For a biological chemist, possessing atomic-level knowledge of all of the molecules inside a livi ng cell would be nirvana. Until recently, however, biological macromolecules were almost always removed from cells before study at the atomic level. Such reductionism has taught us a lot—just try lifting a general biochemistry text—but the approach is inherently limiting because information about biological context is lost. With the advent of in-cell NMR, it is now possible to study atoms in biological macromolecules while they remain in living cells (1). An article by Selenko et al. (2) in this issue of PNAS marks an important advance in these endeavors by providing the first high-resolution glimpse into how the cytoplasm of a higher eukaryotic cell can affect the properties of a folded protein.
Unraveling the effects of biological context is important only if the intracellular environment can impact the properties of proteins. The most obvious difference between a typical in vitro sample and the inside of the cell is the solute concentration. Macromolecular solutes reach concentrations of hundreds of grams per liter in cells (3) and other biological fluids, but most in vitro studies are performed in buffered solution with <1% of the cellular macromolecule concentration. These conditions give optimal signals but may lack biological relevance.
Theories of macromolecular crowding have been exquisitely crafted (4) to address diffusion, stability, and structure, but let us focus here on a simple analogy, umbrellas. When it rains, many people open their umbrellas. What happens as a crowd builds up? Individuals have less space, and the fraction of closed umbrellas is likely to increase. Now consider those closed umbrellas as folded proteins and the open umbrellas as denatured proteins, and you can visualize why macromolecular crowding should stabilize proteins. In short, crowded conditions generally favor species that minimize their surface area.
NMR spectroscopy is the only routine method that provides atomic-level information about proteins in solution. The hydrogen nucleus is very NMR-sensitive, and, of course, proteins are chock full of hydrogen atoms. This surfeit of hydrogens is also a problem, making it difficult to distinguish individual hydrogens. The problem was solved over the last 30 years by developing experiments that spread the information across several independent dimensions and by enriching the protein with other NMR-active nuclei, for instance 15N. Isotopic enrichment allows sorting of the protons because there is only one backbone nitrogen in each amino acid residue.
Isotopic enrichment is also the key to in-cell NMR. The atoms of the protein being studied are enriched in the isotope relative to the atoms in other molecules in the cell, thereby separating their signal from those of the unenriched molecules. The first in-cell protein NMR experiments were performed in yeast by enriching with 19F-labeled amino acids (5). Volker Dötsch and colleagues (6) then did the seminal work to bring high-resolution in-cell 15N-NMR and 13C-NMR to the workhorse bacterium Escherichia coli (1). Their work opened the door to an exciting new world where pH effects (7), metal binding (8), structure formation (9), protein–protein interactions (10), and dynamics (11) can be measured inside cells. This is but the beginning of the story.
There remains a lot that in-cell NMR will tell us about the environment in prokaryotes such as E. coli, but the next goal is to apply in-cell NMR to higher eukaryotic cells. Selenko et al. (2) take this step by using oocytes, which are immature eggs. Frog oocytes are used extensively in biological research because, for cells, they are gigantic. Their ≈1-mm diameter is 10 times that of a human oocyte, 50 times that of a typical somatic cell, and 200 times longer than rod-shaped E. coli cells.
