Abstract
Rab proteins constitute the largest branch of the Ras GTPase superfamily. Rabs use the guanine nucleotide-dependent switch mechanism common to the superfamily to regulate each of the four major steps in membrane traffic: vesicle budding, vesicle delivery, vesicle tethering, and fusion of the vesicle membrane with that of the target compartment. These different tasks are carried out by a diverse collection of effector molecules that bind to specific Rabs in their GTP-bound state. Recent advances have not only greatly extended the number of known Rab effectors, but have also begun to define the mechanisms underlying their distinct functions. By binding to the guanine nucleotide exchange proteins that activate the Rabs certain effectors act to establish positive feedback loops that help to define and maintain tightly localized domains of activated Rab proteins, which then serve to recruit other effector molecules. Additionally, Rab cascades and Rab conversions appear to confer directionality to membrane traffic and couple each stage of traffic with the next along the pathway.
Keywords: Rab GTPases
Intracellular organelles are a defining feature of eukaryotic cells. Each organelle must maintain its characteristic structure, biochemical composition, and function, which represents a formidable challenge for the organelles of the exocytic and endocytic pathways given the continuous flow of protein and membrane along these pathways. The exocytic pathway sorts newly synthesized proteins from the endoplasmic reticulum, through the Golgi apparatus to their final destination at the lysosome/vacuole or plasma membrane. Conversely, the endocytic pathway is required for the uptake of nutrients and the internalization of receptors. Newly internalized material is transported to the early endosome, a tubulo-vesicular network localized to the cell periphery. Proteins destined for recycling are sorted to recycling endosomes and then to the plasma membrane, whereas proteins destined for degradation are transported to late endosomes (also called prelysosomes or prevacuoles) and subsequently to the lysosome/vacuole (reviewed in ref. 1).
The mechanisms underlying membrane traffic can be divided into four essential steps (Fig. 4, which is published as supporting information on the PNAS web site). Cargo is selected, and transport intermediates in the form of vesicles or tubules are formed (2–4). For simplicity, we will refer to the transport intermediates as vesicles. These vesicles are delivered to their target membrane, often using molecular motors to transport vesicles along the cell’s microtubule or actin filament system. Tethering then brings the vesicle and the target membrane into close proximity. The final step is the fusion of those vesicles with the target membrane. As we will discuss, Rab GTPases have been implicated in the regulation of each of these steps in membrane traffic.
Rabs are a ubiquitously expressed family of small (20–29 kDa) monomeric Ras-like GTPases (reviewed in ref. 5). To date, 11 Rabs have been identified in yeast (including Sec4p and the Ypt proteins) and >60 in mammalian cells (6). The much larger number of Rabs in mammals reflects the higher complexity of transport events in higher eukaryotes, as indicated by the fact that several mammalian Rab proteins are expressed only in certain tissues and differentiated cell types, where they participate in specialized transport pathways (7–13).
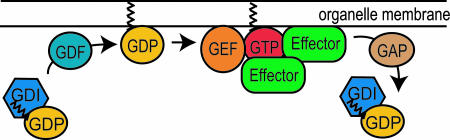
Rab GTPases function as molecular switches, cycling between GTP-bound and GDP-bound states (Fig. 1). This switch is controlled by guanine nucleotide exchange factors (GEFs), which trigger the binding of GTP, and GTPase-activating proteins (GAPs), which accelerate hydrolysis of the bound GTP to GDP (for review see refs. 14 and 15). Rabs also undergo a membrane insertion and extraction cycle, which is partially coupled to the nucleotide cycle (Fig. 1). Membrane insertion requires the irreversible modification of two carboxyl-terminal cysteines with isoprenyl lipid (geranylgeranyl) moieties (16). A protein called GDP dissociation inhibitor (GDI) binds to prenylated Rabs in their GDP-bound form (17–19), masking their isoprenyl anchor (20) and thereby maintaining the Rab protein in the cytosol (reviewed in ref. 21). Membrane attachment of Rab proteins therefore requires the function of a GDI displacement factor (GDF; reviewed in ref. 22). Once dissociated from GDI the Rabs are available for GEF-stimulated GTP binding. The active, membrane-bound Rabs are then able to fulfill their various functions in membrane traffic by binding to their specific effectors. After inactivation by their specific GAPs, the GDP-bound Rabs can be extracted from the membrane by GDI and recycled back to the cytosol (17–19, 23, 24).
Fig. 1.
The nucleotide and membrane attachment/detachment cycles of Rab GTPases. Inactive (GDP-bound) prenylated Rab GTPases are bound to GDI, which masks their isoprenyl anchor and thereby keeps the Rab in a soluble cytosolic form (17–20, 23). Membrane attachment of Rabs requires the function of a GDF that dissociates the GDI–Rab complex and allows the prenyl anchor to be inserted into the membrane. Subsequently, specific GEFs exchange the bound GDP for GTP, thereby activating the Rab GTPases. The active, membrane-bound Rabs are then able to fulfill their various functions in membrane traffic by binding to their specific effector proteins. Finally, specific GAPs inactivate the Rabs by accelerating the hydrolysis of the bound GTP into GDP. The inactive, GDP-bound Rabs can then be extracted from the membrane by GDI and recycled for another round of function (reviewed in ref. 22).
The term effector implies a protein that responds to a specific Rab and mediates at least one element of its downstream effects (for review see refs. 14, 25, and 26). They have been operationally defined through their ability to bind to a specific Rab, selectively in its GTP-bound state, and have been identified through a variety of approaches, such as the yeast two-hybrid system, genetic screens, and affinity purification. We will review the rapidly growing collection of Rab effectors and describe what is known about Rab effector recognition. As we will describe, each Rab appears to signal through a variety of different effectors that together act to translate the signal from one Rab protein to several diverse aspects of membrane transport. We will also describe how effectors help establish membrane domains marked by a specific Rab and how these domains mature by a Rab cascade mechanism. These principles are fundamental to understanding how Rabs and their effectors contribute to specificity in membrane traffic.
Specific Roles of Rab Effectors in Membrane Traffic
Although the list of known Rab effectors is growing ever longer (see Tables 1 and 2, which are published as supporting information on the PNAS web site), because of space limitations, we must focus on selected examples chosen from a variety of different systems.
