy and image reconstruction techniques. The closely similar structures show 60 icosahedrally organized trimeric spikes on the particle surface. Each spike consists of three prM:E heterodimers, where E is an envelope glycoprotein and prM is the precursor to the membrane protein M. The pre-peptide components of the prM proteins in each spike cover the fusion peptides at the distal ends of the E glycoproteins in a manner similar to the organization of the glycoproteins in the alphavirus spikes. Each heterodimer is associated with an E and a prM transmembrane density. These transmembrane densities represent either an EE or prMprM antiparallel coiled coil by which each protein spans the membrane twice, leaving the C-terminus of each protein on the exterior of the viral membrane, consistent with the predicted membrane-spanning domains of the unprocessed polyprotein.
Keywords: conformational changes/dengue virus/EM reconstruction/immature particles
Introduction
The Flaviviridae are a family of icosahedral enveloped viruses that infect vertebrates and frequently cause serious, often lethal, infections in humans. The family has been divided into the flavivirus, pestivirus and hepacivirus genera. The flaviviruses, which include dengue virus, West Nile virus, yellow fever virus and tick-borne encephalitis virus (TBEV), rely on insects for their transmission between vertebrate hosts. Although flaviviruses have a similar structural organization to that of the alphaviruses (family Togaviridae), they have a different gene order and utilize a different strategy for genome replication and virion assembly (Burke and Monath, 2001; Lindenbach and Rice, 2001). The four serotypes of dengue virus collectively cause more than 50 million human infections per year. Uncomplicated dengue fever is predominantly a self-limiting febrile illness, but in a small proportion of cases there are more serious manifestations called dengue hemorrhagic fever and dengue shock syndrome. No effective vaccine or antiviral drug therapy is currently available against dengue virus. Although there is an excellent vaccine against yellow fever virus (Monath, 1996), it is still endemic and epidemic in many parts of the world.
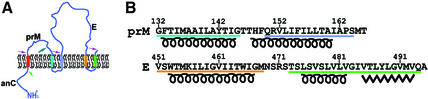
Mature infectious flaviviruses are icosahedral, ∼500 Å in diameter (Kuhn et al., 2002) and contain a positive-strand RNA genome of 10.7 kb with a single long open reading frame that is translated as a polyprotein of about 3388 amino acids. Signal and stop-transfer sequences direct the translocation of the polyprotein back and forth across the membrane. The structural proteins are in the N-terminal region and are anchored in the endoplasmic reticulum by multiple membrane-spanning amino acid sequences (Figure 1A). The polypeptide is subsequently cleaved by cellular and virally encoded proteinases and glycosylated by cellular glycosyltransferases to yield three structural proteins: anchored capsid (anC) consisting of (for dengue virus) 113 residues, pre-membrane (prM) consisting of 166 residues and glycoprotein (E) consisting of 495 residues.
Fig. 1. The transmembrane region of dengue polyprotein. (A) The predicted threading of the structural proteins across the membrane. The different transmembrane regions are color coded. Arrows indicate post-translational enzymatic cleavages, with specific enzymes indicated by different colors. The maturation cleavage by furin is shown with a large green arrow (Strauss and Strauss, 2002). (B) Transmembrane amino acid sequences of the prM and E dengue virus proteins color coded to correspond to (A). Secondary structural predictions based on the PROF program (Rost, 1996) are also shown.
The nucleocapsid core of the mature virion consists of the genomic RNA surrounded by multiple copies of the capsid protein C. This core is enveloped by a 40 Å thick lipid bilayer derived from the endoplasmic reticulum of the host cell. Outside the membrane envelope is a layer of 180 copies of the E glycoprotein organized into a herringbone pattern (Kuhn et al., 2002) plus 180 copies of the M protein. Both the E and M proteins are anchored in the membrane by their C-terminal domains (Lindenbach and Rice, 2001).
The structure of the 395 residue N-terminal tryptic fragment of the TBEV E glycoprotein (Heinz et al., 1991; Allison et al., 1995) has been determined by X-ray crystallography (Rey et al., 1995). This E fragment crystallizes as a dimer, consistent with the expectation that TBEV assembly results in the formation of homodimers. Each monomer consists of three domains: the structurally central N-terminal domain I, followed by the dimerization domain II and finally the C-terminal Ig-like domain III. The dimerization domain contains the hydrophobic ‘fusion’ peptide (residues 98–110) essential for virus–cell fusion (Allison et al., 2001). It has been proposed that domain III functions as the binding site for cellular receptors (Rey et al., 1995; Bhardwaj et al., 2001). In addition, domain III has been recognized as the receptor attachment site in competition experiments with monoclonal antibodies (Beasley and Aaskov, 2001; Crill and Roehrig, 2001; Lok et al., 2001). The 101 residue C-terminal end of the TBEV E glycoprotein, which was not part of the X-ray structure, is called the ‘stem anchor’ region, with the stem being composed of residues 396 to ∼449 and the hydrophobic transmembrane anchor region composed of residues 450–496. The stem and transmembrane anchor regions have each been predicted to consist of two α-helices (Stiasny et al., 1996; Allison et al., 1999) (Figure 1B).
The structure of the mature dengue virus has been determined to 24 Å resolution (Kuhn et al., 2002) using cryoelectron microscopy (cryoEM) and image reconstruction techniques. The cryoEM density was interpreted in terms of 90 copies of an E:E dimer, homologous with the TBEV E:E dimer crystal structure. The dengue virus structure did not have the anticipated T = 3 symmetry (Caspar and Klug, 1962) expected when there are 3 × 60 monomers in the glycoprotein shell. However, it was proposed (Kuhn et al., 2002) that acid pH would weaken E–E adhesion within the E:E dimer and allow reorganization of the E molecules to generate a particle with classical T = 3 symmetry. By extrapolation (Kuhn et al., 2002) from the cryoEM structure of a recombinant fusogenic T = 1 subviral particle (Ferlenghi et al., 2001) of E and M proteins, such a T = 3 structure would have E:E:E homotrimers, as expected for fusogenic particles (Heinz and Allison, 2000), and sufficient exposed membrane areas to allow fusion with the host cell.
