Abstract
Genes involved in a viral resistance response in common bean (Phaseolus vulgaris cv. Othello) were identified by inoculating a geminivirus reporter (Bean dwarf mosaic virus expressing the green fluorescent protein), extracting RNA from tissue undergoing the defense response, and amplifying sequences with degenerate R gene primers. One such gene (a TIR-NBS-LRR gene, RT4-4) was selected for functional analysis in which transgenic Nicotiana benthamiana were generated and screened for resistance to a range of viruses. This analysis revealed that RT4-4 did not confer resistance to the reporter geminivirus; however, it did activate a resistance-related response (systemic necrosis) to seven strains of Cucumber mosaic virus (CMV) from pepper or tomato, but not to a CMV strain from common bean. Of these eight CMV strains, only the strain from common bean systemically infected common bean cv. Othello. Additional evidence that RT4-4 is a CMV R gene came from the detection of resistance response markers in CMV-challenged leaves of RT4-4 transgenic plants, and the identification of the CMV 2a gene product as the elicitor of the necrosis response. These findings indicate that RT4-4 functions across two plant families and is up-regulated in a non-virus-specific manner. This experimental approach holds promise for providing insights into the mechanisms by which plants activate resistance responses against pathogens.
Keywords: avirulence gene, Cucumber mosaic virus, geminivirus, host defense response, systemic necrosis
Plants defend themselves against pathogen invasion through the action of specific resistance (R) genes and various nonspecific host responses (1, 2). Genetic studies have established that dominant R genes generally function in a gene-for-gene manner, whereby resistance is afforded based on an interaction between the R gene encoded protein and the cognate pathogen elicitor, commonly referred to as an avirulence (Avr) factor or effector protein (3). Such interactions trigger a cascade of defense responses (4), leading to pathogen confinement within the initial zone of infection. This defense cascade can also be associated with the development of a localized or, in some cases systemic, hypersensitive response (HR) (5).
The most well characterized R gene proteins are comprised of an N-terminal coil-coil (CC) or Toll-interleukin-1 receptor (TIR) homology domain, a centrally located nucleotide-binding site (NBS), and C-terminal leucine-rich repeats (LRRs) (6, 7). Dominant viral R genes characterized to date encode either TIR-NBS-LRR (TNL) (8) or CC-NBS-LRR (CNL)-type proteins (9–13). Studies of these genes have been focused primarily on members of the Solanaceae (e.g., tobacco, tomato, and potato) and Arabidopsis (7, 14).
Genomic studies have established that TNL and CNL R genes are present in the plant genome as multigene families. Arabidopsis and rice have ≈150 and 480 such genes, respectively (15, 16). The size of these gene families may reflect the diversity of pathogen challenge, during plant evolution, as well as the role of these genes in plant signaling processes. Thus, this complexity may reflect the mechanism by which new genes arise in response to various selection pressures. In any event, this combination of genetic conservation and complexity presents a formidable challenge in terms of the identification and characterization of any given member of an R gene family.
In this study, we identified genes involved in a viral resistance response in common bean (Phaseolus vulgaris cv. Othello) using a combination of a viral reporter and reverse transcription (RT)-PCR with degenerate R gene primers. A candidate R gene, RT4-4, a member of the TNL family, was selected for detailed analysis. Screening of RT4-4 transgenic Nicotiana benthamiana lines for viral resistance revealed that, although RT4-4 did not confer resistance to the reporter virus, it activated a resistance-like response (systemic necrosis) to Cucumber mosaic virus (CMV). Further evidence that RT4-4 is a CMV R gene came from our identification of the CMV 2a gene product as the elicitor of the necrosis response. The finding that RT4-4 functions across two plant families and is up-regulated in a non-virus-specific manner, provides insights into the mechanisms by which plants activate the pathogen resistance response.
Results
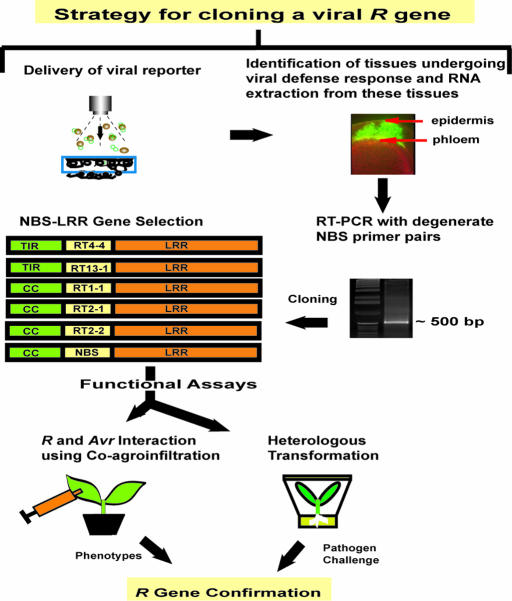
The general strategy developed to clone viral R genes is outlined in Fig. 1. A geminiviral-GFP reporter [Bean dwarf mosaic virus (BDMV)-GFP], (17) was used to identify tissues undergoing a defense response in the resistant common bean cv. Othello (18). Poly(A)+ RNA, extracted 4 days after inoculation, was used in the RT-PCR with degenerate primers to amplify domains of putative R genes expressed in these tissues. Degenerate primers were designed based on conserved NBS domains of previously characterized TNL and CNL R genes and included three forward primers and a single reverse primer. These primer pairs directed the amplification of DNA fragments of ≈350–500 bp. One such fragment had amino acid sequence identity (≈43%) with the Tobacco mosaic virus (TMV) N gene and was selected for further characterization.