A Potential Sorting Function for Rab Effectors in Vesicle Formation.
In forming a transport vesicle, the correct cargo and the appropriate transport and fusion machinery must be incorporated into the vesicle before scission from the donor membrane. Several lines of evidence implicate Rabs in cargo selection and vesicle formation (27–31). However, only one effector with a clearly defined role in vesicle formation has been found to date, TIP47 (Fig. 5, which is published as supporting information on the PNAS web site).
Mannose 6-phosphate receptors (MPRs) transport newly synthesized lysosomal hydrolases bearing mannose 6-phosphate from the trans-Golgi network to late endosomes (prelysosomes), but the receptor must be recycled back to the Golgi for additional rounds of transport. Diaz and Pfeffer (32) identified a protein required for recycling and established that it binds to the cytoplasmic tail of MPRs, hence its name, tail-interacting protein of 47 kDa (TIP47). TIP47 also binds GTP-Rab9, thus it is a Rab9 effector (31). Rab9 is principally localized to late endosomes and, like TIP47, is required for the recycling of MPRs (33, 34). GTP–Rab9 increases the association of TIP47 with late endosomes and, furthermore, increases the affinity of TIP47 for MPRs (31). Therefore, GTP–Rab9 stimulates the capture of MPRs through its effector TIP47, which leads to enrichment of the MPRs within vesicles that carry the appropriate Rab GTPase for their transport back to the Golgi (Fig. 5). Additional studies will be needed to determine whether this type of mechanism is used at other stages of membrane traffic.
Motors or Motor Adapters Are Rab Effectors in Intracellular Transport.
Vesicles are often actively transported through the cytoplasm toward their target membrane by using either actin-dependent motors (myosins) or microtubule-dependent motors (kinesins or dyneins) (reviewed in ref. 35). A number of studies have implicated Rabs and their effectors in the regulation of this transport step.
A screen for yeast mutants that block secretion (36) identified one of the first Rab GTPases, Sec4p (37, 38) and its specific GEF, Sec2p. Further studies showed that sec2 mutants display an accumulation of vesicles randomly distributed throughout the cell, implying that the activation of Sec4p by Sec2p is needed for the polarized delivery of secretory vesicles (39). Supporting the proposal that activated GTP–Sec4p promotes myosin-dependent movement of secretory vesicles along actin cables, Sec4p was found to coimmunoprecipitate with the type V myosin, Myo2p (40). However, it is not yet known how Sec4-containing vesicles are attached to the myosin motor.
In some cases, it appears that a motor protein is directly attached to a Rab (see Table 2). For instance, yeast two-hybrid data suggest that active Rab6 binds the microtubule motor Rabkinesin-6 and could thereby promote the delivery of vesicles from the Golgi to the endoplasmic reticulum (41). More typically, Rabs interact with motors via an intermediary protein (see Table 2). Perhaps the best-studied example is the recruitment of myosin-Va, to melanosomes by Rab27a (Fig. 6, which is published as supporting information on the PNAS web site). Rab27a is localized to the pigment-containing melanosome granules and is essential for their retention at the cell periphery of melanocytes (42). The Rab27a effector melanophilin links Rab27a-positive melanosomes to the actin motor myosin-Va (43–45). Without myosin-Va-dependent capture of these organelles on actin filaments, melanosomes are not retained in the cell periphery and, subsequently, cannot be transferred to neighboring keratinocytes (46). As a result, mouse mutants of Rab27, its effector melanophilin, or myosin-Va display a pigmentary dilution of their skin hence their names, ashen, leaden, and dilute mice, respectively (47, 48). Genetic alterations of human Rab27a lead to Griscelli syndrome that is manifest as light skin and hair color. Additionally, Griscelli patients display various immunodeficiencies because Rab27a and its effectors are also required for various exocytic transport events (reviewed in ref. 49).
Rab Effectors in Vesicle Tethering.
The next step of membrane traffic is the tethering of transport vesicles to the target membrane, a process that brings the vesicle and target membrane into close proximity. Tethering factors can be divided into two groups: long coiled-coil proteins and large multisubunit complexes (reviewed in ref. 50). The former includes p115 (in yeast, Uso1p), early endosome antigen 1 (EEA1), and the Golgins, and the latter group includes the exocyst (Sec6/8 complex), TRAPP-I and TRAPP-II, the conserved oligomeric Golgi complex (Sec34/35p complex), and the homotypic fusion and vacuole protein sorting (HOPS)/vacuole protein sorting (VPS) complex.
Coiled-coil tethers are mostly long, rod-like molecules (51) thought to function as a bridge between the vesicle and target membranes. Bipartite or tripartite complexes of these proteins form the required membrane tethers (reviewed in ref. 50). For example, the interaction of the vesicle-associated protein p115 with a Golgi-residing GM130- and GRASP65-containing complex is thought to tether endoplasmic reticulum-derived vesicles to the Golgi (reviewed in ref. 50). Interestingly, both tethers, vesicle-associated p115 (52) and the GM130/GRASP65 complex at the Golgi (53), have been shown to be effectors of Rab1 (refs. 53–55 and Fig. 7A, which is published as supporting information on the PNAS web site). These data are consistent with studies showing a requirement for Rab function on both donor and acceptor organelles for fusion events (56–59). Active Rab1 is required for recruitment of p115 into transport vesicles (ref. 52 and Fig. 7A), and the yeast Rab1 homologue, Ypt1p similarly increases the membrane association of the p115 homologue, Uso1p (60). The role of Rab1 on the Golgi may be in the regulation of assembly and/or activity of the GM130/GRASP65 complex (53). Recent evidence suggests a regulatory role for some of these coiled-coil tethers. Vesicle-localized Giantin and Golgi-localized GM130 positively influence the binding of p115 to Rab1 (61). This regulation might ensure specific activation of p115 and prevent its mistargeting to other Rab1-positive membranes because Rab1 displays a much broader distribution along the exocytic pathway than does Giantin and GM130 (62).