The final step in virion assembly is cleavage of the prM glycoprotein by furin or a furin-like protease into an N-terminal pre-peptide 91 amino acids long (for dengue virus) and an M protein 75 residues long. Furin is a cellular subtilisin-like endoproteinase that recognizes an (R/K)– X–(R/K)–R motif (Randolph et al., 1990; Stadler et al., 1997; Thomas, 2002). Cleavage leads to the dissociation of prM:E heterodimers (Wengler and Wengler, 1989; Allison et al., 1995) and the formation of E:E homodimers (Rey et al., 1995; Stiasny et al., 1996). The glycosylated pre-peptide is released from the maturing particle, altering the accessibility of domain II of the E glycoprotein to antibody binding (Stiasny et al., 1996). Unlike the mature virion, the prM particles are unable to fuse with cells in acid conditions (Guirakhoo et al., 1992). In contrast, when mature particles become fusogenic, as in acidified endosomes during the normal infectious process (Heinz et al., 1994), they undergo conformational changes that alter the antigenic properties once again and result in the formation of E:E:E homotrimers (Figure 2A) (Stiasny et al., 1996). Thus the pre-peptide probably functions to prevent the E protein from prematurely undergoing conformational changes that trigger fusion in endosomal vesicles (Heinz et al., 1994), analogous to the function of PE2 in alphaviruses (Strauss and Strauss, 1994).
Fig. 2. Maturation. (A) Maturation of immature prM particle to mature infectious virus and formation of the initial fusogenic particle. (B) The most probable rearrangement required for the immature particles to become mature infectious virions. The Cα backbones of the three independent E molecules per icosahedral asymmetric unit are colored red, blue and green. The three domains in each monomer are identified by Roman numerals: I, central; II, dimerization; III, Ig-like.
We report here similar cryoEM structures of dengue and yellow fever virus immature particles, determined to 16 Å and 25 Å, respectively. The samples were produced by growing the virus in medium containing acidotropic (NH4)Cl, thus inhibiting furin cleavage of prM (Heinz et al., 1994; Stadler et al., 1997). The resultant image reconstructions of these particles show that the predicted prM:E heterodimers are arranged in asymmetric trimeric spikes in which the pre-peptides cover and hide the fusion peptides on the E glycoproteins. The spikes in these immature particles are similar to the trimeric spikes in mature Sindbis virus in which the E2 glycoproteins cover the fusion peptides in the E1 glycoproteins (Zhang et al., 2002). The structure of the prM particles shows that the flavivirus E glycoproteins must undergo large quaternary structural rearrangements during maturation.
Results and discussion
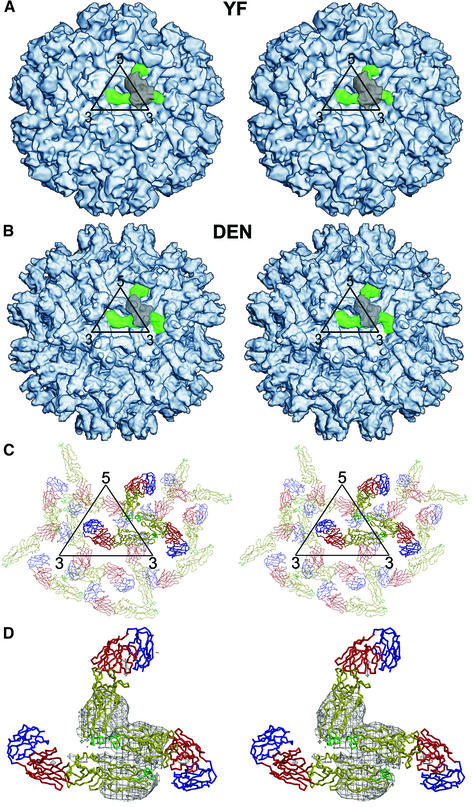
CryoEM image reconstructions were computed to 16 and 25 Å resolution from micrographs of dengue virus and yellow fever virus immature particles, respectively (Figure 3). They show very similar density distributions for both viruses. This is the first three-dimensional image reconstruction of any yellow fever virus particle and suggests a close structural similarity between divergent flaviviruses. However, as the reconstruction of the dengue virus immature particles is at a considerably higher resolution, the bulk of this article refers exclusively to the dengue virus results.
Fig. 3. Structure of immature flavivirus particles. (A and B) Stereo views showing the surfaces of dengue and yellow fever prM particles at 16 and 25 Å, respectively. An icosahedral asymmetric unit is outlined in black. One of the 60 spikes in each particle is identified in color: prM is gray and E is green. (C) Stereo top view of the organization of the E glycoproteins within and surrounding one icosahedral asymmetric unit (black). Each monomer is shown as a Cα backbone with domains I, II, and III in red, yellow and blue, respectively. The monomers associated with one trimer are in bold. (D) Stereo top view of a spike also showing the density (contoured at a 1σ level) associated with the prM molecule covering the spike structure shown in the same orientation as in (A), (B) and (C). The pairing of the E and prM molecules as a heterodimer is apparent.
The E glycoprotein
The surface of the immature particle is characterized by the presence of 60 fairly prominent projections or spikes (Figure 3). This contrasts with the previously determined structure of the mature virus, which has a rather smooth surface (Kuhn et al., 2002). Both the well formed external features and the convergence of the reconstruction process validate the assumed icosahedral symmetry used in the image processing. The spikes cause the immature particles to have a considerably larger diameter (600 Å) than mature virions (500 Å). The base of each spike has a radius of ∼80 Å. The obvious signature of the lipid bilayer (Figure 4) established that the ectodomains of the E and prM proteins both lie outside a 205 Å radius.
