Abstract
Mitotic homologous recombination (HR) is critical for the repair of double-strand breaks, and conditions that stimulate HR are associated with an increased risk of deleterious sequence rearrangements that can promote cancer. Because of the difficulty of assessing HR in mammals, little is known about HR activity in mammalian tissues or about the effects of cancer risk factors on HR in vivo. To study HR in vivo, we have used fluorescent yellow direct repeat mice, in which an HR event at a transgene yields a fluorescent phenotype. Results show that HR is an active pathway in the pancreas throughout life, that HR is induced in vivo by exposure to a cancer chemotherapeutic agent, and that recombinant cells accumulate with age in pancreatic tissue. Furthermore, we developed an in situ imaging approach that reveals an increase in both the frequency and the sizes of isolated recombinant cell clusters with age, indicating that both de novo recombination events and clonal expansion contribute to the accumulation of recombinant cells with age. This work demonstrates that aging and exposure to a cancer chemotherapeutic agent increase the frequency of recombinant cells in the pancreas, and it also provides a rapid method for revealing additional factors that modulate HR and clonal expansion in vivo.
Keywords: aging, homologous recombination, mutation, chemotherapy
Cells are constantly exposed to endogenous and exogenous DNA-damaging agents that can lead to double-strand breaks, either by causing breaks in both strands of DNA or by causing replication fork breakdown (1). Homologous recombination (HR) is critical for repairing double-strand breaks in mammalian cells. By using homologous DNA sequences present on the sister chromatid or homologous chromosome, damage can be repaired accurately without loss of sequence information (2, 3). Thus, the frequency of HR reflects both the levels of double-strand breaks and the ability of cells to use HR during DNA repair.
Although HR is generally error-free, recombination between misaligned sequences can cause insertions, deletions, and translocations. Furthermore, recombination between homologous chromosomes can lead to loss of heterozygosity (4), and HR has been estimated to be the underlying cause of loss of heterozygosity 25–80% of the time in mammalian cells (e.g., see ref. 5). Germ-line mutations in genes that modulate the frequency of HR are associated with an increased risk of cancer. For example, inherited mutations in the HR helicases BLM and WRN lead to increased rates of HR (6, 7) and increase the risk of cancer (8).
Whereas too much HR can be problematic, too little HR can also destabilize the genome, possibly as a result of nonhomologous end-joining of DNA ends created at broken replication forks (4, 9). In the pancreas, inherited mutations in BRCA1 (8), BRCA2 (10), and FANCC (11) increase the risk of pancreatic cancer, and loss of function of these genes suppresses HR (12–14), causing an increased frequency of tumorigenic sequence rearrangements (15, 16). Although these findings suggest that HR is critical for maintaining genomic integrity in the pancreas, it had not been shown that HR is an active pathway in mature pancreatic cells and no studies had explored the effects of cancer risk factors on potentially mutagenic HR events in the pancreas.
With a 5-year survival rate of <5%, pancreatic cancer remains one of the deadliest cancers in the United States (17, 18). One important risk factor for pancreatic cancer is aging (19). To our knowledge, mutation frequency has not been reported in the pancreas (20), so the effect of age on pancreatic mutation frequency was not known. However, a number of studies have investigated the impact of age on mutation frequency in other cell types. For example, the frequency of loss of heterozygosity increases by ≈10-fold with increasing age in lymphocytes (21, 22). Furthermore, a significant fraction of these loss of heterozygosity events are because of mitotic recombination, suggesting that HR contributes to gene inactivation during aging (21–23).
In this study, we have explored the effects of aging and exposure to a cancer chemotherapeutic agent on the frequency of HR in the mouse pancreas by exploiting mice in which HR at an integrated transgene yields a fluorescent phenotype (24). Furthermore, we describe methodology for rapid and sensitive detection of recombinant cells within intact pancreata.
Results
Flow Cytometry Analysis of HR Events in Adult Pancreatic Cells.