Fig. 1.
A general strategy to identify and characterize viral resistance (R) genes. Infectious cloned DNA of the Bean dwarf mosaic virus (BDMV)-GFP reporter was bombarded into hypocotyl tissues of resistant common bean cv. Othello. Tissues undergoing the pathogen defense response were identified by using GFP fluorescence and then excised, and poly(A)+ RNA was extracted. Putative R gene fragments were amplified by using RT-PCR with degenerate R gene primers, cloned, and characterized by using standard molecular and genomic approaches. Functional analysis involved transformation of N. benthamiana with candidate R genes (e.g., RT4-4) and screening for resistance to a range of viruses. Cognate pathogen avirulence (Avr) factor(s) were identified through transient expression experiments.
The complete sequence of this gene, referred to as RT4-4, was determined from 5′ and 3′ RT4-4 fragments generated with RACE PCR, and a full-length clone was generated by PCR with RT4-4-specific primers. Sequence analysis of the RT4-4 gene revealed a 3565 bp ORF encoding a predicted protein of 1,133 aa (Table 1 and Fig. 6, which are published as supporting information on the PNAS web site). As some R genes are up-regulated during pathogen challenge, expression of RT4-4 was next examined in hypocotyl tissues of resistant (cv. Othello) and susceptible (cv. Topcrop) common bean cultivars inoculated with BDMV-GFP. RT-PCR and Northern blot analyses of poly(A)+ RNA, extracted from infected cv. Othello tissues, revealed up-regulation of RT4-4 expression beginning 2 days after inoculation (Fig. 2A and B). In contrast, little or no expression was detected in hypocotyl tissues of infected cv. Topcrop (Fig. 2C) or those of cvs. Othello and Topcrop bombarded with gold particles only (data not shown). Southern blot hybridization analysis (high-stringency) of total genomic DNA, with an RT4-4 LRR domain probe, established that cvs. Othello and Topcrop each contained a single copy of this gene (data not shown). These results indicate that RT4-4 is specifically up-regulated in cv. Othello tissues undergoing the viral defense response.
Fig. 2.
Expression of the RT4-4 gene. (A) Expression of the RT4-4 gene detected by RT-PCR with an RT4-4-specific primer pair. Total RNA was extracted from common bean (cv. Othello) hypocotyl tissues at 0, 2, and 4 days postbombardment (dpb) of a Bean dwarf mosaic virus (BDMV)-GFP reporter. An equivalent analysis of actin gene expression was used as a control for levels of constitutive gene expression. (B) Expression of the RT4-4 gene in BDMV-resistant common bean cv. Othello detected by Northern blot hybridization analysis. One microliter of poly(A)+ RNA was analyzed in each lane, and the blot was hybridized with a 32P-labeled RT4-4 DNA fragment including a portion of the LRR region. (C) Expression of the RT4-4 gene in the BDMV-susceptible cv. Topcrop (TC) and cv. Othello (O) analyzed by RT-PCR with an RT4-4-specific primer pair.
Role of RT4-4 Gene in Viral Defense.
To determine whether RT4-4 is the dominant R gene mediating BDMV resistance in cv. Othello (19), agroinfiltration (20) was used for expression in the susceptible cv. Topcrop. Primary leaves were infiltrated with Agrobacterium strains carrying RT4-4 and/or BDMV in the following combinations: RT4-4 together with BDMV, RT4-4 followed by BDMV (24 h later) or BDMV alone. Irrespective of the treatment, most of the plants developed symptoms typical of BDMV infection (data not shown).
As RT4-4 did not appear to confer resistance to BDMV, it may represent an R gene up-regulated, in a nonspecific manner, during the geminivirus defense response. To test this hypothesis, a functional analysis was next performed by using 35S promoter-driven RT4-4 transgenic N. benthamiana lines to screen for viral resistance. This solanaceous host was chosen because of its susceptibility to a wide range of viruses, including BDMV. Eighteen single-copy lines were selected, based on PCR detection of RT4-4 in T0 plants and T1 progeny having a 3:1 ratio of kanamycin-resistant/-susceptible seedlings. The phenotype of these transgenic plants was indistinguishable from that of the nontransformed controls.
Viruses representing four different families/groups were inoculated onto kanamycin-resistant T1 RT4-4 transgenic plants. These viruses included a single-stranded (ss) DNA virus (the geminivirus, BDMV), a tripartite ssRNA virus (CMV, a bromovirus), and two different types of monopartite ssRNA viruses [the potyviruses Bean common mosaic virus (BCMV) and Bean yellow mosaic virus (BYMV), and TMV, a tobamovirus]. At least 10 T1 plants of each of the 18 transgenic lines were sap-inoculated with each virus, and disease development assessed 3 weeks after inoculation. Most transgenic and nontransgenic control plants inoculated with BDMV, BCMV, BYMV, or TMV developed typical disease symptoms (data not shown), indicating no RT4-4-mediated resistance response in the transgenic plants.