The exocyst was the first large, multisubunit tethering complex to be identified as a Rab effector (63). It is an octameric complex required for tethering secretory vesicles to the plasma membrane in yeast (63, 64). One of its subunits, Sec15p, directly interacts with the Rab Sec4p it its GTP-bound form (ref. 63 and Fig. 7B). Sec8p, another exocyst subunit, was recently found to coimmunoprecipitate with Sec4p, suggesting that the entire exocyst complex acts downstream of Sec4p (65). Mammalian Sec15 was recently identified as a Rab11 effector, suggesting a conserved interaction between Rabs and the exocyst (66). The current view is that Rabs, in coordination with other GTPases, such as Rho1, Rho3, and Cdc42 in yeast and RalA in mammals (67–73), regulate vesicle tethering by promoting rearrangements within, or assembly of, the exocyst complex. In agreement with this view, preliminary data indicate that Sec4p is required for the assembly of the exocyst complex (ref. 63 and Fig. 7B). Further studies will be necessary to investigate the exact mechanism.
One interesting idea about the role of Rabs in tethering is that they might link the functions of coiled-coil tethers with those of the multisubunit tethering complexes. For instance, the yeast tethering complex TRAPP-I displays exchange activity toward the Rab GTPase Ypt1p (74), and active Ypt1p would then be able to recruit its potential effector Uso1p (60, 75).
Rab Effectors in Membrane Fusion.
Rabs also influence vesicle fusion, through their effects on members of the family of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). Vesicle-associated SNAREs and SNAREs at the target membrane (t-SNAREs) form trans-SNARE complexes leading to the opening of a fusion pore and final fusion of vesicle and organelle membranes (76). Most evidence indicates indirect regulation of SNARE function by Rabs. For example, interaction of the Rab5 effector and coiled-coil tethering factor, EEA1, with the t-SNARE, syntaxin-13, is required for homotypic early endosome fusion (ref. 77 and Fig. 8A, which is published as supporting information on the PNAS web site). The class C VPS/HOPS tethering complex, an effector of the yeast Rab Ypt7p, also binds SNAREs required for vacuole fusion (78). When bound to Ypt7p, HOPS interacts with unpaired SNAREs and, subsequently, Ypt7p appears to influence SNARE complex assembly via HOPS (79).
We recently identified Sro7p, the yeast lethal giant larvae family member, as a Sec4p effector (80). Sro7p also binds to Sec9p, a plasma membrane SNARE at the target membrane (81). Furthermore, GTP–Sec4p, Sro7p, and Sec9p can form a ternary complex (80). Sro7p does not contain coiled-coil regions, nor is it a component of a large complex, therefore it does not fit either profile of a tethering factor. However, genetic data demonstrate that Sro7p acts downstream of Sec4p in an exocyst-related function (80). Furthermore, it binds to the exocyst subunit Exo84p (82). Therefore, Sec4p might influence SNARE function via the ternary complex with Sro7p and Sec9p, in conjunction with Sec4p’s established function in vesicle tethering through its other effector, the exocyst (Fig. 8B).
Another group of Rab effectors or effector-interacting proteins that influence SNARE function are members of the SM (Sec1/Munc18) family of proteins (83–85). However, the precise role of these proteins is still unclear (reviewed in ref. 86).
N-ethylmaleimide-sensitive factor (NSF), an ATPase that disassembles preformed cis–SNARE complexes before tethering, is required for Rab effector–SNARE interaction and effector complex formation (77, 87). Interaction of the class C VPS/HOPS tethering complex, an effector of the yeast Rab Ypt7p, and SNAREs depends on the presence of the NSF homologue Sec18p (87). In mammals, the association of the Rabex5–Rabaptin5 complex with the tethering factor EEA1 on endosomal membranes also depends on NSF (ref. 77 and Fig. 8A). These actions of NSF may couple cis–SNARE complex disassembly to later stages of the reaction, tethering, and trans-SNARE pairing.
Taken together, Rab GTPase signaling is required for fusion to the target membrane as indicated by the finding that SNARE-interacting proteins are Rab effectors. However, the exact functions of these interactions are still somewhat elusive.
Achieving Specificity in Membrane Traffic Via Rab Effector Interactions
How Does an Effector Find its Rab?
Because of space limitation we can only briefly summarize the structural studies on Rab effector recognition and refer the reader to the excellent review by S. R. Pfeffer (88), which covers the topic in detail.
Specific Rab signal transmission requires specificity of effector recognition. This is particularly important because some Rabs display partially overlapping localizations within certain organelles, such as Rab5 and Rab4 on early endosomes. Conversely, some Rab effectors are able to recognize more than one Rab protein as exemplified by Rabaptin5, which appears to link endocytosis and recycling by its specific interaction with both Rab4 and Rab5 (89). Rab proteins are highly homologous and share a common structure with relatively subtle variations. In contrast, Rab effectors are highly divergent, which reflects the needs for different types of proteins to mediate the diverse pathways downstream of Rab GTPases. How then is specific binding of structurally conserved Rabs to structurally divergent effectors achieved?
Effectors, by definition, must recognize the GTP-bound form of a Rab GTPase. The presence of GTP is reflected by changes in the Rab switch region (switch I and switch II), and available crystal structures and biochemical binding studies confirm that effectors mainly interact with these conserved regions (90–94). Although highly conserved among Rab proteins, recent evidence suggests that this region can confer specificity. Small differences, i.e., changes in only a few amino acids, within the switch region of different Rab proteins account for the observed Rabenosyn5 binding specificity (94). Additionally, conservation in sequence does not necessarily imply similar binding specificity. The crystal structure of the Rab3–Rabphilin3 complex showed that a conserved triad of hydrophobic amino acids within the switch region of Rab3 is required for Rabphilin3 binding (90). Although Rab5 has the identical sequence, it is unavailable for Rabphilin3 binding because changes in the amino acid composition between the switch regions dramatically alters the conformation of this hydrophobic triad (95). Thus, despite the very similar overall shape of the Rab switch regions, subtle variations within and outside these regions appear to confer very distinctive surfaces that permit the required specificity. Finally, effectors are also found to interact with nonconserved regions of Rab proteins, further increasing binding specificity.
Because of their structural diversity, Rab effectors generally do not appear to have a common structural motif for interaction with their Rab GTPases. However, in case of existing similar Rab-binding motifs, like, for instance, the zinc-finger domains of the Rab5 effectors EEA1 and Rabaptin5 and the Rab3 effector Rabphilin3, specificity is achieved by relatively small differences in their structure (90, 91, 93).