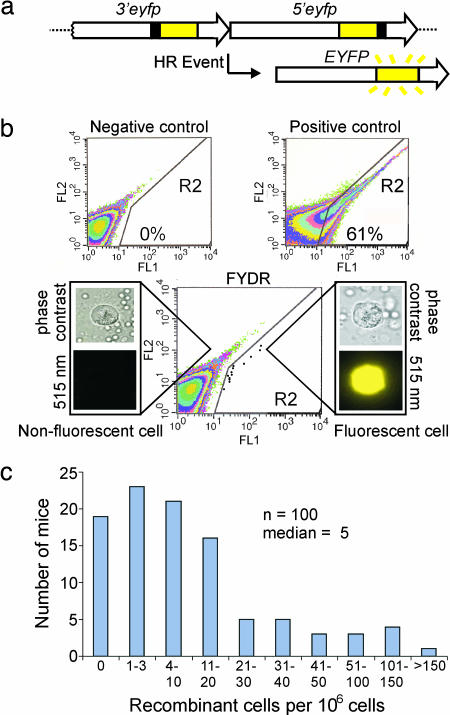
The fluorescent yellow direct repeat (FYDR) mice carry a direct repeat recombination substrate in which an HR event can restore full-length enhanced yellow fluorescent protein (EYFP) coding sequence (24) (Fig. 1a). Because HR had not been studied in primary pancreatic cells, we first set out to determine whether HR is an active process in the pancreas. To establish flow cytometry parameters, we compared fluorescence intensities in disaggregated whole pancreata from negative control and positive control mice (Fig. 1b). To quantify fluorescent recombinant cells, a region (R2) was created that excludes negative control cells (Fig. 1b). We analyzed >34 million cells from 33 negative control pancreata by flow cytometry, and only one cell appeared in the R2 region, indicating an extremely low background. Analysis of 100 mice aged 4–10 weeks shows that the median frequency of recombinant pancreatic cells is five per million (Fig. 1c). In addition, the recombinant cell frequency is highly variable among mice (Fig. 1c), which is consistent with the possibility that recombinant cells that arise at different times during growth can clonally expand.
Fig. 1.
FYDR system and analysis of pancreatic cells by flow cytometry. (a) Arrangement of the FYDR recombination substrate. Large arrows indicate expression cassettes; yellow boxes show coding sequences; black boxes show positions of deleted sequences (deletion sizes not to scale). For a more complete description of HR mechanisms that can restore full-length EYFP coding sequences, see Jonnalagadda et al. (39). (b) Flow cytometry results of disaggregated pancreatic cells. Axes indicate relative fluorescence intensity at 515–545 nm (FL1) vs. 562–588 nm (FL2). R2 region delineates EYFP-positive cells. Representative data are shown for a negative control mouse, a positive control FYDR-recombined mouse, and an FYDR mouse. For clarity, data for individual cells (dots) have been darkened in the FYDR R2 region. Percentages of fluorescent cells identified in the R2 region are indicated for the positive and negative controls. Cell images are representative of disaggregated pancreatic cells from an FYDR mouse taken at ×40 with phase contrast or an EYFP filter (515 nm). Image of a fluorescent cell under an EYFP filter is colorized. (c) Spontaneous frequency of recombinant pancreatic cells per 106 as determined by flow cytometry for 100 4- to 10-week-old FYDR mice. n = number of independent samples.
In Situ Detection of Recombinant Pancreatic Cells.
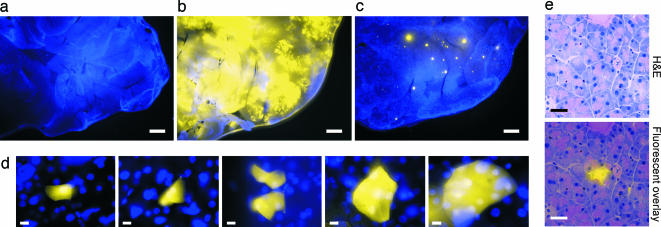
To learn more about the timing of HR events, and to reveal the cell types in which they occur, methodology was developed for direct detection of recombinant cells in situ. Under an epifluorescent microscope, negative control pancreata are essentially nonfluorescent when viewed with an EYFP filter, whereas Hoechst 33342-stained nuclei fluoresce under UV (Fig. 2a). In contrast, in positive control pancreata, much of the tissue is brightly fluorescent (Fig. 2b). In FYDR pancreata, yellow fluorescent foci are readily apparent (Fig. 2c). At a higher magnification, it becomes clear that these fluorescent foci are actually single isolated cells and small clusters of cells (Fig. 2d), indicating that recombinant cells can be directly detected within intact tissue by microscopic examination of intact pancreata.
Fig. 2.