A very different situation was observed for CMV. Here, nontransgenic control plants infected with CMV developed typical systemic symptoms, including stunted growth and leaf epinasty, crumpling, and mosaic. In contrast, RT4-4 transgenic plants inoculated with CMV developed a systemic necrosis response, which started in the inoculated leaves and progressed throughout the plant, eventually resulting in death (Fig. 7, which is published as supporting information on the PNAS web site). RT-PCR analysis was used to confirm both RT4-4 expression in selected transgenic plants and CMV infection in symptomatic nontransgenic plants (Fig. 7). Genetic analysis of the RT4-4/CMV interaction revealed a 3:1 ratio of systemic necrosis: mosaic phenotypes, consistent with RT4-4 segregating as a single dominant gene.
RT4-4 Displays Specificity in Recognizing CMV Strains.
Specificity of the RT4-4/CMV interaction was studied by inoculating kanamycin-resistant T1 RT4-4 transgenic plants with CMV strains representing the three recognized subgroups: Ia, Ib, and II (21). Strains 113B, 116B, C94T5, Fny, HR2, M, and NB originated from pepper and tomato, whereas strain 67 originated from common bean. All pepper and tomato strains, regardless of subgroup, induced mosaic symptoms in nontransgenic N. benthamiana, and systemic necrosis in the RT4-4 transgenic plants (Fig. 3 A–D and Table 2, which is published as supporting information on the PNAS web site). In contrast, the common bean-infecting strain induced mosaic symptoms in both nontransgenic and RT4-4 transgenic N. benthamiana plants (Fig. 3 E and F).
Fig. 3.
RT4-4 recognizes Cucumber mosaic virus (CMV) in a strain-specific manner. Disease symptoms were induced by various CMV strains in nontransgenic or kanamycin-resistant T1 RT4-4 transgenic N. benthamiana (line RT6) and common bean (cv. Othello) plants. Symptoms were recorded 14 days after sap inoculation. (A) Systemic mosaic symptoms induced by strain Fny (subgroup Ia) in a nontransgenic N. benthamiana plant. (B) Systemic necrosis symptoms induced by strain Fny in a T1 RT4-4 transgenic plant (RT6 line). (C) Leaf epinasty and systemic mosaic symptoms induced by strain M (subgroup II) in a nontransgenic N. benthamiana plant. (D) Systemic necrosis symptoms induced by strain M in a T1 RT4-4 transgenic plant (RT6 line). (E and F) Systemic mosaic symptoms induced by the bean-infecting strain 67 (subgroup Ia) in a nontransgenic N. benthamiana and a T1 RT4-4 transgenic plant (RT6 line), respectively. (G) Symptomless phenotype in a common bean plant (cv. Othello) inoculated with strain Fny. (H) Mild mosaic, leaf crumpling, and distortion symptoms induced in a common bean plant (cv. Othello) by strain 67.
An equivalent test for CMV strain specificity was next performed with cv. Othello. Here, no visible symptoms developed in plants inoculated with the pepper and tomato strains, whereas the bean-infecting strain induced mild systemic mosaic symptoms (Fig. 3 G and H and Table 2). The results of these assays confirmed that both RT4-4 transgenic plants and cv. Othello plants exhibited the same differential CMV strain-specificity, although the symptom phenotype of the resistance response differed (systemic necrosis vs. no symptoms). These findings support the hypothesis that RT4-4 functions as a CMV R gene.
Defense Response Markers Associated with RT4-4-Mediated Systemic Necrosis.
The typical dominant resistance response is associated with several defense-related events, including rapid induction of active oxygen species, phytoalexin accumulation, and activation of salicylic acid (SA) biosynthesis and pathogenesis-related (PR) genes (4). To determine whether RT4-4-mediated CMV systemic necrosis represents a bona fide defense response, Fny and the bean-infecting CMV strains were inoculated onto leaves of 2- to 3-week-old kanamycin-resistant T1 RT4-4 transgenic and nontransgenic N. benthamiana plants. Leaves were collected 48 h after inoculation and analyzed for a set of defense response markers.
RT-PCR analysis revealed a strong induction of PR1a, PR2, and chitinase (PR4) gene expression in leaves of RT4-4 transgenic plants inoculated with strain Fny. In contrast, induction was not detected in leaves of transgenic plants inoculated with the bean-infecting strain, nontransgenic plants inoculated with either strain, or in mock-inoculated transgenic or nontransgenic plants (Fig. 4A). A biochemical assay for H2O2 revealed accumulation only in leaves of RT4-4 transgenic plants inoculated with CMV strain Fny; H2O2 was not detected in leaves of transgenic plants inoculated with the bean-infecting strain, nontransgenic plants inoculated with either strain, or in mock-inoculated transgenic or nontransgenic plants (Fig. 4 B–E). Parallel studies in which we measured SA accumulation revealed up-regulation only in leaves of Fny-inoculated RT4-4 transgenic plants (Fig. 4F). Taken together, these results support the hypothesis that the systemic necrosis response, induced by CMV in transgenic RT4-4 N. benthamiana, reflects an incompatible host–pathogen defense response.
Fig. 4.