Given the great structural diversity of Rab effectors and potentially also their Rab-interacting domains, further studies will be necessary to investigate the full range of effector–Rab binding interactions.
Rab Effector Interactions in Membrane Organization and Maturation.
Each Rab must be localized to, and activated at, a specific organelle to fulfill its function. Furthermore, some organelles can carry distinct Rab-containing subdomains, each of which participate in different transport steps. How is this very exquisite localization achieved?
Many factors are needed for the correct targeting of Rab proteins (reviewed in ref. 96). GDI binds the prenylated, GDP-bound Rabs and keeps them in the cytosol awaiting membrane attachment (reviewed in ref. 22). GDFs are integral membrane proteins that displace GDI from the Rab proteins and thereby allow them to be inserted into the membrane (ref. 97 and reviewed in ref. 96). However, GDI displays no specificity in Rab binding, and the only well characterized GDF, Yip3, displays only limited specificity (19, 22). Therefore, GDI and GDF are not sufficient to generate the necessary specificity in Rab localization.
It is, however, important to remember that until a Rab is activated it is subject to back extraction by GDI. Therefore, GEFs can contribute to Rab localization specificity. Effectors can also contribute to the localization of Rabs. By binding to the activated Rab, an effector may directly contribute to its localization by limiting its diffusion in the membrane or indirectly block Rab inactivation by a GAP, which would otherwise lead to extraction by GDI. Thus, Rabs recruit effectors to the appropriate membrane, but at the same time effectors can also contribute to the localization of the Rab. As discussed below, by physically coupling a GEF to an effector, these mechanisms can all work in concert to achieve a very specific localization.
GEF–Rab effector complexes create positive feedback loops that couple Rab localization and activation to downstream effector function.
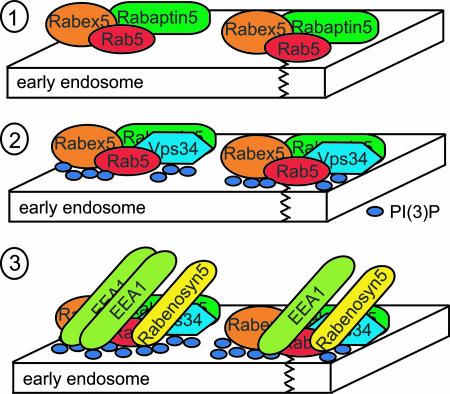
One mechanism to specifically counteract inactivation once the Rab has reached the correct location is through the formation of a GEF–Rab effector complex. A well studied example is the Rabex5–Rab5–Rabaptin5 complex (Fig. 2). Rab5 is the endocytic Rab required for transport and heterotypic fusion of plasma membrane-derived endocytic vesicles to early endosomes and for homotypic early endosome fusion (59, 98–102). For its functions, active Rab5 must be localized to both endocytic vesicles and early endosomes. This is achieved in part through the actions of the Rab5–GEF, Rabex5, and one of its effectors, Rabaptin5 (59). The initial membrane recruitment of Rab5 is followed by Rabex5-mediated activation (103, 104). GTP–Rab5 then is able to interact with its various effectors, including Rabaptin5 (105). Rabaptin5 in turn binds to Rabex5 and, additionally, increases the exchange activity of Rabex5 on Rab5 (ref. 106 and Fig. 2 Top). These interactions generate a positive feedback loop, which counteracts GAP inactivation and GDI-mediated membrane extraction, thereby ensuring that Rab5 stays in its activated, GTP-bound form attached to endocytic vesicles or the early endosome.
Fig. 2.
Rab–GEF effector complexes stabilize activated Rabs on membranes and allow the installment of Rab-specific membrane domains. (Top) After recruitment to the membrane of the early endosome, Rab5 is activated by its GEF Rabex-5 (103, 104). GTP–Rab5 then interacts with its effector Rabaptin5 (105). Rabaptin5 in turn binds to Rabex-5 and, additionally, increases the exchange activity of Rabex-5 on Rab5 (106), thereby stabilizing Rab5 in its GTP-bound state. This positive feedback loop counteracts GAP inactivation and GDI-mediated membrane extraction, thereby ensuring that GTP–Rab5 stays attached to the early endosome as long as necessary to recruit other necessary effectors. (Middle) The PI-3-OH kinase hVPS34/p150 (VPS34) is a Rab5 effector recruited to the Rabex-5–Rab5–Rabaptin5 platform (110). VPS34 generates the lipid PI(3)P (111), which is subsequently enriched on the early endosomal membrane (112). (Bottom) Both signals, PI(3)P and GTP–Rab5, are required to recruit more Rab5 effectors, such as EEA1 (91, 110, 115–118) and Rabenosyn5 (85). EEA1 and Rabenosyn5 are factors required for homotypic early endosome fusion and fusion of vesicles to the early endosome (85, 110). Therefore, the specificity of early endosomal membrane traffic is ensured by the specific recruitment of the key proteins into Rab-defined membrane domains.
Similar positive feedback loops have also been found in other systems. Vps33p, an effector of the Rab GTPase Ypt7p, is a subunit of the yeast class C VPS/HOPS complex (84), whereas another subunit of the same complex, Vps39p, serves as a Ypt7p GEF (107). Our data indicate that there is a complex between the yeast exocytic Rab, Sec4p (37), one of its effectors, the exocyst (63), and the Sec4p GEF, Sec2p (39, 108). Thus, the principle of GEF–Rab effector complexes appears to be a recurring, if not general, mechanism.
Rab effector interactions achieve specificity in membrane traffic by defining specific membrane domains.
Organelle- and Rab-specific membrane domains on the endocytic pathway have been intensively characterized. Early endosomes (also called sorting endosomes) are found to either harbor only Rab5 or a combination of Rab4 and Rab5, whereas recycling endosomes carry distinct domains of Rab4 and Rab11. Rab7 and Rab9 similarly share the late endosome (98–100, 109). The establishment of such specific membrane domains appears to be achieved by the interplay between Rabs and their effectors.