Fig. 4. Cross-section of prM particle. (A) The ectodomain is shown in blue, the lipid bilayer in green and the nucleocapsid in orange. Plots of maximum height and average height against radius are shown in blue and red, respectively. (B) The equatorial cross-section showing the roughly polygonal shape of the lipid and nucleocapsid. Note the gap between the inner surface of the membrane and the nucleocapsid. This gap is crossed by the nucleocapsid density only at positions corresponding to the base of the external spikes (arrow).
Three E monomers per icosahedral asymmetric unit could easily be positioned into the cryoEM density of the immature particle reconstruction (Table I). The organization of the three monomers does not conform to T = 3 quasi-symmetry (Caspar and Klug, 1962) (Figure 3C and D), as is also the case for the mature virus (Kuhn et al., 2002). Presumably, the association between heterodimers within an immature particle spike is sufficiently strong, compared with the association between spikes, to cause a spike to be treated as a single assembly unit. Hence the organization of the particles could be described as being T = 1 with an asymmetric spike as the basic building block.
Table I. Fitting of the E glycoprotein X-ray structure into the cryoEM map of dengue prM particles using the EMfit program.
| Mola | sumf1b (%) | sumf2b (%) | sumf3b (%) | -denc (%) | clash (%) | centxd (Å) | centyd (Å) | centzd (Å) | θ1e (°) | θ2e (°) | θ3e (°) | Order of fittingf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blue | 50.8 | 55.8 | 42.3 | 1.0 | 0.0 | 32.0 | –7.7 | 220.9 | 15.0 | 61.0 | 349.2 | First |
| Blue | 49.7 | 56.4 | 44.0 | 1.0 | 0.0 | 31.0 | –6.7 | 221.4 | 21.0 | 61.5 | 345.0 | Second |
| Blue | 50.9 | 56.0 | 40.5 | 0.8 | 0.0 | 31.5 | –6.7 | 220.4 | 10.8 | 61.5 | 355.2 | Third |
| Red | 48.4 | 57.6 | 42.9 | 1.0 | 0.0 | 72.1 | 8.2 | 210.6 | 38.0 | 64.5 | 162.5 | Third |
| Red | 49.8 | 57.4 | 41.8 | 0.8 | 0.0 | 71.6 | 8.2 | 210.6 | 34.8 | 63.5 | 164.8 | First |
| Red | 49.7 | 57.4 | 41.9 | 0.8 | 0.0 | 72.1 | 7.7 | 210.6 | 37.5 | 64.0 | 163.5 | Second |
| Green | 48.9 | 54.7 | 42.1 | 0.8 | 0.0 | 10.5 | 48.3 | 217.0 | 19.8 | 58.0 | 240.2 | Second |
| Green | 49.2 | 53.1 | 41.3 | 1.0 | 0.0 | 9.0 | 47.8 | 217.0 | 22.2 | 58.5 | 238.5 | Third |
| Green | 49.5 | 54.9 | 42.8 | 0.8 | 0.0 | 10.0 | 48.3 | 217.0 | 18.2 | 57.0 | 241.8 | First |
For more details of the EMfit program parameters, see Rossmann et al., 2001.
aMol, red, blue and green correspond to the E monomers shown in Figure 2B.
bsumf1, sumf2, and sumf3 are the average densities of Cα atoms for domains I, II and III, respectively, expressed as a percentage of the highest density in the map.
c-den is the percentage of atoms in negative density.
dcentx, centy and centz are the refined coordinates of the center of mass of the E monomer in the map. Note the consistency of the results, independent of the order of fitting.
eθ1, θ2 and θ3 are the refined Eulerian angles required to rotate the atomic coordinates of E into position. Note the consistency of the results.
fPossible fitting procedures: (1) blue molecule (first), followed by green molecule (second), followed by red molecule (third); (2) red molecule (first), followed by blue molecule (second), followed by green molecule (third); (3) green molecule (first), followed by red molecule (second), followed by blue molecule (third).
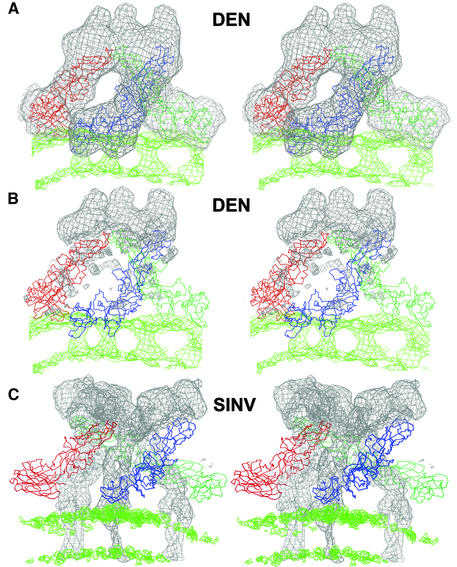
The three E monomers within each spike of the immature particles are tilted with respect to the viral surface such that their long axes make an angle of ∼25° with the surface (Figure 5A and B). The distal ends of the three monomers approach each other, making a short superhelix, in the formation of a spike. The fusion peptides of the three E monomers are at the tip of each spike. The twist of the E monomers around each other within one spike has the same handedness as the twist of the E1 glycoproteins forming the spike of Sindbis virus (Figure 5C). However, unlike Sindbis virus spikes which have strict or quasi-threefold symmetry, the monomers in the immature particle spikes are only very approximately related by a threefold axis.