Expression of pathogenesis-related (PR) genes and accumulation of H2O2 and SA. (A) Expression of PR genes was detected by RT-PCR with PR gene-specific primer pairs. Total RNA was extracted from nontransgenic and kanamycin-resistant T1 RT4-4 transgenic N. benthamiana (RT-6 line) leaves at 2 days after sap inoculation of CMV strains Fny and 67. An equivalent analysis of actin gene expression was used as a control for levels of constitutive gene expression. (B–E) Detection of H2O2 accumulation in third- or fourth-order veins of CMV-inoculated leaves was visualized with a light microscope at ×200 magnification. (F) Detection of SA accumulation from CMV-inoculated leaves.
CMV 2a Protein Is the Elicitor of the RT4-4-Induced Necrosis Response.
Identification of the viral elicitor (Avr factor) of the CMV-induced systemic necrosis response in RT4-4 transgenic plants would provide another line of evidence that RT4-4 is indeed a CMV R gene. To this end, Agrobacterium-mediated transient expression was used to test CMV-encoded proteins (Fig. 5A) for elicitation of the necrosis response. No obvious symptoms were observed in leaves of nontransgenic plants infiltrated with Agrobacterium strains carrying the empty vector (EV), capsid protein (CP), movement protein (MP), or 2a constructs (Fig. 5B). Infiltration of the 2a strain into leaves of RT4-4 transgenic plants resulted in cell collapse and death in the infiltrated areas, whereas no such response was observed in equivalent areas infiltrated with the EV, CP, or MP strains (Fig. 5C). When leaves of nontransgenic plants were coinfiltrated with the RT4-4 Agrobacterium strain and each of the viral gene-containing strains, necrosis was observed only in areas coinfiltrated with the RT4-4 and CMV 2a strains (Fig. 5 D and E).
Fig. 5.
Cucumber mosaic virus (CMV) 2a protein is the elicitor of the necrosis phenotype in RT4-4 transgenic N. benthamiana. Agroinfiltration was used to transiently express CMV strain 113B proteins in N. benthamiana leaves. (A) Schematic diagrams of the T-DNA constructs of pBV:35S:2a gene (2a), pBV:35S:capsid protein gene (CP), pBV:35S:movement protein gene (MP), and pGA643:35S:RT4-4. Empty vector (EV) is the pBV vector only. RB and LB indicate the right and left borders of the T-DNA, respectively. (B) Infiltration of A. tumefaciens LBA 4404 strains containing the EV, 2a, CP, or MP constructs into leaves of nontransgenic N. benthamiana plants. (C) Infiltration of A. tumefaciens LBA 4404 strains containing the EV, 2a, CP, or MP constructs into a leaf of a kanamycin-resistant T1 RT4-4 transgenic plant (RT6 line). (D) Coinfiltration of A. tumefaciens LBA 4404 strain containing the RT4-4 construct with A. tumefaciens strains with the 2a, CP, or MP constructs, into young leaves of nontransgenic N. benthamiana plants. (E) Coinfiltration of A. tumefaciens LBA 4404 strain containing the RT4-4 construct with A. tumefaciens strains with the 2a, CP, or MP constructs, into mature leaves of nontransgenic N. benthamiana plants.
CMV 2a Motif Is Necessary for the Necrosis Response.
Having identified the CMV 2a protein as the elicitor of the RT4-4-mediated necrosis response, we next sought to identify the amino acid residue(s) involved in this recognition. In a previous study, Kim and Palukaitis (22) identified amino acid residues 631 (Phe) and 641 (Ala) of the Fny 2a protein as determinants of a CMV-mediated HR in cowpea. To test the hypothesis that these same residues are involved in our observed CMV-mediated necrosis response, the 2a sequences of the eight CMV strains used in the present study were determined. All of the systemic necrosis-inducing strains had the Phe-631 and Ala-641 residues; however, the bean-infecting strain had Tyr-631 and Ser-641 (Table 2). Because the bean-infecting strain failed to elicit the systemic necrosis response in RT4-4 transgenic plants, the role of these two residues in this resistance response was next explored by mutational analysis.
Mutations were introduced into the cloned 2a gene of the necrosis-inducing CMV strain 113B, and each mutant construct was agroinfiltrated into leaves of kanamycin-resistant T1 RT4-4 transgenic plants. The Ala-641 → Ser-641 mutant elicited a necrosis response that was indistinguishable from that induced by infiltration of the wild-type 113B 2a, whereas the Phe-631 → Tyr-631 mutant did not induce necrosis (Fig. 8, which is published as supporting information on the PNAS web site). The double mutant, Phe-631 → Tyr-631 and Ala-641 → Ser-641, in which both residues were changed to those in the 2a of the bean-infecting strain, also did not elicit the necrosis response. Finally, a phosphorylation mimic mutant (Phe-631 → Asp-631) failed to elicit any necrosis response. These results indicate that the Phe-631 residue is important in RT4-4-mediated recognition of the CMV 2a.
Discussion
In this report, we identified a viral R gene, RT4-4, from common bean (P. vulgaris) using an approach that optimizes the probability of isolating genes that are up-regulated during pathogen infection. Through the use of a GFP-tagged viral reporter, tissues undergoing the early stages of a viral defense response were identified and used to obtain poly(A)+ RNA enriched for transcripts of genes expressed during viral infection. Degenerate primers, designed based on conserved NBS domains of known pathogen R genes (including viral TNL/CNL R genes), allowed for the amplification of R gene analogues (RGAs) potentially involved in this response. A number of candidate TNL/CNL RGAs were cloned (Table 1) and one candidate gene (RT4-4) was selected for further characterization, based on having homology with a known viral R gene. A functional analysis of this gene was performed in the heterologous host, N. benthamiana, because common bean remains recalcitrant to routine transformation (Fig. 1).