Analysis of fluorescent foci in mouse pancreata. For analysis of freshly excised tissue, images show overlays of EYFP- (510–560 nm) and UV- (420 nm) filtered images. Nuclei are stained with Hoechst 33342. (a–c) Portions of negative control (a), positive control (b), and FYDR (c) mouse pancreata imaged at ×1. (Scale bar, 1 mm.) Brightness and contrast for UV-filtered images were adjusted equivalently. For EYFP images, brightness and contrast for negative control and FYDR (5-s exposure) images were adjusted equivalently. To avoid overexposure of the positive control, a shorter exposure time was used (1 s) and brightness and contrast were not adjusted. (d) Images of fluorescent recombinant cells in freshly excised tissue at ×40. (Scale bar, 10 μm.) Brightness and contrast were optimized for each image. (e) Histological images were collected at ×40. (Scale bar, 30 μm.) (Upper) H&E-stained section. (Lower) Overlay of H&E and fluorescence imaged under an EYFP filter (515 nm). Brightness and contrast of fluorescence were optimized. EYFP fluorescence is colorized.
To explore which cell types recombine, frozen sections of pancreata were analyzed. By overlaying fluorescence and H&E-stained images, histological analysis of pancreata from positive control mice revealed that acinar, islet, and ductal cells can fluoresce (data not shown). In pancreata of FYDR mice, EYFP fluorescence is confined within cell boundaries and recombinant cells are acinar cells (no islet or ductal cells were detected among 43 independent foci) (Fig. 2e). Therefore, the fluorescent foci seen within the pancreas of FYDR mice are most often due to fluorescent acinar cells.
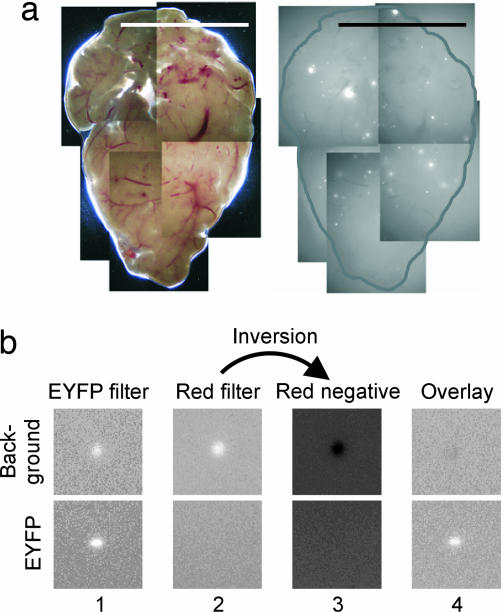
To standardize conditions for foci quantification, pancreata were uniformly compressed to a thickness of 0.5 mm, and composite images that cover one side of the pancreatic surface area were created (Fig. 3a). Comparison of both sides of the same pancreas showed that similar frequencies of recombinant cells were detected on both sides (data not shown; note that most foci detected on one side could not be detected on the other side because of sample thickness). In negative control mice, background fluorescence was occasionally observed and generally emitted under multiple filters. To reduce background fluorescence, images collected under a red filter were inverted, and these negatives were then merged with images taken with an EYFP filter (Fig. 3b). Using this subtraction methodology, we analyzed 24 negative control animals, and no foci were detected. These methods make it possible to specifically detect EYFP fluorescence and thus provide a means for rapid quantification of recombinant cells in situ.
Fig. 3.
Imaging methodology for quantifying recombinant cells in whole pancreata. (a) Compiled image of 9-week-old FYDR pancreas at ×1 taken under visible light (Left) and an EYFP filter (510–560 nm) (Right). (Scale bar, 1 cm.) The edge of the pancreatic tissue is outlined. (b) Method for reducing background fluorescence. (Upper) Background fluorescence appears brightly under EYFP (510–560 nm, column 1) and red (605/55 nm, column 2) filters. The red-filtered image is inverted (column 3) and merged with the EYFP-filtered image (column 4). (Lower) Similar analysis of an EYFP focus.
DNA Damage-Induced Recombination in Pancreatic Cells.
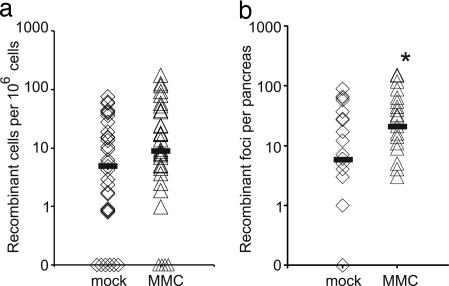
To determine whether HR can be induced in postnatal pancreatic cells, 5- to 6-week-old FYDR mice were injected with the recombinogenic interstrand cross-linking agent mitomycin-C (MMC). The median frequencies of recombinant cells by flow cytometry (Fig. 4a) and of recombinant foci by in situ imaging (Fig. 4b) were higher among the MMC-treated mice (note the logarithmic scale). It is formally possible that the increased frequency of recombinant cells after MMC treatment is because of increased EYFP expression. To explore this possibility, we exploited positive control animals in which all cells carry the recombined substrate (full-length EYFP). Flow cytometry of pancreatic cells from mock- and MMC-treated positive control mice revealed that there was no statistically significant difference in EYFP expression between the cohorts (data not shown). Thus, we conclude that the increase in recombinant cell frequency after MMC treatment is the result of an induction of recombinant cells, indicating that the FYDR model specifically detects HR events, and that HR is an active repair process in the postnatal pancreas.