Fig. 5. Comparison of the spike structure of immature dengue particles (A and B) and mature Sindbis virus (C). The Cα backbones of the E and E1 glycoprotein trimers (in blue, red and green) are shown for the dengue and Sindbis particles, respectively. (A) The fit of the three E glycoprotein Cα backbones into the spike density (gray) of the immature dengue particle cryoEM density. (B) The density corresponding to the fitted E molecule shown in (A) has been zeroed out, leaving only the density corresponding to the three prM molecules. (C) For comparison, the E1 Cα backbone in the mature Sindbis virus cryoEM density is shown with the density for E1 zeroed out, leaving only the density for E2. The density corresponding to the lipid bilayer is shown in green. The slab used for depicting the lipid bilayer is thinner than that used to define the ectodomain trimers.
A difference map, in which the density corresponding to the fitted E monomers was set to zero, shows the residual density representing the prM protein and the stem region of the E glycoprotein in the ectodomain. The prM proteins cover the three E fusion peptides of domain II (Figure 5B), consistent with the observation that the prM proteins inhibit fusion (Guirakhoo et al., 1992). Furthermore, the difference map shows a thin density trace, attributed to prM, that follows the length of each E monomer in a spike down to the membrane (Figure 5B). The molecular weight of the 91 amino acids associated with the pre-peptide is in excellent agreement with the volume of the prM density in the difference map when the fitted E structure is used to calibrate the ratio of volume to molecular weight (Table II). Thus the E molecules in immature particles do not associate as homodimers like those observed in crystals of TBEV glycoprotein E (Rey et al., 1995), in mature dengue virus (Kuhn et al., 2002) and in recombinant subviral T = 1 particles (Ferlenghi et al., 2001). Rather, the prM:E heterodimers observed here are consistent with previous biochemical studies of immature flavivirus particles (Wengler and Wengler, 1989; Heinz et al., 1994). The similarity to alphaviruses, where the E2 glycoproteins protect the fusion peptides of the E1 glycoproteins within a trimeric spike, is striking (Figure 5C). The dominant contacts between the prM:E heterodimers within a spike are between the pre-peptides at the extremities of the spikes, similar to the contact between the E2 glycoproteins within alphavirus spikes (Zhang et al., 2002). Thus cleavage and loss of the pre-peptide would be likely to disrupt and dissociate the trimeric spike and possibly induce the formation of dimers as found in the mature virus.
Table II. Comparison of the molecular weights of E, pr peptide and prM with their designated volumes at a contour level above 0.5σ in the cryoEM mapa.
| E | pr | prM | Glycan | |
|---|---|---|---|---|
| Volume (Å3) | 74 624 | 21 089 | 32 319 | 4510 |
| Molecular weight (kDa) | 43.7 | 12.3 | 18.9 | 2.6 |
aDifferentiation between E and prM was established by fitting the structure of three Es into the map. The pre-peptide volume was assumed to be all the globular volume covering the top of E. The volumes are the average of three copies of each protein.
Small density protrusions on the distal ends of the immature particle spikes are reminiscent of similar features on the surface of the E2 Sindbis virus glycoproteins, which correspond to carbohydrate sites (Pletnev et al., 2001). As there is a potential glycosylation site on the prM glycoprotein of dengue virus at Asn69 (Hahn et al., 1988), it was presumed that this protrusion was due to carbohydrate. The volume of this feature corresponded to 2.6 kDa (Table II).
The lipid bilayer
The characteristic lipid bilayer is recognizable between 165 and 205 Å radius (Figure 4) in the immature dengue virus reconstruction. However, the envelope is markedly polygonal, rather than spherical, with constrictions where proteins cross the membrane (Figure 4B). The deviations from a simple sphere in the shape of the lipid envelope demonstrate that the external icosahedral glycoprotein assembly influences the membrane structure.
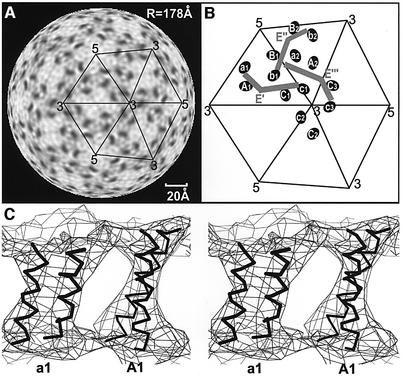
The center of the transmembrane region, corresponding to the lower density of the aliphatic chains, is traversed by six elliptical higher-density regions per icosahedral asymmetric unit (Figure 6B). These were interpreted to correspond to the transmembrane regions of each of the three E and the three prM proteins (Figure 1A). A dimeric antiparallel coiled coil, taken from the structure of colicin E3 (PDB accession code 1JCH), could be readily fitted into each transmembrane region. Furthermore, the elliptical cross-section rotates as it traverses the membrane, generating a twist consistent with the left-handed superhelix of the coiled coil (Figure 6C).
Fig. 6. The transmembrane structure of the immature particles. (A) Radial section (radius of 178 Å), corresponding roughly to the center of the membrane, showing six elliptical transmembrane regions of higher density arranged in pairs separated by 20 Å. (B) Diagrammatic representation of the transmembrane regions (Aiai, Bibi and Cici, i = 1, 2, 3) and the associated E′, E′′ and E′′′ glycoproteins. The symmetry relationships of E′ to E′′ to E′′′ exactly correspond to the relationships of A1a1 to B2b2 to C3c3, respectively. (C) Stereo view of antiparallel coiled coils for the E and M proteins passing through and back across the membrane at position A1a1. The center of the particle lies below the figure.