Sequence analysis revealed that RT4-4 was a typical member of the TNL gene family; however, the functional analysis revealed a number of interesting features. First, RT4-4 was up-regulated in tissues undergoing the viral defense response (Fig. 2), but results of coagroinfiltration studies in common bean indicated that it was not the dominant BDMV R gene from cv. Othello (19). A similar finding came from BDMV infection studies performed in RT4-4 transgenic N. benthamiana. This was not totally unexpected, because it has been established that various factors, such as SA treatment, pathogen challenge, or constitutive expression of transcriptional coactivator binding factor 1c, can nonspecifically activate expression of R genes (23–25). Systemic acquired resistance is another example of how a single pathogen can induce broad-spectrum activation of defense pathways (26).
To test the hypothesis that RT4-4 represents an R gene up-regulated in a nonspecific manner, the transgenic N. benthamiana plants were inoculated with a range of viruses. These experiments revealed that RT4-4 triggered a defense response (systemic necrosis) upon infection with CMV (Fig. 3). Thus, these results provide further evidence for nonspecific activation of R genes during pathogen attack. Additional support for this concept came from our identification of a spectrum of TNL and CNL RGAs expressed during the BDMV defense response (Table 1). Thus, our results suggest that activation of an R gene by a pathogen may lead to the nonspecific activation of other R genes (e.g., a quorum sensing-type phenomenon). This activation may result in an enhanced defense response, possibly via multiple defense pathways.
Several lines of evidence indicate that RT4-4 is a common bean R gene that recognizes CMV. First, hallmarks of the classical pathogen defense response were detected in leaves of RT4-4 transgenic N. benthamiana plants inoculated with CMV (Fig. 4). Because CMV typically induces mottle/mosaic symptoms in this host, these results demonstrate that RT4-4 triggers a CMV defense response in the transgenic plants, which is manifested as systemic necrosis. The specificity of the systemic necrosis for certain CMV strains indicates that it is not a generalized response to viral infection or expression of a heterologous R gene. The latter result is also supported by previous reports of functional expression of heterologous R genes in N. benthamiana in the absence of systemic necrosis (9, 27, 28).
The capacity to trigger a defense response in N. benthamiana reveals that this legume R gene is functional in a solanaceous genetic background, and that the RT4-4 protein interacts with host factors in the activation of this response. To date, the capacity of R genes to act across family boundaries has been the exception rather than the rule (7, 14), with most examples of heterologous R gene function being in species within a single family (20, 29). Our results add to the emerging picture that some R genes can function across plant families (28, 30, 31), implying that family boundaries may not necessarily impose limits on R gene function(s).
The CMV-induced systemic necrosis response observed in RT4-4 transgenic N. benthamiana contrasts with the symptomless phenotype associated with CMV resistance in common bean cv. Othello. This difference may reflect the nature of the interaction of RT4-4 with host factors and/or the defense pathways involved. In fact, a range of resistance phenotypes can be observed for a single R gene within a single plant species. For example, in the homozygous condition, the dominant I gene in common bean confers extreme resistance (i.e., a symptomless phenotype) to BCMV; whereas in the heterozygous condition, resistance is associated with necrotic lesions (32). In contrast, bean plants homozygous or heterozygous for the I gene develop a systemic necrosis resistance response when infected with the closely related potyvirus, Bean common mosaic necrosis virus. Interestingly, a TNL gene family has been associated with the I locus (33). Similarly, resistance to Soybean mosaic virus mediated by the Rsv1 gene is represented by a range of phenotypes, including systemic necrosis (34). Furthermore, in N gene-mediated resistance to TMV, a local lesion phenotype typically develops; however, expression of various N gene mutants, in transgenic N. tabacum, triggers a systemic hypersensitive response upon infection with TMV (35). Hence, the systemic necrosis in RT4-4 transgenic N. benthamiana may reflect a delayed defense response to CMV infection. This hypothesis is supported by the detection of CMV in newly emerging leaves undergoing systemic necrosis, consistent with long-distance spread of the virus. Alternatively, overexpression of RT4-4 in these transgenic plants may overactivate this defense response pathway (36). In the cognate background of common bean cv. Othello, the symptomless phenotype associated with CMV resistance indicated confinement of the virus to the inoculated leaves and, thus, an extreme resistance phenotype.
Another line of evidence indicating that RT4-4 is a CMV R gene was the strain specificity of the systemic necrosis response in transgenic N. benthamiana. This type of differential response to pathogen strains, or races, is a typical property of single dominant R genes. The finding of the same CMV strain specificity in the capacity to systemically infect cv. Othello supports the hypothesis that RT4-4 mediates CMV resistance in common bean. Additional evidence that RT4-4 functions as a CMV R gene was provided by the identification of the CMV 2a protein as the elicitor (Avr factor) of the RT4-4-mediated necrosis response (Fig. 5) and the identification of Phe-631 as a determinant of 2a-mediated necrosis (Fig. 8). Consistent with this result, the Phe-631 → Tyr-631 mutation, identified in the 2a gene of the bean-infecting strain 67, abolished the 2a-mediated necrosis phenotype. Thus, the capacity of strain 67 to systemically infect cv. Othello and not elicit systemic necrosis in the RT4-4 N. benthamiana plants likely reflects the inability of RT4-4 to recognize the 2a protein of this CMV strain. Finally, a role for posttranslational modification in 2a protein recognition, by RT4-4, came from the finding that the CMV 2a phosphorylation mimic mutant failed to elicit the necrosis phenotype (37). Together, these findings are consistent with results of previous studies that identified the 2a protein as the elicitor of a CMV defense response in cowpea (22).