As described in the previous section, the Rabex5–Rab5–Rabaptin5 complex serves as a positive feedback loop to specifically concentrate activated Rab5 at the early endosome. Among the Rab5 effectors recruited to this GTP–Rab5 platform is the phosphatidylinositol (PI) 3-OH kinase hVPS34/p150 (ref. 110 and Fig. 2 Middle). This kinase catalyzes the phosphorylation of PI to PI 3-phosphate [PI(3)P] (111). The recruitment of this kinase by Rab5 therefore leads to the enrichment of PI(3)P on early endosomes (ref. 112 and Fig. 2 Middle). Rab5 also recruits another effector, EEA1 (refs. 91 and 110 and Fig. 2 Bottom), which contains a FYVE domain that specifically binds to PI(3)P (113–115). Specific binding of EEA1 to early endosomes requires interaction of this FYVE domain with PI(3)P in addition to binding to GTP–Rab5 (116–118). The necessity for these two recruitment signals is demonstrated by the observation that endocytic vesicles, which contain active Rab5 but lack hVPS34/p150 and therefore cannot produce PI(3)P, fail to recruit EEA1 (29, 59, 119). Other Rab5 effectors, such as Rabenosyn5, display the same dual requirement for their recruitment to their sites of function (ref. 85 and Fig. 2). In yeast, recruitment of Vac1p, an effector of the yeast Rab5 homologue Vps21/Ypt51p (83), to endosomes also requires synthesis of PI(3)P by the hVPS34/p150 homologue Vps34p and subsequent binding of its FYVE domain to this lipid (113, 120, 121). Signals other than lipids may also cooperate with Rab binding. The Rab9 effector p40 may require phosphorylation by PIKfyve in addition to active Rab9 for efficient membrane attachment (122, 123).
Thus, localized clustering of active Rabs ensures specific recruitment and regulation of proteins required for membrane traffic. These Rab membrane domains appear to be highly flexible. As in the case of Rab5, factors are recruited that are necessary for tethering/fusion of endocytic vesicles to the early endosome (heterotypic fusion) and also for homotyic early endosome fusion (85, 110). Reaching an even higher level of complexity, Rab membrane domains might be structurally and functionally linked. For example, Rabaptin4, Rabaptin5, Rabenosyn5, and Rabip4′ are each able to interact with both Rab5 and Rab4 (89, 124–126), which might allow coordination of endocytic traffic (via Rab5) with recycling traffic (via Rab4).
In summary, active Rabs are enriched in microdomains on specific membranes by effector- and GEF-mediated positive feedback loops. Active Rabs can recruit additional effectors into these domains, which then accomplish their function in membrane traffic. The high specificity of effector localization is achieved by use of two parallel signals for efficient membrane recruitment.
Rab cascades and Rab conversions couple trafficking events and lead to membrane maturation.
Each organelle carries its own set of Rabs, which ensures the specificity of intracellular membrane transport, and Rabs appear to be required for all steps of membrane traffic. Given the need for efficient membrane traffic in a cell, it would make sense to couple the different transport steps to ensure transport continuity and specificity. This might, at least in part, be accomplished by so-called Rab cascades and Rab conversions. The concept of Rab cascades and conversions postulates that the GEF of a downstream Rab GTPase will also serve as an effector of an upstream Rab protein.
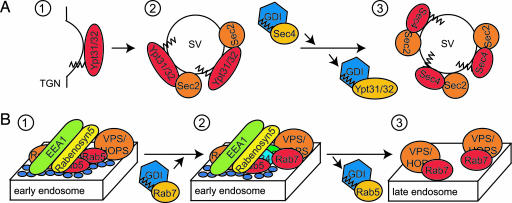
The first Rab cascade was identified through the analysis of the final stage of the yeast exocytic pathway (Fig. 3A). The redundant and highly homologous Rabs Ypt31p and Ypt32p are involved in several trafficking events at the Golgi (30, 127). Genetic and biochemical characterization revealed that Sec2p is an effector of these Rabs (128). Sec2p is also the GEF for Sec4p (39), a Rab required for transport of secretory vesicles from the trans-Golgi network to the plasma membrane in yeast (37). Thus, Ypt31/32p and Sec4p are functionally linked in a regulatory Rab cascade through the exchange protein Sec2p (Fig. 3A). Such cascades might ensure that distinct trafficking events, like, for instance, vesicle formation and vesicle delivery, are functionally coupled, affording greater specificity and continuity in membrane traffic. The widespread use of such a mechanism is suggested by the identification of Rab cascades in mammals (129) and other potential Rab cascades in yeast (74, 130).
Fig. 3.
Rab cascades/Rab conversion. (A) Yeast Ypt31/32 form a Rab cascade with Sec2p and Sec4p. The redundant yeast Rab GTPases Ypt31p and Ypt32p are involved in several Golgi-related trafficking steps, including the exit of secretory vesicles from the trans-Golgi network (TGN) (1) (30, 127). Sec2p is the GEF for the Rab GTPase Sec4p (39). Both Sec2p and Sec4p are required for the transport of secretory vesicles (SV) from the TGN to the plasma membrane (37). Biochemical characterization revealed that Sec2p is an effector of Ypt31/32p (128). Ypt31/32p are at least partially responsible for the recruitment of Sec2p to secretory vesicles (2) (128), leading to the activation of Sec4p (3). (B) Rab conversion of Rab5 to Rab7 might drive early to late endosome maturation. Early endosomes are partially marked by the presence of Rab5 and its effectors (1; see Fig. 2). One Rab5 effector is Vps11p, a subunit of the conserved class C VPS/HOPS complex (1) (110, 129). A second subunit of this complex, Vps39p, displays GEF activity toward the yeast Rab7 ortholog, Ypt7p (107). Therefore, Rab5, class C VPS/HOPS, and Rab7 appear to form a Rab cascade on early endosomes (2). Gradual inactivation and replacement of Rab5 and its effectors concomitant with recruitment and activation of Rab7 and its effectors might lead to maturation of early endosomes into late endosomes (3) as has been observed (129).
A recent study puts an interesting twist on the function of Rab cascades (Fig. 3B). By using fast, quantitative, live-cell imaging, Rink et al. (129) observed that over time individual Rab5-containing early endosomes grew in size and moved toward the center of the cell. This movement was accompanied by an eventually complete loss of Rab5 and the Rab5 effector EEA1 (129). Strikingly, coincident with the loss of Rab5, Rab7 was recruited onto these endosomes. Late endosome-specific cargo appeared within the same time scale of Rab exchange, and the observed endosomes acquired degradative properties typical of late endosomes (129). Thus, it appears that the Rab5-containing early endosomes were converted into Rab7-containing late endosomes (ref. 129 and Fig. 3B).