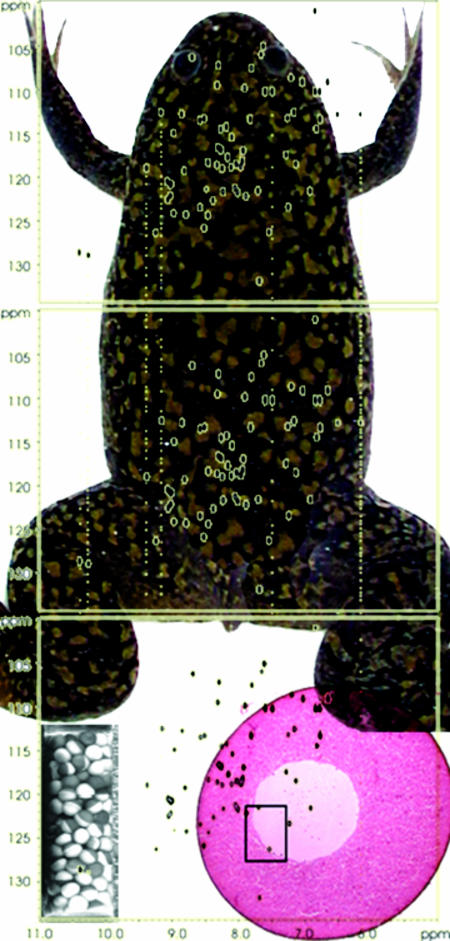
Their significant size makes frog oocytes an easy target for injecting exogenous compounds. Instead of producing their 15N-enriched protein inside the oocytes, Selenko et al. used a robot to inject ≈200 cells with a 15N-enriched recombinant protein purified from E. coli. Such treatment eliminates any background caused by the incorporating of the isotope into other molecules. Fig. 1 shows an NMR sample tube containing enough oocytes for an in-cell NMR experiment. The cells fill ≈1 cm of the tube. Next to the tube is the heteronuclear single quantum correlation spectrum obtained from the sample.
Fig. 1.
Heteronuclear single quantum correlation spectra of 15N-enriched GB1 acquired in dilute solution (Top), egg extracts (Middle), and oocytes (Bottom). An example of the frog, Xenopus laevis, is shown in the background. (Bottom) An NMR tube filled with 200 oocytes injected with the 15N-enriched protein is shown on the left, and a single oocyte is shown on the right.
The subject of the study is a small (7 kDa) stable protein called GB1. This protein is an excellent choice because we know a lot about its structure, dynamics, and stability from years of dilute-solution studies (12). As it turns out, GB1 gives an outstanding spectrum in oocytes. The positions of the peaks in Fig. 1 do not change between dilute solution and the inside of cells. This observation means that the interior of the cells does not change the protein's structure. That's good; crowding should not change the structure of a stable protein because stable proteins are already maximally compact. Selenko et al. (2) did notice, however, that certain resonances are not as intense in cells as they are in dilute solution. The affected residues are all from regions of defined secondary structure. Although dynamic changes in the cell seem to be the likely explanation, proof will require further experiments. Whatever the explanation, however, these data demonstrate that information is missed when we focus only on dilute-solution data.
Next, Selenko et al. (2) asked whether this intensity effect was unique to the inside of cells. To answer this question, they added 15N-enriched GB1 to frog egg extracts. Such extracts are popular for ex vivo experiments because the large size of frog eggs makes them a ready source of cytoplasm. The same intensity effects were observed in cytosol alone. Finally, Selenko et al. asked whether the intensity effects were the result of simple macromolecular crowding by examining GB1 in concentrated solutions of BSA. Again, they obtained the same result. Together, these data show that expertise from several areas—from NMR to cell physiology—will need to be combined to understand protein chemistry in cells.
Although in-cell NMR has opened doors to exciting opportunities, the technique is still in its infancy, and several potential problems need to be addressed. For one, NMR is insensitive, requiring sample concentrations in the micromolar to millimolar range. Do such high concentrations compromise the physiological significance of the results? Early indications are that the dynamics of proteins, at least, are not drastically affected (11). The ability of eukaryotic cells to express proteins of interest is another concern. E. coli can crank out hundreds of milligrams of protein per liter of culture. Similar yields have been reported from higher eukaryotic cells (13). Will these systems yield enough protein while giving a low background from extraneous enriched nuclei? Health and welfare are another challenge. E. coli are robust and survive for days in the spectrometer at room temperature (11). Higher eukaryotic cells are not as hearty. Bioreactor technology (14) will have to be adapted to keep higher eukaryotic cells “healthy and happy” in the spectrometer. Again, several levels of expertise will be required to fulfill the potential of in-cell NMR.