In summary, the experimental approach described in this study resulted in the identification and characterization of a viral R gene from common bean that functions across two plant families and is up-regulated in a non-virus-specific manner. Based on its interaction with the CMV 2a protein and its elicitation of a CMV-specific defense response, this gene was named P. vulgaris CMV RESISTANCE 1 (PvCMR1). This experimental approach holds promise for providing insights into the mechanisms by which plants activate resistance responses to pathogens, and could facilitate the identification of R genes from a broad spectrum of plant species.
Materials and Methods
RNA Extraction and RT-PCR.
Total RNA was extracted from common bean hypocotyl tissues bombarded with BDMV-GFP or gold particles as described (38). Poly(A)+ RNA was purified from total RNA (500 μg) with the Oligotex mRNA isolation kit according to manufacturer's instructions (Qiagen, Valencia, CA). The RT reactions were performed with Superscript reverse transcriptase (Gibco BRL, Grand Island, NY) and 5 μl of poly(A)+ RNA. The PCR was performed with 2–5 μl of RT reaction in a total volume of 50 μl, for 30–35 cycles of 94°C, 1 min; 55°C, 2 min; and 72°C, 3 min. The negative control reaction was poly(A)+ RNA, not subjected to RT, used in the PCR (note that contaminating DNA was not detected in these reactions). PCR products were analyzed by agarose gel electrophoresis in 1.0% Tris-borate EDTA buffer (TBE).
Design of Degenerate Primers for R Gene Analogs.
Three forward primers were generated [kinase-1a (5′-GGIGGRRTAGGTAARACRAC-3′), kinase-2 (5′-CTTRTYGTTCTYGATGATGT-3′), and kinase-3a (5′-AGTARRATYATTATIACIACAMG-3′)] and paired with a reverse primer generated from the hydrophobic motif (HM, 5′-RARRGCCAAWGGAAGTCC-3′).
5′ and 3′ RACE.
RACE was performed with the SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions. For 5′ RACE, the nested gene-specific primer was 5′-CTTGTTGTTCTTGATGATGTAAGTGAC-3′; whereas for 3′ RACE, the nested gene-specific primer was 5′-AGCCAAAGGAAGTCCTCTTGCATATTC-3′. The PCR products of each RACE reaction were cloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA), and representative clones were sequenced.
Plant Transformation.
The full-length RT4-4 sequence was amplified from cDNA prepared from BDMV-GFP-infected cv. Othello tissues, undergoing the defense response (≈4 days after inoculation), by PCR with an RT4-4-specific primer pair (5′-GTTAACGGAACCAGTGTTATTGCAG-3′ and 5′-ATCGATCAAGACACATAATTACATGAA-3′). This ≈3.6-kb fragment, which contains the RT4-4 sequence and 35 and 180 bp of the 5′ and 3′ nontranslated sequences, respectively, was cloned into pCRII-TOPO to generate pCR-RT4-4. The integrity of the RT4-4 sequence was confirmed by sequencing. The RT4-4 DNA fragment was released from pCR-RT4-4 by digestion with BamHI and HpaI and cloned into the binary vector pGA643 (kindly provided by G. An), digested with BglII and HpaI. This process places the RT4-4 gene under the transcriptional control of the 35S promoter and the NOS terminator. The recombinant binary plasmid (pGA643:RT4-4) was transformed into A. tumefaciens LBA4404, and transgenic N. benthamiana plants were generated by the leaf disk method (39). Regenerated plantlets were rooted, planted in soil, grown in a greenhouse, and allowed to self-pollinate. Seeds were collected and tested for kanamycin resistance by germinating on agar plates containing 300 μg/ml kanamycin. The presence of the RT4-4 gene in kanamycin-resistant plants was determined by PCR with the RT4-4 primer pair, pA6 (5′-GCACTCATCATTCTCTCACC-3′) and pB4 (5′-CTTGCCTTACCTATGCCTCC-3′), which directs the amplification of a 460-bp RT4-4 DNA fragment.
Identification of Defense Response Markers.
Expression of PR protein genes was detected by RT-PCR with the following primers: PR1a, 5′-AATATCCCACTCTTGCCG-3′ and 5′-CCTGGAGGATCATAGTTG; PR2, 5′-ACCATCAGACCAAGATGT-3′ and 5′-TGGCTAAGAGTGGAAGGT-3′; and chitinase (PR4), 5′-ATGGAGTTTTCTGGATCACC-3′ and 5′-CTAGCCCTGGCCGAAGTT. Hydrogen peroxide generation was detected as described (40). Changes in the level of free SA were determined in the leaves by using a modified spectrophotometric method (41). Leaves were ground in liquid nitrogen with a mortar and pestle and extracted with 2 ml of 50% ethanol. The supernatant was centrifuged (5,000 × g for 15 min), filtered through two layers of gauze, and then 0.5 ml of 6 M HCl was added for SA hydrolysis. To extract SA, 10 ml of tetrachloride was added to each sample, and the extract was mixed with 5 ml of ferric nitrate solution for 2 min. After centrifugation, the aqueous phase was analyzed by spectrophotometry (530 nm). For quantitative analysis, a standard curve was established with commercial SA (S-3007; Sigma, St. Louis, MO) suspended in 50% ethanol.