Fig. 4.
MMC-induced HR in mouse pancreata. Medians are indicated by black bars. Points on the x axis indicate individual mice with zero recombinant cells. (a) Frequency of recombinant cells per million as determined by flow cytometry for mock-treated (n = 35) and MMC-treated (n = 34) FYDR mice (P = 0.06). (b) Recombinant foci per pancreas detected by in situ image analysis for mock-treated (n = 24) and MMC-treated (n = 23) FYDR mice. ∗, MMC-treated cohort is statistically significantly higher than mock-treated cohort (P < 0.05).
Interestingly, MMC induction is statistically significant only when analyzed by in situ imaging. It is noteworthy that mice with a similar number of recombinant foci can show a broad range of recombinant cell frequencies when analyzed by flow cytometry, possibly because a single focus may contain many recombinant cells. Thus, when studying the effects of an environmental exposure by flow cytometry, it should be noted that large foci can potentially mask the induction of multiple smaller foci. Although both flow cytometry and in situ imaging detect recombinant cells, in situ imaging may be a more sensitive method for detecting exposure-induced recombinant cells (e.g., independent HR events).
Effect of Aging on Recombinant Cell Frequency.
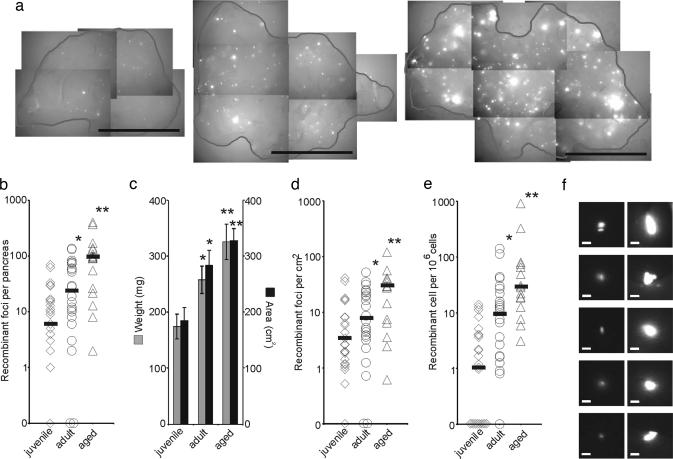
Age is an important risk factor for cancer. To explore the effects of aging, recombinant foci were quantified in three different age groups: juvenile (4 weeks old), adult (9 weeks old), and aged (67–74 weeks old). Whereas the number of foci detected by in situ imaging varied among individual animals within each age group, the median clearly increased with age (Fig. 5b). This increase was especially evident when examining images of pancreata with the highest frequency of recombinant foci from each age group (Fig. 5a). Relative to juvenile mice, the median number of recombinant foci increased ≈4- and ≈16-fold in adult and aged mice, respectively (Fig. 5b), demonstrating that recombinant cells accumulate with age.
Fig. 5.
Effects of aging on the frequency of recombinant cells in the pancreas. (a) Compiled images for juvenile (4 weeks old) (Left), adult (9 weeks old) (Center), and aged (64–72 weeks old) (Right) FYDR mice. Examples of pancreata with the highest number of recombinant foci for each cohort are shown. Images were collected at ×1 with an EYFP filter (510–560 nm). (Scale bar, 1 cm.) The edge of the pancreatic tissue is outlined. (b) Recombinant foci per pancreas detected by in situ image analysis for juvenile (n = 25), adult (n = 24), and aged (n = 16) mice. (c) Weight (mg) and area (cm2) of mouse pancreata for juvenile (n = 25), adult (n = 25), and aged (n = 17) cohorts. (d) Recombinant foci per squared centimeter for juvenile (n = 25), adult (n = 24), and aged (n = 16) mice. (e) Frequency of recombinant cells per million as determined by flow cytometry for juvenile (n = 24), adult (n = 24), and aged (n = 16) mice. (b–e) ∗, adult cohort is significantly higher statistically than juvenile cohort (P < 0.05). ∗∗, aged cohort is significantly higher statistically than juvenile and adult (P < 0.05) cohorts. Medians are indicated by black bars. Points on the x axis indicate individual mice with zero recombinant cells detected. (f) Images of the five largest foci among all juvenile (Left) and all aged (Right) mice taken under an EYFP filter (510–560 nm) at ×1. (Scale bar, 100 μm.) Brightness and contrast were not adjusted.