The six transmembrane regions are organized into three pairs (Aiai, Bibi and Cici in icosahedral asymmetric units, i = 1, 2, …, 60) (Figure 6B) with a 20 Å separation between the two regions within a pair. The A1a1, B2b2 and C3c3 pairs are related by the same rotations and translations as exist between the fitted E glycoproteins E′, E′′ and E′′′, all belonging to the same spike. Thus the A1a1, B2b2, and C3c3 transmembrane regions could be fitted simultaneously with the program EMfit (Rossmann et al., 2001) by assuming the relationship between the independently fitted E′, E′′ and E′′′ glycoproteins (Table I). Therefore it is likely that the A1a1, B2b2 and C3c3 transmembrane pairs might be associated with E′, E” and E′′′, respectively. However, the distance between the C-termini of the fitted E glycoproteins and their corresponding transmembrane helices was ∼22 Å and 31 Å. This distance is spanned by the helical stem region of about 56 residues, and is ∼42 Å in the mature virus (our unpublished data). An alternative solution was to associate the transmembrane regions A2a2, B1b1 and C1c1 with the E″, E′ and E′′′ glycoproteins, respectively. Although in this arrangement the relationship of A2a2 to B1b1 is the same as that of E′′ to E′, the relationship of B1b1 to C1c1 is not exactly the same as that of E′ to E′′′, resulting in a misplacement of the C1c1 transmembrane helices by ∼8 Å. Nevertheless, the distance of the transmembrane regions to the E glycoprotein is ∼35 Å and ∼37 Å, rather closer to that observed in the mature virus than obtained with the previous solution, albeit lacking an accurate symmetry relationship.
Inspection of the E and prM amino acid sequences shows that they both have polar residues in the middle of their hydrophobic transmembrane regions that are unlikely to be buried within the central hydrophobic region of the lipid bilayer (Figure 1B). Thus it is probable that the six transmembrane densities are each composed of an antiparallel coiled-coil structure of either E or M proteins (Figure 6C). This is consistent with the prM:E heterodimer structure in which the C-terminal ends of both proteins are near the outer leaflet of the lipid bilayer and with the interpretation of an 11 Å resolution map of mature dengue virus (our unpublished data). Thus, within each pair of transmembrane regions, one region corresponds to an antiparallel coiled coil of consecutive helices in the E protein and the other to an antiparallel coiled coil of consecutive helices in the prM protein. The linking residues between the transmembrane helices do not appear to extend into the cytoplasmic domain, and each prM:E pair of densities maintains a similar relationship to the prM:E ectodomain heterodimer. The cryoEM density representing the membrane outer leaflet of the immature dengue particle is higher than that of the inner leaflet. Furthermore, the C-termini of the E and prM molecules are near the surface of the outer membrane. However, there are 56 residues in the stem region of the E glycoprotein that are not accounted for in the fitted E fragment. The stem region of E has been predicted to be two α-helices (Stiasny et al., 1996; Allison et al., 1999). This suggests that these two helices are buried in the outer leaflet of the membrane, giving rise to the higher density, in agreement with results for mature dengue virus (our unpublished results). In contrast, the E1 glycoprotein C-termini in alphaviruses are well separated from the membrane surface with no indication of the stem region being buried in the outer membrane leaflet (Figure 5C) (Zhang et al., 2002).
Although α-helices are a common feature of membrane proteins, it is less common to see transmembrane helices associated as coiled coils, as occurs in dengue virus and alphaviruses (Zhang et al., 2002). In alphaviruses, the viral membrane is traversed by 240 parallel coiled coils, each consisting of a segment from the E1 and the E2 glycoproteins (Zhang et al., 2002). These polypeptides traverse the membrane only once without returning, in contrast with what is demonstrated here to occur in flaviviruses. The PE2 glycoprotein of alphaviruses, like the prM protein of flaviviruses, is synthesized as a molecule that spans the lipid bilayer twice. The present studies show that both transmembrane segments of prM remain within the lipid bilayer. In contrast, the C-terminal segment of E2 in alphaviruses is retracted into the cytoplasm during maturation where it interacts with the nucleocapsid (Strauss et al., 1995; Zhang et al., 2002). The latter process requires phosphorylation and dephosporylation (Liu and Brown, 1993a,b). The orientation of viral glycoproteins in viral envelopes has previously been deduced from amino acid sequence and limited biochemical studies. Visualization of the transmembrane domains in dengue virus prM particles as shown here, as well as in Sindbis virus (Zhang et al., 2002), directly confirms the orientations of these transmembrane helices.
The nucleocapsid core
In both the dengue and yellow fever virus immature particle reconstructions, the inner membrane layer is separated from the nucleocapsid (outer radius of ∼150 Å) by a gap of ∼30 Å. However, immediately underneath the center of each spike there is density that reaches from the nucleocapsid across this gap to contact the inner layer of the membrane. This density distorts the inner membrane layer and gives rise to the polygonal morphology of the membrane and nucleocapsid (Figure 4B). Unlike the nucleocapsid shell of alphaviruses, or indeed of most viruses, there is no distinction between an anticipated protein shell and the RNA.
The maximum height of the nucleocapsid density is about a quarter of that observed in the outer glycoprotein shell (Figure 4A). Similarly, the height of the nucleocapsid cryoEM density in the mature virions was only about half the height of the outer glycoprotein (Kuhn et al., 2002). In contrast, the capsid protein in a Sindbis virus cryoEM map at 11 Å resolution was at least as high as the outer glycoprotein and was readily interpretable in terms of the known X-ray structure (Zhang et al., 2002). This, together with the short capsid amino acid sequence (99 residues in the dengue virus capsid protein compared with 264 residues in Sindbis virus) and its internal basic regions, supports the hypothesis (Kuhn et al., 2002) that either the core of flaviviruses is partially disordered or its icosahedral orientation is not synchronized with the larger and dominant external structure. In alphaviruses, at least two of the 33 cytoplasmic residues of each of the 240 copies of the E2 protein bind to a corresponding capsid protein, thus ensuring that the icosahedral symmetry of the external glycoprotein assembly is synchronized with that of the internal core structure (Lee et al., 1996; Skoging et al., 1996). However, in dengue virus, neither the E nor the M protein appear to extend into the cytoplasmic side of the lipid membrane, thus allowing the capsid to be randomly positioned relative to the outer glycoprotein shell.