Site-Directed Mutagenesis of the CMV (Strain 113B) 2a Gene.
To generate CMV 2a gene mutants, mutagenesis was performed in pBluescript KS+ (pKS; Stratagene, La Jolla, CA) containing an ≈750 bp HindIII–BamHI 2a gene fragment [pKS-2a (HindIII–BamHI)], which was subcloned from pCR2.1–2a (E. Maciel-Zambolim and R.L.G., unpublished data). Site-specific mutations were introduced into the 2a gene by using the GeneTailor site-directed mutagenesis system (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions (sequences of primers are given in Supporting Text, which is published as supporting information on the PNAS web site). After mutations were introduced into target sites and confirmed by sequencing, the mutated HindIII–BamHI 2a gene fragment was exchanged with that of the full-length wild-type 2a gene in pKS (pKS-2a).
Agrobacterium-Mediated Transient Expression.
Constructs for the transient assays were made in the binary vector pBV (42), except for the previously described RT4-4 construct. Binary plasmids containing the CMV CP, MP, 2a, and RT4-4 genes were each transformed into A. tumefaciens strain LBA4404; CMV-2a gene mutants were transformed into A. tumefaciens strain C58C1. The generation of a BDMV agroinoculation system has been described (43). Agroinfiltration was performed as described (44).
Virus Isolates and Inoculation and Detection of CMV.
CMV strains M (subgroup II), 67 (subgroup Ia), 113B, HR2, and NB (subgroup Ib) were previously characterized in our laboratory (E. Maciel-Zambolim and R.L.G., unpublished data), whereas strains 116B and Fny (subgroup Ia) and C94T5 (subgroup Ib) were kindly provided by B. W. Falk (University of California, Davis, CA). All CMV strains were maintained in pumpkin (Cucurbita pepo L., cv. Small sugar). Sap inoculum was prepared by grinding young systemically infected pumpkin leaves in 0.1 M potassium phosphate buffer (pH 7.8), and sap inoculation involved rubbing celite-dusted leaves of pumpkin (cotyledon leaf stage), common bean (primary leaf stage), and N. benthamiana (five- to seven-leaf stage) with a pestle. Inoculated plants were maintained in a greenhouse at 25–30°C and examined for symptoms 14–21 days after inoculation. CMV was detected in plants by RT-PCR with the primer pair pF3 (5′-AACACGGAATCAGACTGG-3′) and pF4 (5′-TTGAGTCGAGTCATGGACAAATC-3′), which directs the amplification of a 710-bp CP gene fragment. In some cases, CMV was detected by indirect ELISA (45) with polyclonal CMV antisera.
Supplementary Material
Acknowledgments
We thank Dr. B. W. Falk for providing several CMV strains and the CMV PCR primer pair and Dr. G. An (Pohang University of Science and Technology, Pohang, Republic of Korea) for providing the pGA643 binary vector. This research was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants 9901578 and 2002-01418 (to R.L.G. and W.J.L.).
Abbreviations
- HR
hypersensitive response
- CMV
Cucumber mosaic virus
- BDMV
Bean dwarf mosaic virus
- SA
salicylic acid
- TMV
Tobacco mosaic virus.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Glazebrook J., Rogers E. E., Ausubel F. M. Annu. Rev. Genet. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- 2.Heath M. C. Curr. Opin. Plant Biol. 2000;3:315–319. doi: 10.1016/s1369-5266(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 3.Flor H. Annu. Rev. Phytopathol. 1971;9:275–296. [Google Scholar]
- 4.Hammond-Kosack K. E., Jones J. D. G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilchrist D. G. Annu. Rev. Phytopathol. 1998;36:393–414. doi: 10.1146/annurev.phyto.36.1.393. [DOI] [PubMed] [Google Scholar]
- 6.Dangl J. L., Jones J. D. G. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 7.Hulbert S. H., Webb C. A., Smith S. M., Sun Q. Annu. Rev. Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 8.Whitham S., Dinesh-Kumar S. P., Choi D., Hehl R., Corr C., Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 9.Bendahmane A., Kanyuka K., Baulcombe D. C. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brommonschenkel S. H., Frary A., Frary A., Tanksley S. D. Mol. Plant–Microbe Interact. 2000;13:1130–1138. doi: 10.1094/MPMI.2000.13.10.1130. [DOI] [PubMed] [Google Scholar]