During aging, the mouse pancreas continues to grow (Fig. 5c) (25, 26), which raises the possibility that the increase in the number of foci is because of the increase in surface area of pancreata (Fig. 5c). After correcting for the total surface area, the median frequencies of recombinant foci per cm2 in adult and aged cohorts are still significantly higher than juvenile mice (≈2- and ≈9-fold higher, respectively; Fig. 5d), suggesting that new recombination events contribute to the accumulation of recombinant cells with age.
As an alternative approach for analyzing the effects of aging on HR, disaggregated pancreatic cells were quantified by flow cytometry (Fig. 5e). Similar trends in the frequency of recombinant cells were observed with age. Relative to juvenile mice, the median frequency of fluorescent cells per million increased in adult and aged mice by ≈8- and ≈26-fold, respectively. A comparison of the mice with the highest frequencies from the young vs. the aged cohorts shows there are 14 vs. 914 recombinant cells per million, respectively (note the logarithmic scale). Together, these data indicate that as mice age, the frequency of cells harboring DNA sequence rearrangements increases significantly.
To determine whether the increased frequency of recombinant cells with age is because of an increase in EYFP expression, we analyzed pancreata from positive control mice. Although EYFP expression varies greatly among individual positive control mice (which undoubtedly contributes to variation in the frequency of fluorescent cells among the FYDR mice), there were no statistically significant differences in expression levels among the young, adult, and aged cohorts (data not shown). Because expression of the FYDR locus does not increase with age, we conclude that the increase in recombinant cell frequency with age is the result of the accumulation of recombinant cells in the pancreas.
Clonal Expansion of Recombinant Cells.
Clonal expansion of mutant cells is an important precursor to the development of cancer (27). To explore the possibility that recombinant cells clonally expand with age, images from all juvenile and aged animals were carefully examined to identify the five largest foci from within each cohort. Comparison of the foci images revealed that foci are clearly much larger in aged animals (Fig. 5f). Given that the geometric mean fluorescence intensity of recombinant cells is not significantly different among cohorts (data not shown), the observed increase in foci sizes cannot be due to increased brightness. It is formally possible that the increase in recombinant foci sizes is the result of multiple independent recombination events occurring in neighboring cells. However, because the frequency of recombinant cells in the pancreas is ≈5 per million, the probability that two adjacent recombinant cells occurred from independent events is ≈1 in 1010 (assuming that each cell touches ≈10 neighbors), making it virtually impossible that multiple adjacent recombinant cells in large foci occurred independently. These data indicate that recombinant cells can clonally expand during aging, which suggests that there is clonal expansion throughout the pancreas that can be visualized in cases where the progenitor cell is fluorescent.
Discussion
With a 5-year survival rate of <5% (17, 18), pancreatic cancer remains a fatal disease. Pancreatic cancer is caused by the accumulation of genetic mutations in a single cell lineage (e.g., activation of K-ras and inactivation of p16, p53, Smad4, and BRCA2) (28). The probability that multiple mutations occur in the same lineage depends on both the mutation rate and the total number of cells that harbor tumorigenic mutations (27, 29). Therefore, increasing either the mutation rate or the number of mutant cells (by clonal expansion) concomitantly increases the risk of acquiring subsequent, and possibly transforming, mutations in the same cell lineage.
Among >2,000 rodent experimental records in which mutation frequencies have been assessed by using transgenic animals, none describe the mutation frequency in the pancreas (20). Here, we have used FYDR mice to study one important class of mutations, HR events. When both in situ imaging and flow cytometry are used, results show that the number of recombinant cells increases with age, and a comparison of the sizes of recombinant foci in juvenile and aged mice shows that pancreata of aged mice have larger foci. Therefore, as mice age, both de novo HR events and clonal expansion contribute to the overall increase in the number of pancreatic cells harboring rearranged DNA.
Within all cohorts, a wide range of recombinant cell frequencies is observed. This intermouse variation may result from differences in recombination rates, fluctuations in foci sizes caused by clonal expansion, or variation in expression of EYFP. Although we have not yet tested for differences in recombination rates among individual mice, it is clear that clonal expansion can contribute to variation in the total number of recombinant cells. In addition, the positive control mice indicate that variable levels of EYFP expression also contribute to intermouse variation. Although we do not yet know the cause for the variegated expression in the positive control mice, recombination can still be studied by ascertaining whether a variable of interest affects expression in a cohort of positive control animals. For example, in these studies, EYFP fluorescence does not increase with age, indicating that the increased frequency is due to HR. Indeed, expression may even decrease with age, so the effect of age on the accumulation of recombinant cells may actually be underestimated.