The Rab5–Rab7 conversion appears to be a result of a Rab cascade. Vps11p, one subunit of the conserved class C VPS/HOPS complex (84, 131–134), is a Rab5 effector (110, 129), whereas a second subunit, Vps39p, displays GEF activity toward the yeast Rab7 ortholog (107). Therefore, these data indicate that a Rab cascade (Rab5–class C VPS/HOPS complex–Rab7) is the basis of the Rab5 (early endosomes) to Rab7 (late endosomes) conversion.
Both principles, Rab cascades and conversions, require that the upstream Rab must be deactivated and extracted from the membranes containing the downstream Rab GTPase. For that reason, we can speculate that the GAPs that act on the upstream Rabs might be effectors of the downstream Rabs. The process of Rab conversion also adds mechanistic detail to the paradigm of organelle maturation (135), by explaining one aspect of the maturation of an early endosome into a late endosome.
Outlook
Recent years have brought the identification of numerous effectors of Rab GTPases. Analysis of their function has significantly improved our understanding of how Rab GTPases help to achieve specificity and directionality in intracellular membrane traffic. Although some Rab effectors share common principles of function, the majority of them appear to have adapted to their unique requirements with great structural diversity. Ongoing characterization of the role of these effectors and identification and analysis of additional Rab effectors will help to elucidate the mechanisms underlying membrane transport specificity.
Supplementary Material
Abbreviations
- GEF
guanine nucleotide exchange factor
- GAP
GTPase-activating protein
- GDI
GDP dissociation inhibitor
- GDF
GDI displacement factor
- MPR
mannose 6-phosphate receptor
- TIP47
tail-interacting protein of 47 kDa
- EEA1
early endosome antigen 1
- VPS
vacuole protein sorting
- HOPS
homotypic fusion and vacuole protein sorting
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- PI
phosphatidylinositol
- PI(3)P
PI 3-phosphate
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Geli M. I., Riezman H. J. Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- 2.Palade G. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 3.Rothman J. E. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 4.Kartberg F., Elsner M., Froderberg L., Asp L., Nilsson T. Biochim. Biophys. Acta. 2005;1744:351–363. doi: 10.1016/j.bbamcr.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Chavrier P., Goud B. Curr. Opin. Cell. Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 6.Schultz J., Doerks T., Ponting C. P., Copley R. R., Bork P. Nat. Genet. 2000;25:201–204. doi: 10.1038/76069. [DOI] [PubMed] [Google Scholar]
- 7.Padfield P. J., Balch W. E., Jamieson J. D. Proc. Natl. Acad. Sci. USA. 1992;89:1656–1660. doi: 10.1073/pnas.89.5.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senyshyn J., Balch W. E., Holz R. W. FEBS Lett. 1992;309:41–46. doi: 10.1016/0014-5793(92)80735-y. [DOI] [PubMed] [Google Scholar]
- 9.Oberhauser A. F., Monck J. R., Balch W. E., Fernandez J. M. Nature. 1992;360:270–273. doi: 10.1038/360270a0. [DOI] [PubMed] [Google Scholar]
- 10.Baldini G., Hohl T., Lin H. Y., Lodish H. F. Proc. Natl. Acad. Sci. USA. 1992;89:5049–5052. doi: 10.1073/pnas.89.11.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elferink L. A., Anzai K., Scheller R. H. J. Biol. Chem. 1992;267:5768–5775. [PubMed] [Google Scholar]
- 12.Lutcke A., Jansson S., Parton R. G., Chavrier P., Valencia A., Huber L. A., Lehtonen E., Zerial M. J. Cell Biol. 1993;121:553–564. doi: 10.1083/jcb.121.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olkkonen V. M., Dupree P., Killisch I., Lutcke A., Zerial M., Simons K. J. Cell Sci. 1993;106:1249–1261. doi: 10.1242/jcs.106.4.1249. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer S. R. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 15.Segev N. Curr. Opin. Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 16.Kinsella B. T., Maltese W. A. J. Biol. Chem. 1992;267:3940–3945. [PubMed] [Google Scholar]
- 17.Garrett M. D., Zahner J. E., Cheney C. M., Novick P. J. EMBO J. 1994;13:1718–1728. doi: 10.1002/j.1460-2075.1994.tb06436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro A. D., Pfeffer S. R. J. Biol. Chem. 1995;270:11085–11090. doi: 10.1074/jbc.270.19.11085. [DOI] [PubMed] [Google Scholar]
- 19.Shisheva A., Chinni S. R., DeMarco C. Biochemistry. 1999;38:11711–11721. doi: 10.1021/bi990200r. [DOI] [PubMed] [Google Scholar]
- 20.Rak A., Pylypenko O., Durek T., Watzke A., Kushnir S., Brunsveld L., Waldmann H., Goody R. S., Alexandrov K. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 21.Goody R. S., Rak A., Alexandrov K. Cell. Mol. Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer S., Aivazian D. Nat. Rev. Mol. Cell. Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 23.Araki S., Kikuchi A., Hata Y., Isomura M., Takai Y. J. Biol. Chem. 1990;265:13007–13015. [PubMed] [Google Scholar]
- 24.Ullrich O., Stenmark H., Alexandrov K., Huber L. A., Kaibuchi K., Sasaki T., Takai Y., Zerial M. J. Biol. Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 25.Zerial M., McBride H. Nat. Rev. Mol. Cell. Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 26.Munro S. Curr. Opin. Cell Biol. 2002;14:506–514. doi: 10.1016/s0955-0674(02)00350-2. [DOI] [PubMed] [Google Scholar]
- 27.McLauchlan H., Newell J., Morrice N., Osborne A., West M., Smythe E. Curr. Biol. 1998;8:34–45. doi: 10.1016/s0960-9822(98)70018-1. [DOI] [PubMed] [Google Scholar]
- 28.Pagano A., Crottet P., Prescianotto-Baschong C., Spiess M. Mol. Biol. Cell. 2004;15:4990–5000. doi: 10.1091/mbc.E04-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Hoop M. J., Huber L. A., Stenmark H., Williamson E., Zerial M., Parton R. G., Dotti C. G. Neuron. 1994;13:11–22. doi: 10.1016/0896-6273(94)90456-1. [DOI] [PubMed] [Google Scholar]