- 11.Cooley M. B., Pathirana S., Wu H. J., Kachroo P., Klessig D. F. Plant Cell. 2000;12:663–676. doi: 10.1105/tpc.12.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kachroo P., Yoshioka K., Shah J., Dooner H. K., Klessig D. F. Plant Cell. 2000;12:677–690. doi: 10.1105/tpc.12.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi H., Miller J., Nozaki Y., Takeda M., Shah J., Hase S., Ikegami M., Ehara Y., Dinesh-Kumar S. P. Plant J. 2002;32:655–667. doi: 10.1046/j.1365-313x.2002.01453.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin G. B., Bogdanove A. J., Sessa G. Annu. Rev. Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 15.Meyers B. C., Kozik A., Griego A., Kuang H., Michelmore R. W. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T., Wang Y., Chen J. Q., Araki H., Jing Z., Jiang K., Shen J., Tian D. Mol. Genet. Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 17.Sudarshana M. R., Wang H. L., Lucas W. J., Gilbertson R. L. Mol. Plant–Microbe Interact. 1998;11:277–291. doi: 10.1094/MPMI.2000.13.11.1184. [DOI] [PubMed] [Google Scholar]
- 18.Wang H. L., Sudarshana M. R., Gilbertson R. L., Lucas W. J. Mol. Plant–Microbe Interact. 1999;12:345–355. doi: 10.1094/MPMI.2000.13.11.1184. [DOI] [PubMed] [Google Scholar]
- 19.Seo Y. S., Gepts P., Gilbertson R. L. Theor. Appl. Genet. 2004;108:786–793. doi: 10.1007/s00122-003-1504-9. [DOI] [PubMed] [Google Scholar]
- 20.Tai T. H., Dahlbeck D., Clark E. T., Gajiwala P., Pasion R., Whalen M. C., Stall R. E., Staskawicz B. J. Proc. Natl. Acad. Sci. USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roossinck M. J. Mol. Plant Pathol. 2001;2:59–63. doi: 10.1046/j.1364-3703.2001.00058.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim C. H., Palukaitis P. EMBO J. 1997;16:4060–4068. doi: 10.1093/emboj/16.13.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirano Y., Kachroo P., Shah J., Klessig D. F. Plant Cell. 2002;14:3149–3162. doi: 10.1105/tpc.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V., Mittler R. Plant Physiol. 2005;139:1313–1322. doi: 10.1104/pp.105.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D. G., Felix G., Boller T. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 26.Dong X. N. Curr. Opin. Plant Biol. 2001;4:309–314. doi: 10.1016/s1369-5266(00)00178-3. [DOI] [PubMed] [Google Scholar]
- 27.Leister R. T., Dahlbeck D., Day B., Li Y., Chesnokova O., Staskawicz B. J. Plant Cell. 2005;17:1268–1278. doi: 10.1105/tpc.104.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao S. Y., Charoenwattana P., Holcombe L., Turner J. G. Mol. Plant–Microbe Interact. 2003;16:289–294. doi: 10.1094/MPMI.2003.16.4.289. [DOI] [PubMed] [Google Scholar]
- 29.Whitham S., McCormick S., Baker B. Proc. Natl. Acad. Sci. USA. 1996;93:8776–8781. doi: 10.1073/pnas.93.16.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodds P. N., Lawrence G. J., Catanzariti A. M., Ayliffe M. A., Ellis J. G. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B. Y., Lin X. H., Poland J., Trick H., Leach J., Hulbert S. Proc. Natl. Acad. Sci. USA. 2005;102:15383–15388. doi: 10.1073/pnas.0503023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collmer C. W., Marston M. F., Taylor J. C., Jahn M. Mol. Plant–Microbe Interact. 2000;13:1266–1270. doi: 10.1094/MPMI.2000.13.11.1266. [DOI] [PubMed] [Google Scholar]
- 33.Vallejos C. E., Astua-Monge G., Jones V., Plyler T. R., Sakiyama N. S., Mackenzie S. A. Genetics. 2005;172:1229–1242. doi: 10.1534/genetics.105.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajimorad M. R., Eggenberger A. L., Hill J. H. J. Virol. 2005;79:1215–1222. doi: 10.1128/JVI.79.2.1215-1222.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinesh-Kumar S. P., Tham W. H., Baker B. J. Proc. Natl. Acad. Sci. USA. 2000;97:14789–14794. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao Y., Yuan F., Leister R. T., Ausubel F. M., Katagiri F. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S. H., Palukaitis P., Park Y. I. EMBO J. 2002;21:2292–2300. doi: 10.1093/emboj/21.9.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty D. R. Maize Gen. Coop. Lett. 1986;60:61. [Google Scholar]
- 39.Jefferson R. A., Kavanagh T. A., Bevan M. W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald H. A., Chern M.-S., Navarre R., Ronald P. C. Mol. Plant–Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 41.Farid N. A., Born G. S., Kessler W. V., Shaw S. M., Lange W. E. Clin. Chem. 1975;21:1167–1168. [PubMed] [Google Scholar]
- 42.Lee J. Y., Yoo B. C., Rojas M. R., Gomez-Ospina N., Staehelin L. A., Lucas W. J. Science. 2003;299:392–396. doi: 10.1126/science.1077813. [DOI] [PubMed] [Google Scholar]
- 43.Hou Y.-M., Paplomatas E. J., Gilbertson R. L. Mol. Plant–Microbe Interact. 1998;11:208–217. [Google Scholar]
- 44.Bendahmane A., Querci M., Kanyuka K., Baulcombe D. C. Plant J. 2000;21:73–81. doi: 10.1046/j.1365-313x.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- 45.Clark M. F., Lister R. M., Bar-Joseph M. Methods Enymol. 1986;118:742–766. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.