What can we look forward to from in-cell NMR? First, there is a great deal to be learned about so-called natively disordered proteins (15). Unlike stable proteins, this recently identified class of proteins is not maximally compact in dilute solution, and therefore, its properties are expected to be extremely sensitive to crowding. Early studies indicate that some natively disordered proteins can gain structure in cells, whereas others lose structure in cells compared with dilute solution (9, 16). Second, the cytosol is not only crowded, it is also organized (17). NMR provides a method for studying such organization by using residual dipolar couplings (18). It will be interesting to see whether these couplings can be exploited inside cells. Third, the ability to study larger proteins and membrane proteins will also be key. Again, the required tools can be recruited from dilute-solution NMR studies (19). Perhaps the most exciting advance will be the capacity to monitor signal transduction. In-cell NMR could be applied after adding an external stimulus to observe the machinations, be they protein–protein interactions or posttranslational modifications, that turn the stimulus into a biological response. These are ambitious goals, but biological chemists may yet reach their nirvana because high-resolution NMR has been exceeding expectations for >50 years.
Acknowledgments
We thank Elizabeth Pielak and Matthew Redinbo for helpful comments and Philipp Selenko and Peter S. Pielak for making Fig. 1. Our research is supported by the National Science Foundation and the Petroleum Research Fund of the American Chemical Society.
Footnotes
Conflict of interest statement: No conflicts declared.
See companion article on page 11904.
References
- 1.Reckel S., Löhr F., Dötsch V. Chem. Biol. Chem. 2005;6:1601–1606. doi: 10.1002/cbic.200500076. [DOI] [PubMed] [Google Scholar]
- 2.Selenko P., Serber Z., Gadea B., Ruderman J., Wagner G. Proc. Natl. Acad. Sci. USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luby-Phelps K. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 4.Minton A. P. Biophys. J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams S. P., Haggie P. M., Brindle K. M. Biophys. J. 1997;72:490–498. doi: 10.1016/S0006-3495(97)78690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serber Z., Keatinge-Clay A. T., Ledwidge R., Kelly A. E., Miller S. M., Dötsch V. J. Am. Chem. Soc. 2001;123:2446–2447. doi: 10.1021/ja0057528. [DOI] [PubMed] [Google Scholar]
- 7.Shimba N., Serber Z., Ledwidge R., Miller S. M., Craik C. S., Dötsch V. Biochemistry. 2003;42:9227–9234. doi: 10.1021/bi0344679. [DOI] [PubMed] [Google Scholar]
- 8.Hubbard J. A., MacLachlan L. K., King G. W., Jones J. J., Fosberry A. P. Mol. Microbiol. 2003;49:1191–1200. doi: 10.1046/j.1365-2958.2003.03628.x. [DOI] [PubMed] [Google Scholar]
- 9.McNulty B. C., Young G. B., Pielak G. J. J. Mol. Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Burz D. S., Dutta K., Cowburn D., Shekhtman A. Nat. Methods. 2006;3:80–81. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant J. E., Lecomte J. T., Lee A. L., Young G. B., Pielak G. J. Biochemistry. 2005;44:9275–9279. doi: 10.1021/bi050786j. [DOI] [PubMed] [Google Scholar]
- 12.Goehlert V. A., Krupinska E., Regan L., Stone M. J. Protein Sci. 2004;13:3322–3330. doi: 10.1110/ps.04926604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strauss A., Bitsch F., Fendrich G., Graff P., Knecht R., Meyhack M., Jahnke W. J. Biomol. NMR. 2005;31:343–349. doi: 10.1007/s10858-005-2451-3. [DOI] [PubMed] [Google Scholar]
- 14.Gmati D., Chen J., Jolicoeur M. Biotechnol. Bioeng. 2005;89:138–147. doi: 10.1002/bit.20293. [DOI] [PubMed] [Google Scholar]
- Daughdrill G. W., Pielak G. J., Uversky V. N., Cortese M. S., Dunker A. K. In: Protein Folding Handbook. Kiefhaber T., editor. Weinheim, Germany: Wiley; 2005. pp. 275–357. [Google Scholar]
- 16.Dedmon M. M., Patel C. N., Young G. B., Pielak G. J. Proc. Natl. Acad. Sci. USA. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovadi J., Saks V. Mol. Cell Biochem. 2004;256/257:5–12. doi: 10.1023/b:mcbi.0000009855.14648.2c. [DOI] [PubMed] [Google Scholar]
- 18.Lipsitz R. S., Tjandra N. Annu. Rev. Biophys. Biomol. Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 19.Tzakos A. G., Grace C. R. R., Lukavsky P. J., Riek R. Annu. Rev. Biophys. Biomol. Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]