- 30.Jedd G., Mulholland J., Segev N. J. Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll K. S., Hanna J., Simon I., Krise J., Barbero P., Pfeffer S. R. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 32.Diaz E., Pfeffer S. R. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 33.Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riederer M. A., Soldati T., Shapiro A. D., Lin J., Pfeffer S. R. J. Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallik R., Gross S. P. Curr. Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 36.Novick P., Field C., Schekman R. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 37.Salminen A., Novick P. J. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 38.Goud B., Salminen A., Walworth N. C., Novick P. J. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 39.Walch-Solimena C., Collins R. N., Novick P. J. J. Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner W., Bielli P., Wacha S., Ragnini-Wilson A. EMBO J. 2002;21:6397–6408. doi: 10.1093/emboj/cdf650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Echard A., Jollivet F., Martinez O., Lacapere J. J., Rousselet A., Janoueix-Lerosey I., Goud B. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 42.Bahadoran P., Aberdam E., Mantoux F., Busca R., Bille K., Yalman N., de Saint-Basile G., Casaroli-Marano R., Ortonne J. P., Ballotti R. J. Cell Biol. 2001;152:843–850. doi: 10.1083/jcb.152.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda M., Kuroda T. S., Mikoshiba K. J. Biol. Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- 44.Nagashima K., Torii S., Yi Z., Igarashi M., Okamoto K., Takeuchi T., Izumi T. FEBS Lett. 2002;517:233–238. doi: 10.1016/s0014-5793(02)02634-0. [DOI] [PubMed] [Google Scholar]
- 45.Strom M., Hume A. N., Tarafder A. K., Barkagianni E., Seabra M. C. J. Biol. Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 46.Wu X., Bowers B., Rao K., Wei Q., Hammer J. A., III J. Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore K. J., Swing D. A., Rinchik E. M., Mucenski M. L., Buchberg A. M., Copeland N. G., Jenkins N. A. Genetics. 1988;119:933–941. doi: 10.1093/genetics/119.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matesic L. E., Yip R., Reuss A. E., Swing D. A., O’Sullivan T. N., Fletcher C. F., Copeland N. G., Jenkins N. A. Proc. Natl. Acad. Sci. USA. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Izumi T., Gomi H., Kasai K., Mizutani S., Torii S. Cell Struct. Funct. 2003;28:465–474. doi: 10.1247/csf.28.465. [DOI] [PubMed] [Google Scholar]
- 50.Sztul E., Lupashin V. Am. J. Physiol. 2006;290:C11–C26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 51.Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. Proc. Natl. Acad. Sci. USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allan B. B., Moyer B. D., Balch W. E. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 53.Moyer B. D., Allan B. B., Balch W. E. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 54.Plutner H., Schwaninger R., Pind S., Balch W. E. EMBO J. 1990;9:2375–2383. doi: 10.1002/j.1460-2075.1990.tb07412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tisdale E. J., Bourne J. R., Khosravi-Far R., Der C. J., Balch W. E. J. Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peter F., Nuoffer C., Pind S. N., Balch W. E. J. Cell Biol. 1994;126:1393–1406. doi: 10.1083/jcb.126.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbieri M. A., Hoffenberg S., Roberts R., Mukhopadhyay A., Pomrehn A., Dickey B. F., Stahl P. D. J. Biol. Chem. 1998;273:25850–25855. doi: 10.1074/jbc.273.40.25850. [DOI] [PubMed] [Google Scholar]
- 58.Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubino M., Miaczynska M., Lippe R., Zerial M. J. Biol. Chem. 2000;275:3745–3748. doi: 10.1074/jbc.275.6.3745. [DOI] [PubMed] [Google Scholar]
- 60.Cao X., Ballew N., Barlowe C. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beard M., Satoh A., Shorter J., Warren G. J. Biol. Chem. 2005;280:25840–25848. doi: 10.1074/jbc.M503925200. [DOI] [PubMed] [Google Scholar]
- 62.Alvarez C., Garcia-Mata R., Hauri H. P., Sztul E. J. Biol. Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- 63.Guo W., Roth D., Walch-Solimena C., Novick P. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.TerBush D. R., Maurice T., Roth D., Novick P. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 65.Toikkanen J. H., Miller K. J., Soderlund H., Jantti J., Keranen S. J. Biol. Chem. 2003;278:20946–20953. doi: 10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X. M., Ellis S., Sriratana A., Mitchell C. A., Rowe T. J. Biol. Chem. 2004;279:43027–43034. doi: 10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- 67.Adamo J. E., Rossi G., Brennwald P. Mol. Biol. Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson N. G., Guo L., Imai J., Toh E. A., Matsui Y., Tamanoi F. Mol. Cell. Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo W., Tamanoi F., Novick P. Nat. Cell Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- 70.Zhang X., Bi E., Novick P., Du L., Kozminski K. G., Lipschutz J. H., Guo W. J. Biol. Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 71.Brymora A., Valova V. A., Larsen M. R., Roufogalis B. D., Robinson P. J. J. Biol. Chem. 2001;276:29792–29797. doi: 10.1074/jbc.C100320200. [DOI] [PubMed] [Google Scholar]
- 72.Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., Ohta Y. Nat. Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 73.Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. Nat. Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 74.Wang W., Ferro-Novick S. Mol. Biol. Cell. 2002;13:3336–3343. doi: 10.1091/mbc.01-12-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.VanRheenen S. M., Cao X., Lupashin V. V., Barlowe C., Waters M. G. J. Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y. A., Scheller R. H. Nat. Rev. Mol. Cell. Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 77.McBride H. M., Rybin V., Murphy C., Giner A., Teasdale R., Zerial M. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 78.Subramanian S., Woolford C. A., Jones E. W. Mol. Biol. Cell. 2004;15:2593–2605. doi: 10.1091/mbc.E03-10-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Collins K. M., Thorngren N. L., Fratti R. A., Wickner W. T. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grosshans B. L., Andreeva A., Gangar A., Niessen S., Yates J. R., III, Brennwald P., Novick P. J. Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lehman K., Rossi G., Adamo J. E., Brennwald P. J. Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X., Wang P., Gangar A., Zhang J., Brennwald P., TerBush D., Guo W. J. Cell Biol. 2005;170:273–283. doi: 10.1083/jcb.200502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tall G. G., Hama H., DeWald D. B., Horazdovsky B. F. Mol. Biol. Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., Zerial M. J. Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toonen R. F., Verhage M. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 87.Price A., Seals D., Wickner W., Ungermann C. J. Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfeffer S. R. J. Biol. Chem. 2005;280:15485–15488. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- 89.Vitale G., Rybin V., Christoforidis S., Thornqvist P., McCaffrey M., Stenmark H., Zerial M. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ostermeier C., Brunger A. T. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 91.Merithew E., Stone C., Eathiraj S., Lambright D. G. J. Biol. Chem. 2003;278:8494–8500. doi: 10.1074/jbc.M211514200. [DOI] [PubMed] [Google Scholar]