Histological analysis of ≈40 foci revealed that all recombinant fluorescent cells were acinar cells. Although the majority (80–90%) of human pancreatic neoplasms are ductal pancreatic adenocarcinomas (18), there is debate over the actual origin of cells that lead to ductal adenocarcinomas. Interestingly, acinar cells can transdifferentiate into ductal cells both in vitro and in vivo (30–32). Furthermore, expression of activated K-ras in pancreatic acinar cells has been shown to induce preinvasive pancreatic neoplastic lesions (31). Therefore, genetic changes in acinar cells may contribute to tumor formation. Regardless of whether acinar cells are the precursors of ductal adenocarcinomas, genetic and environmental conditions that induce HR in acinar cells may do so in other pancreatic cell types as well. Therefore, detection of HR in acinar cells may be a gauge of genetic insult to the pancreas as a whole, making the FYDR mice potentially useful as sensors of pancreatic genotoxins.
Other mouse models, including BigBlue (33, 34), MutaMouse (35), and LacZ (36), have been used to examine changes in the frequency of point mutations and small deletions with age. The effect of age on these classes of mutations appears to be strongly tissue-dependent. In certain tissues, such as brain and testis, the mutant frequency remains fairly constant (33–35). In contrast, for liver and bladder, mutant cell frequency increases with age similar to what has been observed for the pancreas in these studies (33–35). It is interesting to speculate that for tissues in which spontaneous mutations accumulate with age, mutagenic exposures during adult life may have a greater influence on cancer risk. Indeed, smoking is an important risk factor for liver, bladder, and pancreatic cancer, and has less of an effect on the risk of brain or testicular cancer (37).
HR events are an important class of mutations that are known to promote cancer (4, 38). Here we have shown that detection of recombinant cells in FYDR mice by in situ imaging and flow cytometry can be used to monitor the effects of cancer risk factors on HR. Compared with analysis by flow cytometry, in situ detection improves the sensitivity for detecting new mutation formation (e.g., small foci) that can be masked by previously existing larger foci upon tissue disaggregation. Furthermore, because the accumulation of recombinant cells can be monitored over months and even years, long-term effects of both acute and chronic exposures relevant to cancer can also be studied. Although small differences in recombination rate may not be immediately reflected as differences in mutant cell frequency, these changes in recombination rate may result in large changes in mutant cell frequency over time. In summary, we have explored how a key risk factor for pancreatic cancer, aging, affects the frequency of cells harboring recombined DNA. The results of these studies demonstrate that HR is an active process in the adult pancreas, and that cells harboring sequence rearrangements can persist and clonally expand. Furthermore, the methodology used in these studies can now be applied to explore how additional genetic and environmental risk factors modulate double-strand break formation and repair by HR.
Materials and Methods
Animals.
C57BL/6 FYDR mice have been described in ref. 24. Positive control FYDR-recombined mice arose spontaneously from an HR event in an FYDR parental gamete, and all cells within these mice carried the full-length EYFP coding sequence. FYDR cohorts had an ≈1:1 ratio of males to females (preliminary data suggest that there may be a difference in EYFP expression levels among males and females). Controls were sex- and age-matched, except for the aged negative control C57BL/6 mice, which were 47–85 weeks old, and the aged positive control FYDR-recombined mice, which were 52 weeks old.
Flow Cytometry.
Pancreata were isolated and placed in ice-cold PBS containing 0.01% soybean trypsin inhibitor (Sigma). Almost all samples were analyzed by flow cytometry after imaging. Pancreata were minced and divided into two samples. Samples were shaken (150 cycles per minute) in 5 ml of 2 mg/ml collagenase V (Sigma) in Hanks' buffered salt solution (Invitrogen) at 37°C for 20 min. Triturated tissue was filtered (70 μm), and 10 ml of DMEM-F12 (Sigma) supplemented with 20% FBS (Atlanta Biologicals, Lawrenceville, GA) was added. Cells were pelleted, resuspended in 400 μl of OptiMEM (Invitrogen), filtered (35 μm), and analyzed by flow cytometry as described in ref. 24. On average, ≈1 million cells were analyzed per sample.
Fluorescence Intensity Measurements.
Geometric mean of fluorescence intensity of pancreatic cells in the R2 region of juvenile and aged mice was calculated by CellQuest acquisition and analysis software on the Becton Dickinson FACScan flow cytometer.