The absence of distinct capsid protein density, combined with the unusually low height of the nucleocapsid density, suggests a rather different situation than has previously been observed for viruses studied by X-ray crystallography (Rossmann and Johnson, 1989) or cryoEM (Baker et al., 1999). In many viruses, including alphaviruses, there is a basic N-terminal domain that is associated with the RNA in a non-icosahedral manner. Therefore it is possible that the entire small basic capsid protein of flaviviruses may function like the N-terminal domain of many other viral capsid proteins, leaving no organized capsid structure separating the lipid membrane from the nucleocapsid. Thus the size of the assembled virus might be determined by the size of the genomic RNA, much as the size of the three distinct alfalfa mosaic virus capsids are determined by the RNA sizes of the segmented genome (Hull, 1970).
In a recent study of mature dengue virus, it was found that the earlier assignment (Kuhn et al., 2002) of the radial limits of the lipid bilayer and nucleocapsid had been about 15 Å too small (our unpublished results). The new assignments place the membrane and nucleocapsid at almost the same radial limits as reported here for the prM particles. Although the dimensions of the nucleocapsid core remain unaltered during the maturation process, the positions of the three pairs of transmembrane coiled coils are completely different with respect to the icosahedral axes, while maintaining the 20 Å separation between the E and prM antiparallel coiled coils.
Maturation
The infectivity of TBEV prM particles that are produced by adding NH4Cl to infected cells can be enhanced ∼100-fold when treated with furin (Stadler et al., 1997). Furthermore, about a third of the particles produced by furin treatment of immature dengue virus had a smooth surface and diameters consistent with that of mature particles as opposed to the bumpy surface and larger diameter of the immature particles. In contrast there were no smooth-surfaced particles in untreated immature dengue virus samples (see Materials and methods). The correlation coefficients relating the furin-treated particle projections to the native particle reconstruction (Kuhn et al., 2002) also indicated that the immature particles had been changed to mature virions. Therefore it is apparent that the structure of the prM-containing particles was converted to that of infectious virions by the furin treatment and that the immature particles serve as intermediates in the assembly pathway. The furin treatment results in the conversion of the prM:E heterodimer to an E:E homodimer and releases the pre-peptides from the top of the spikes, thus allowing the E glycoproteins to lie parallel to the membrane. Inspection of the structure shows that the most probable way that this might happen (Figure 2B) is for the ‘red’ (R) and ‘blue’ (B) monomers to associate as an RB dimer and for the ‘green’ (G) monomer to associate with a 2-fold-related green monomer (G′) to form a GG′ dimer. When the structure of the TBEV E:E dimer is viewed from outside the virus, the three domains of each monomer have the order (II, I, III)–(II, I, III) clockwise around the central dimer axis. The RB dimer would have its domains ordered in a clockwise manner, showing that the surface of the dimer known to form the external surface of the virus is correctly facing outwards. However, the GG′ dimer would have an anticlockwise order of domains, which would require it to turn upside down in order to expose the side of the dimer with the (II, I, III) sequence of domains having a clockwise order.
Both alpha- and flaviviruses require the formation of homotrimers to become fusogenic, a property that is required for both class I and class II fusion (Lescar et al., 2001). In the immature flavivirus particle structure, as in alphaviruses, the monomers within a trimer are tilted upwards with their fusion peptides appropriately directed towards the host cell membrane (Figure 5C). Fusion might be expected to proceed once the protective prM pre-peptide in flaviviruses or the E2 glycoprotein in alphaviruses has been removed. Although this is true for alphaviruses, in flaviviruses fusion is inhibited after the pre-peptide has diffused away, presumably by the formation of the E homodimers in which the fusion peptide is protected and by making a densely packed array that completely covers the viral membrane. Further major conformational changes are presumed to occur upon exposure of virions to acidic pH in endosomes, in which the homodimers re-form as homotrimers (Kuhn et al., 2002) and the E monomers resume a tilted orientation relative to the viral surface, thereby directing the fusion peptides towards the host cell membrane and exposing some of the outer leaflet of the viral membrane.
The immature flavivirus particle, with its 60 trimeric spikes, appears to be poised for fusion. However, in a process that is difficult to comprehend, the structure of the immature virus apparently undergoes a major reorganization to produce the mature virus. As the similar dengue and yellow fever prM particles were generated in two separate laboratories (the California Institute of Technology and Purdue University, respectively), it is likely that the immature particle structure is representative of all flaviviruses rather than being the result of some artifact of the propagation procedures. Thus the immature flavivirus particles examined here, and which have been extensively reported in the literature (Wengler and Wengler, 1989; Randolph et al., 1990; Guirakhoo et al., 1991, 1992; Heinz et al., 1994), must undergo a radical rearrangement to produce the mature virions. A similar equally large reorganization has also been predicted for conversion of the mature virus to a fusion active state (Kuhn et al., 2002). Thus flavivirus particles participate in a series of dramatic reorganizations during their life cycle, reflecting the dynamic properties of the virion and its component proteins.