- 92.Fukuda M. J. Biol. Chem. 2003;278:15373–15380. doi: 10.1074/jbc.M212341200. [DOI] [PubMed] [Google Scholar]
- 93.Zhu G., Zhai P., Liu J., Terzyan S., Li G., Zhang X. C. Nat. Struct. Mol. Biol. 2004;11:975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
- 94.Eathiraj S., Pan X., Ritacco C., Lambright D. G. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merithew E., Hatherly S., Dumas J. J., Lawe D. C., Heller-Harrison R., Lambright D. G. J. Biol. Chem. 2001;276:13982–13988. doi: 10.1074/jbc.M009771200. [DOI] [PubMed] [Google Scholar]
- 96.Pfeffer S. R. Biochem. Soc. Trans. 2005;33:627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 97.Dirac-Svejstrup A. B., Sumizawa T., Pfeffer S. R. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trischler M., Stoorvogel W., Ullrich O. J. Cell Sci. 1999;112:4773–4783. doi: 10.1242/jcs.112.24.4773. [DOI] [PubMed] [Google Scholar]
- 99.Sheff D. R., Daro E. A., Hull M., Mellman I. J. Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sonnichsen B., De Renzis S., Nielsen E., Rietdorf J., Zerial M. J. Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gorvel J. P., Chavrier P., Zerial M., Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 102.Li G., Barbieri M. A., Colombo M. I., Stahl P. D. J. Biol. Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- 103.Horiuchi H., Lippe R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 104.Rybin V., Ullrich O., Rubino M., Alexandrov K., Simon I., Seabra M. C., Goody R., Zerial M. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- 105.Stenmark H., Vitale G., Ullrich O., Zerial M. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 106.Lippe R., Horiuchi H., Runge A., Zerial M. Methods Enzymol. 2001;329:132–145. doi: 10.1016/s0076-6879(01)29074-0. [DOI] [PubMed] [Google Scholar]
- 107.Wurmser A. E., Sato T. K., Emr S. D. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Medkova M., France Y. E., Coleman J., Novick P. Mol. Biol. Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barbero P., Bittova L., Pfeffer S. R. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. Nat. Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 111.Siddhanta U., McIlroy J., Shah A., Zhang Y., Backer J. M. J. Cell Biol. 1998;143:1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burd C. G., Emr S. D. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 114.Stenmark H., Aasland R. J. Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- 115.Lawe D. C., Patki V., Heller-Harrison R., Lambright D., Corvera S. J. Biol. Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 116.Stenmark H., Aasland R., Toh B. H., D’Arrigo A. J. Biol. Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- 117.Patki V., Virbasius J., Lane W. S., Toh B. H., Shpetner H. S., Corvera S. Proc. Natl. Acad. Sci. USA. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gaullier J. M., Ronning E., Gillooly D. J., Stenmark H. J. Biol. Chem. 2000;275:24595–24600. doi: 10.1074/jbc.M906554199. [DOI] [PubMed] [Google Scholar]
- 119.Wilson J. M., de Hoop M., Zorzi N., Toh B. H., Dotti C. G., Parton R. G. Mol. Biol. Cell. 2000;11:2657–2671. doi: 10.1091/mbc.11.8.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 121.Volinia S., Dhand R., Vanhaesebroeck B., MacDougall L. K., Stein R., Zvelebil M. J., Domin J., Panaretou C., Waterfield M. D. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Diaz E., Schimmoller F., Pfeffer S. R. J. Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ikonomov O. C., Sbrissa D., Mlak K., Deeb R., Fligger J., Soans A., Finley R. L., Jr., Shisheva A. J. Biol. Chem. 2003;278:50863–50871. doi: 10.1074/jbc.M307260200. [DOI] [PubMed] [Google Scholar]
- 124.Nagelkerken B., Van Anken E., Van Raak M., Gerez L., Mohrmann K., Van Uden N., Holthuizen J., Pelkmans L., Van Der Sluijs P. Biochem. J. 2000;346:593–601. [PMC free article] [PubMed] [Google Scholar]
- 125.de Renzis S., Sonnichsen B., Zerial M. Nat. Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 126.Fouraux M. A., Deneka M., Ivan V., van der Heijden A., Raymackers J., van Suylekom D., van Venrooij W. J., van der Sluijs P., Pruijn G. J. Mol. Biol. Cell. 2004;15:611–624. doi: 10.1091/mbc.E03-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Benli M., Doring F., Robinson D. G., Yang X., Gallwitz D. EMBO J. 1996;15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- 128.Ortiz D., Medkova M., Walch-Solimena C., Novick P. J. Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 130.Eitzen G., Will E., Gallwitz D., Haas A., Wickner W. EMBO J. 2000;19:6713–6720. doi: 10.1093/emboj/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rieder S. E., Emr S. D. Mol. Biol. Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Caplan S., Hartnell L. M., Aguilar R. C., Naslavsky N., Bonifacino J. S. J. Cell Biol. 2001;154:109–122. doi: 10.1083/jcb.200102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim B. Y., Kramer H., Yamamoto A., Kominami E., Kohsaka S., Akazawa C. J. Biol. Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- 134.Richardson S. C., Winistorfer S. C., Poupon V., Luzio J. P., Piper R. C. Mol. Biol. Cell. 2004;15:1197–1210. doi: 10.1091/mbc.E03-06-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bonfanti L., Mironov A. A., Jr., Martinez-Menarguez J. A., Martella O., Fusella A., Baldassarre M., Buccione R., Geuze H. J., Mironov A. A., Luini A. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.