Imaging.
Pancreata were isolated as described. Nuclei were stained with 50 μg/ml Hoechst 33342 (Sigma). Whole pancreata pressed between glass slides separated by 0.5 mm spacers were imaged on a Nikon E600 microscope with a SPOT RT camera (Diagnostic Instruments, Sterling Heights, MI) with a Nikon ×1 objective. Images were manually compiled to cover the entire visible surface area. SPOT Advanced (Diagnostic Instruments) was used to colorize black and white images. Filters included: visible light; UV (excitation, 330–380 nm; emission, 420 nm); red (excitation, 540/25 nm; emission, 605/55 nm); and EYFP (excitation, 460–500 nm; emission, 510–560 nm). Images were collected by using a fixed aperture time. For foci counting, Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA) was used to optimize brightness and contrast. For images collected with the EYFP filter, identical adjustments were made for all images. Similarly, for images collected with the red filter, brightness was optimized and identical adjustments were made for all red fluorescence images. For pancreata with foci in the red channel, images taken under the red filter were inverted to create negatives that were merged with images taken under the EYFP filter. Foci were counted manually. Similar results were obtained in a subset of blinded samples. The area of compiled pancreata images was determined by using Scion Image Beta 4.02 Win (Scion, Frederick, MD) by manually tracing the pancreas edge.
MMC Treatment.
Five- to 6-week-old FYDR mice were i.p. injected with 2 mg/kg of body weight of 0.5 mg/ml MMC (Sigma) in PBS. Mock-treated controls were injected with equal volumes of PBS. Mice were analyzed 3.5 weeks after injection.
Histology.
Frozen sections (5 μm) were imaged at ×40 with an EYFP filter (excitation, 460–500 nm; emission, 515 nm) on a Nikon E600 microscope. SPOT Advanced (Diagnostic Instruments) was used to colorize images. Sections were subsequently stained with H&E, imaged, and manually overlaid. Histology was assessed by Arlin Rogers (Massachusetts Institute of Technology).
Statistics.
Recombinant cell frequency follows a nonnormal distribution (Fig. 1c). Therefore, frequencies among cohorts were compared by using a one-tailed Mann–Whitney test (for both flow cytometry and foci data). When comparing the frequency of fluorescent cells among positive control cohorts, a two-tailed Student's t test was performed.
Acknowledgments
We thank Barry Alpert for microscopy support; Dr. Arlin Rogers for histological analysis; Glenn Paradis (MIT Center for Cancer Research Flow Cytometry Facility) and the MIT Division of Comparative Medicine for their support; and Evelyn Park, Jennifer Sauchuk, Fallon Lin, and Jeff Loh for additional technical assistance. This work was supported by National Institutes of Health Grants ES002109, CA79827, and CA84740, and Department of Energy Grant DE-FG01-04ER04-21. D.M.W.-B. was supported by National Institutes of Health/National Institute of General Medical Sciences Interdepartmental Biotechnology Training Program Grant GM008334, and C.A.H was supported by National Institutes of Health Institutional Training Grant in Environmental Toxicology Award T32-ES07020.
Abbreviations
- HR
homologous recombination
- FYDR
fluorescent yellow direct repeat
- MMC
mitomycin C
- EYFP
enhanced yellow fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Friedberg E., Walker G., Siede W., Wood R., Schultz R., Ellenburger T. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 2005. [Google Scholar]
- 2.Helleday T. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 3.West S. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 4.Bishop A. J., Schiestl R. H. Exp. Mol. Pathol. 2003;74:94–105. doi: 10.1016/s0014-4800(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 5.Morley A. A., Grist S. A., Turner D. R., Kutlaca A., Bennett G. Cancer Res. 1990;50:4584–4587. [PubMed] [Google Scholar]
- 6.Chaganti R. S., Schonberg S., German J. Proc. Natl. Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prince P. R., Emond M. J., Monnat R. J., Jr Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson D., Easton D. F. J. Natl. Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 9.Venkitaraman A. R. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 10.Hahn S. A., Greenhalf B., Ellis I., Sina-Frey M., Rieder H., Korte B., Gerdes B., Kress R., Ziegler A., Raeburn J. A., et al. J. Natl. Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 11.Couch F. J., Johnson M. R., Rabe K., Boardman L., McWilliams R., de Andrade M., Petersen G. Cancer Res. 2005;65:383–386. [PubMed] [Google Scholar]