Materials and methods
Preparation and characterization of prM particles
C6/36 mosquito cells were infected with dengue virus 2 strain PR159-S1 at a multiplicity of infection (MOI) of 1 for 2 h at 30°C. The inoculum was replaced by Eagle’s medium containing 10% fetal bovine serum (FBS), non-essential amino acids, twice the normal glutamine concentration and 20 mM NH4Cl. After 24 h, the medium was replaced by serum-free medium containing 20 mM NH4Cl. The culture fluid was harvested at 72–80 h post-infection. Virus particles were purified by precipitation with polyethylene glycol and sedimentation in potassium tartrate gradients as previously described (Kuhn et al., 2002). The 17D vaccine strain of yellow fever virus was used to infect SW13 cells at an MOI of 0.1 for 2 h at room temperature. The inoculum was then replaced by Minimum Essential Medium containing 5% FBS, non-essential amino acids and twice the normal glutamine concentration. At 48 h post-infection, the medium was replaced by fresh medium supplemented with 20 mM NH4Cl. The cells were washed 5 h later and the medium was again replaced with fresh medium and 20 mM NH4Cl. The supernatant was harvested after incubation for an additional 24 h. The yellow fever virus prM particles were then purified as described above for dengue virus prM particles.
Immature dengue virus was treated by adding 12.5 units (1 pmol of substrate per minute at 30°C) of furin to 10 µl (1 mg/ml) sample at 30°C for 2 h. The treated sample was then applied to EM grids for immediate freezing.
EM analysis
Small aliquots (∼3.5 µl) of purified dengue and yellow fever prM particles were applied to perforated carbon-coated electron microscope grids and vitrified in liquid ethane (Baker et al., 1999). Images were recorded using low-dose conditions (<15 electrons/Å2) on Kodak SO-163 film with a Philips CM300 field emission gun transmission electron microscope. Micrographs from a wide range of defocus values (Table III) were digitized on a Zeiss SCAI scanner using a 7 µm step size and were bin averaged later to give 4.24 Å pixels at the specimen.
Table III. Dengue (DEN2) and yellow fever (YF) virus cryoEM image reconstruction dataa.
| Virus | No. of micrographs | Defocus (µm) | Particle No. (selected/total) | PFT CCb | Resolution (Å) |
|---|---|---|---|---|---|
| DEN2 | 25 | 1.6–3.8 | 4099/6149 | 0.385 | 16.0 |
| YF | 32 | 1.8–4.3 | 186/2056 | 0.274 | 25.0 |
aThe microscope magnification was a nominal 33 000 for all images.
bPFT CC is the polar Fourier transform correlation coefficient (Baker and Cheng, 1996).
The defocus level of each micrograph was determined using the program RobEM, which fits theoretical microscope contrast transfer functions to the incoherent sum of the Fourier transforms of all particle images from each micrograph. A total of 4099 and 186 particles were used to compute the final electron density maps from dengue and yellow fever prM particles, respectively. The common-line method (Baker et al., 1999) was used to generate an initial dengue prM particle model utilized in subsequent polar Fourier transform refinements of the dengue data (Baker and Cheng, 1996). The resulting dengue prM reconstruction acted as the initial model for the yellow fever virus prM data processing.
The resolutions of the final three-dimensional reconstructions were estimated by randomly dividing each of the datasets in half and comparing the structure factors obtained from the separate reconstructions. Phase agreement (<50°) and correlation coefficients (>0.5) indicate resolutions of the reconstructions to be 16 Å for dengue and 25 Å for yellow fever virus.
Fitting E to density
The Cα backbone of the TBEV E monomer (PDB accession code 1SVB) was used to fit into the 16 Å resolution density map of dengue virus using the program EMfit (Rossmann et al., 2001). The pixel separation in the maps was 4.24 Å. One E monomer was fitted at a time by making a complete three-dimensional angular search, but assuming an initial approximate site for placing the center of mass of the search model. The best 25 results were subsequently refined in a six-dimensional process. There was little ambiguity in the final best fit. The density corresponding to this monomer was set to zero before fitting the next monomer in a similar manner. The third monomer was fitted in a map in which the densities for the first two monomers had been zeroed out. As one of the criteria of fit was to minimize the number of atoms falling into zero or negative density, the process of setting density to zero for the previously fitted monomer was equivalent to removing steric clashes between the sequentially fitted monomers. The process was restarted by fitting the monomers in a different order, but the resultant fits remained consistent, independent of the order of fitting (Table I). As the absolute handedness of the cryoEM map was unknown, the fitting procedure was attempted for both possible enantiomorphs. However, only one handedness gave satisfactory results (see Supplementary data available at The EMBO Journal Online). The dengue and yellow fever prM particle structures were found to be very similar, with a root mean square deviation of 3.8 Å between equivalent Cα atoms.
Coordinates of the fitted E proteins have been deposited with the Protein Data Bank (accession codes 1N6G and 1NA4) for immature dengue and yellow fever virus particles, respectively.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Suchetana (Tuli) Mukhopadhyay and Ellen Strauss for many helpful discussions, Rob Ashmore, Chuan (River) Xiao, Yongchang Ji and Dan Marinescu for the use of their various computer programs that were essential for calculating the image reconstruction of the prM particles, and Cheryl Towell and Sharon Wilder for help in preparing the manuscript. We are grateful for an equipment grant from the Keck Foundation that provided the CM300 electron microscope. The work was supported by an NIH Project Program Grant (AI 45976) to M.G.R., R.J.K. and T.S.B., NIH grants to T.S.B. (GM33050) and J.H.S. (AI 10793) and an NSF grant to T.S.B. (DBI 9986316).