- 12.Moynahan M. E., Chiu J. W., Koller B. H., Jasin M. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 13.Moynahan M. E., Pierce A. J., Jasin M. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 14.Niedzwiedz W., Mosedale G., Johnson M., Ong C. Y., Pace P., Patel K. J. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Ban S., Shinohara T., Hirai Y., Moritaku Y., Cologne J. B., MacPhee D. G. Mutat. Res. 2001;474:15–23. doi: 10.1016/s0027-5107(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 16.D'Andrea A. D., Grompe M. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 17.Ries L. A. G., Eisner M. P., Kosary C. L., Hankey B. F., Miller B. A., Clegg L., Mariotto A., Feuer E. J., Edwards B. K., editors. SEER Cancer Statistics Review, 1975–2002. Bethesda, MD: Natl. Cancer Inst; 2005. Available at http://seer.cancer.gov/csr/1975_2002. [Google Scholar]
- 18.Hansel D. E., Kern S. E., Hruban R. H. Annu. Rev. Genomics Hum. Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- 19.Li D., Xie K., Wolff R., Abbruzzese J. L. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 20.Lambert I. B., Singer T. M., Boucher S. E., Douglas G. R. Mutat. Res. 2005;590:1–280. doi: 10.1016/j.mrrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Wijnhoven S. W., Kool H. J., van Teijlingen C. M., van Zeeland A. A., Vrieling H. Mutat. Res. 2001;473:23–36. doi: 10.1016/s0027-5107(00)00163-9. [DOI] [PubMed] [Google Scholar]
- 22.Grist S. A., McCarron M., Kutlaca A., Turner D. R., Morley A. A. Mutat. Res. 1992;266:189–196. doi: 10.1016/0027-5107(92)90186-6. [DOI] [PubMed] [Google Scholar]
- 23.Van Sloun P. P., Wijnhoven S. W., Kool H. J., Slater R., Weeda G., van Zeeland A. A., Lohman P. H., Vrieling H. Nucleic Acids Res. 1998;26:4888–4894. doi: 10.1093/nar/26.21.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendricks C. A., Almeida K. H., Stitt M. S., Jonnalagadda V. S., Rugo R. E., Kerrison G. F., Engelward B. P. Proc. Natl. Acad. Sci. USA. 2003;100:6325–6330. doi: 10.1073/pnas.1232231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taga R., Bispo L. B., Bordin R. A., Hassunuma R. M. Okajimas Folia Anat. Jpn. 1998;74:271–278. doi: 10.2535/ofaj1936.74.6_271. [DOI] [PubMed] [Google Scholar]
- 26.Slack J. M. Development (Cambridge, U.K.) 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 27.Thilly W. G. IARC Sci. Publ. 1988:486–492. [PubMed] [Google Scholar]
- 28.Hruban R. H., Wilentz R. E., Kern S. E. Am. J. Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan R. N., Bielas J. H., Loeb L. A. DNA Repair (Amsterdam) 2005;5:294–302. doi: 10.1016/j.dnarep.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Hall P. A., Lemoine N. R. J. Pathol. 1992;166:97–103. doi: 10.1002/path.1711660203. [DOI] [PubMed] [Google Scholar]
- 31.Grippo P. J., Nowlin P. S., Demeure M. J., Longnecker D. S., Sandgren E. P. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 32.Wagner M., Greten F. R., Weber C. K., Koschnick S., Mattfeldt T., Deppert W., Kern H., Adler G., Schmid R. M. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill K. A., Buettner V. L., Halangoda A., Kunishige M., Moore S. R., Longmate J., Scaringe W. A., Sommer S. S. Environ. Mol. Mutagen. 2004;43:110–120. doi: 10.1002/em.20004. [DOI] [PubMed] [Google Scholar]
- 34.Stuart G. R., Oda Y., de Boer J. G., Glickman B. W. Genetics. 2000;154:1291–1300. doi: 10.1093/genetics/154.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono T., Ikehata H., Nakamura S., Saito Y., Hosoi Y., Takai Y., Yamada S., Onodera J., Yamamoto K. Mutat. Res. 2000;447:165–177. doi: 10.1016/s0027-5107(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 36.Dolle M. E., Snyder W. K., Gossen J. A., Lohman P. H., Vijg J. Proc. Natl. Acad. Sci. USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health . The Health Consequences of Smoking: A Report of the Surgeon General. Washington, DC: Government Printing Office; 2004. [Google Scholar]
- 38.Vilenchik M. M., Knudson A. G. Proc. Natl. Acad. Sci. USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonnalagadda V. S., Matsuguchi T., Engelward B. P. DNA Repair (Amsterdam) 2005;4:594–605. doi: 10.1016/j.dnarep.2005.02.002. [DOI] [PubMed] [Google Scholar]