References
- Allison S.L., Stadler,K., Mandl,C.W., Kunz,C. and Heinz,F.X. (1995) Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol., 69, 5816–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison S.L., Stiasny,K., Stadler,K., Mandl,C.W. and Heinz,F.X. (1999) Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol., 73, 5605–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison S.L., Schalich,J., Stiasny,K., Mandl,C.W. and Heinz,F.X. (2001) Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol., 75, 4268–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.S. and Cheng,R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol., 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Baker T.S., Olson,N.H. and Fuller,S.D. (1999) Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev., 63, 862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley D.M. and Aaskov,J.G. (2001) Epitopes on the dengue 1 virus envelope protein recognized by neutralizing IgM monoclonal antibodies. Virology, 279, 447–458. [DOI] [PubMed] [Google Scholar]
- Bhardwaj S., Holbrook,M., Shope,R.E., Barrett,A.D.T. and Watowich,S.J. (2001) Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol., 75, 4002–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.S. and Monath,T.P. (2001) Flaviviruses. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott-Williams & Wilkins, Philadelphia, PA, pp. 1043–1125.
- Caspar D.L.D. and Klug,A. (1962) Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol., 27, 1–24. [DOI] [PubMed] [Google Scholar]
- Crill W.D. and Roehrig,J.T. (2001) Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to vero cells. J. Virol., 75, 7769–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlenghi I., Clarke,M., Ruttan,T., Allison,S.L., Schalich,J., Heinz,F.X., Harrison,S.C., Rey,F.A. and Fuller,S.D. (2001) Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell, 7, 593–602. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F., Heinz,F.X., Mandl,C.W., Holzmann,H. and Kunz,C. (1991) Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol., 72, 1323–1329. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F., Bolin,R.A. and Roehrig,J.T. (1992) The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology, 191, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y.S., Galler,R., Hunkapiller,T., Dalrymple,J.M., Strauss,J.H. and Strauss,E.G. (1988) Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology, 162, 167–180. [DOI] [PubMed] [Google Scholar]
- Heinz F.X. and Allison,S.L. (2000) Structures and mechanisms in flavivirus fusion. Adv. Virus Res., 55, 231–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F.X., Mandl,C.W., Holzmann,H., Kunz,C., Harris,B.A., Rey,F. and Harrison,S.C. (1991) The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol., 65, 5579–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F.X., Stiasny,K., Püschner-Auer,G., Holzmann,H., Allison,S.L., Mandl,C.W. and Kunz,C. (1994) Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology, 198, 109–117. [DOI] [PubMed] [Google Scholar]
- Hull R. (1970) Studies on alfalfa mosaic virus. III. Reversible dissociation and reconstitution studies. Virology, 40, 34–47. [DOI] [PubMed] [Google Scholar]
- Kuhn R.J. et al. (2002) Structure of the dengue virus: implications for flavivirus organization, maturation and fusion. Cell, 108, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Owen,K.E., Choi,H.K., Lee,H., Lu,G., Wengler,G., Brown,D.T., Rossmann,M.G. and Kuhn,R.J. (1996) Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure, 4, 531–541. [DOI] [PubMed] [Google Scholar]
- Lescar J., Roussel,A., Wein,M.W., Navaza,J., Fuller,S.D., Wengler,G., Wengler,G. and Rey,F.A. (2001) The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell, 105, 137–148. [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D. and Rice,C.M. (2001) Flaviviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott-Williams & Wilkins, Philadelphia, PA, pp. 991–1041.
- Liu N. and Brown,D.T. (1993a) Phosphorylation and dephosphorylation events play critical roles in Sindbis virus maturation. Virology, 196, 703–711. [DOI] [PubMed] [Google Scholar]
- Liu N. and Brown,D.T. (1993b) Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J. Cell Biol., 120, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok S.M., Ng,M.L. and Aaskov,J.G. (2001) Amino acid and phenotypic changes in dengue 2 virus associated with escape from neutralisation by IgM antibody. J. Med. Virol., 65, 315–323. [DOI] [PubMed] [Google Scholar]
- Monath T.P. (1996) Milestones in the conquest of yellow fever. In Koprowski,H. and Oldstone,M.B.A. (eds), Microbe Hunters—Then and Now. Medi-Ed Press, Bloomington, IN, pp. 95–111.
- Pletnev S.V., Zhang,W., Mukhopadhyay,S., Fisher,B.R., Hernandez,R., Brown,D.T., Baker,T.S., Rossmann,M.G. and Kuhn,R.J. (2001) Locations of carbohydrate sites on Sindbis virus glycoproteins show that E1 forms an icosahedral scaffold. Cell, 105, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph V.B., Winkler,G. and Stollar,V. (1990) Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology, 184, 450–458. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Heinz,F.X., Mandl,C., Kunz,C. and Harrison,S.C. (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature, 375, 291–298. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G. and Johnson,J.E. (1989) Icosahedral RNA virus structure. Annu. Rev. Biochem., 58, 533–573. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Bernal,R. and Pletnev,S.V. (2001) Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol., 136, 190–200. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Skoging U., Vihinen,M., Nilsson,L. and Liljeström,P. (1996) Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure, 4, 519–529. [DOI] [PubMed] [Google Scholar]
- Stadler K., Allison,S.L., Schalich,J. and Heinz,F.X. (1997) Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol., 71, 8475–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K., Allison,S.L., Marchler-Bauer,A., Kunz,C. and Heinz,F.X. (1996) Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol., 70, 8142–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H. and Strauss,E.G. (1994) The alphaviruses: gene expression, replication and evolution. Microbiol. Rev., 58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H. and Strauss,E.G. (2002) Viruses and Human Disease. Academic Press, San Diego, CA.
- Strauss J.H., Strauss,E.G. and Kuhn,R.J. (1995) Budding of alphaviruses. Trends Microbiol., 3, 346–350. [DOI] [PubMed] [Google Scholar]
- Thomas G. (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol., 3, 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G. and Wengler,G. (1989) Cell-associated West Nile flavivirus is covered with E + pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol., 63, 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Mukhopadhyay,S., Pletnev,S.V., Baker,T.S., Kuhn,R.J. and Rossmann,M.G. (2002) Placement of the structural proteins in Sindbis virus. J. Virol., 76, 11645–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]