Abstract
Mammalian telomeric proteins function through dynamic interactions with each other and telomere DNA. We previously reported the formation of a high-molecular-mass telomeric complex (the mammalian telosome) that contains the six core proteins TRF1, TRF2, RAP1, TIN2, POT1, and TPP1 (formerly named PTOP/PIP1/TINT1) and mediates telomere end-capping and length control. In this report, we sought to elucidate the mechanism of six-protein complex (or shelterin) formation and the function of this complex. Through reconstitution experiments, we demonstrate here that TIN2 and TPP1 are key components in mediating the six-protein complex assembly. We demonstrate that not only TIN2 but also TPP1 are required to bridge the TRF1 and TRF2 subcomplexes. Specifically, TPP1 helps to stabilize the TRF1–TIN2–TRF2 interaction and promote six-protein complex formation. Consistent with this model, overexpression of TPP1 enhanced TIN2–TRF2 association. Conversely, knocking down TPP1 reduced the ability of endogenous TRF1 to associate with the TRF2 complex. Our results suggest that coordinated interactions among TPP1, TIN2, TRF1, and TRF2 may ensure robust assembly of the telosome, telomere targeting of its subunits, and, ultimately, regulated telomere maintenance.
Keywords: protein complex, telomere, telosome
Mammalian telomeres are regulated by the telomerase and telomeric proteins (1–6). Among the telomere-associated proteins important for mammalian telomere homeostasis, POT1 is likely the major regulator of telomere length control (7–9). POT1 binds the 3′ G-rich telomere overhangs through its oligonucleotide-binding folds (7, 10, 11) and may regulate telomerase access (12–14). The telomere recruitment of POT1 thus constitutes an important step in telomere end-capping and length control. Recently, a new telomeric protein TPP1 (previously PTOP/PIP1/TINT1) was identified as a regulator of POT1 (9, 15, 16). The telomeric targeting of POT1 depends on its interaction with TPP1 (9). It remains to be determined how TPP1 interacts with other telomeric proteins and whether TPP1 has any function other than targeting POT1.
In contrast to POT1, TRF1 and TRF2 directly bind double-stranded telomere DNA and interact with a number of proteins to maintain telomere structure and length (1–5). It has been shown that TRF1 counts and controls the length of telomere repeats, probably through its interaction with TIN2, Tankyrase, PINX1, TPP1, and POT1 (7, 9, 12, 15, 17–24). In comparison, TRF2 has an essential role in end protection and the telomeric recruitment of several proteins, including the BRCA1 C-terminal domain-containing protein RAP1, the nucleotide excision repair protein ERCC1/XPF, BLM, and the DNA repair MRN complex (24–31). Because of their abilities to interact with multiple proteins, TRF1 and TRF2 are by definition hubs of protein–protein interaction at the telomeres (32). Recent studies have established multiple pairwise interactions among the six telomeric proteins (TRF1, TRF2, RAP1, TIN2, POT1, and TPP1), including the association of TIN2 with both TRF1 and TRF2 (16, 33–35). In fact, the six proteins could be copurified in mammalian cells in a large-molecular-mass complex referred to as the mammalian telosome/shelterin (16, 32–36)
The physical link between TRF1 and TRF2 suggests coordinated and integrated signals in telomere maintenance. The connectivity between TRF1 and TRF2 appeared vital for telomere maintenance, because reducing TRF1 levels also resulted in decreased TRF2 telomere localization (37). Moreover, POT1 is most likely one of the initiators of telomere maintenance activity mediated by the telosome. Information regarding the state and length of the telomere ends can be transmitted from TRF1 and TRF2 to POT1 through their interaction within the complex. The assembly and function of the mammalian telosome, and, in particular, the roles and contribution of individual core proteins remain largely unknown. Here we investigated the mechanism of telosome assembly through reconstitution and fractionation experiments. Our findings indicate that the six telomeric proteins are sufficient to form a large complex. Both TPP1 and TIN2 are essential mediators of this process. In addition, the TPP1–TIN2 interaction regulates the bridging between TRF1 and TRF2 and promotes and stabilizes the assembly of high-order telomeric complexes.
Results
TIN2 and TPP1 Are Key Components that Mediate the Six-Protein Complex Assembly.
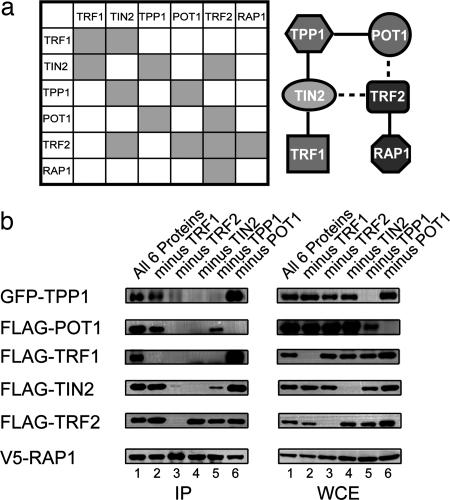
Among the six telomeric proteins, multiple pairwise interactions have been demonstrated (summarized in Fig. 1a). What is the contribution and significance of each interaction to telosome assembly?
Fig. 1.
TPP1 and TIN2 are key components that mediate six-protein complex formation. (a) A schematic representation of known pairwise interactions between known mammalian telomere proteins (Left) and a possible model of interactions in the six-protein complex (Right). Known pairwise interactions: TRF1–TRF1 (44), TRF1–TIN2 (18), TIN2–TPP1 (9, 16), POT1–TPP1 (9, 15), TRF2–TIN2 (16, 33–35), TRF2–TRF2 (45), TRF2–RAP1 (25), and POT1–TRF2 (46). Dashed lines indicate weak interactions. (b) TPP1 and TIN2 regulate six-protein complex assembly. Expression constructs encoding V5-RAP1 were cotransfected into 293T cells with either all five expression vectors encoding FLAG-POT1, FLAG-TIN2, FLAG-TRF1, FLAG-TRF2, and GFP-TPP1 or any four of the five vectors. Approximately 3 μg of DNA was transfected for each construct. Two days after transfection, whole-cell extracts were prepared and immunoprecipitated (IP) with anti-V5 antibodies. Coimmunoprecipitated proteins were detected by Western blotting by using anti-TPP1 antibodies (for GFP–TPP1) and the appropriate anti-epitope tag antibodies (FLAG and V5).
To address this question, we assessed the individual contribution of the six proteins in a reconstitution experiment by transiently expressing epitope-tagged telomere proteins in 293T cells. In this system, transiently expressed exogenous proteins were in vast excess (10- to 100-fold) compared with endogenous proteins (data not shown). As a result, the protein–protein interactions assayed occurred primarily between exogenously expressed proteins.
Consistent with the six-protein complex model, immunoprecipitation of V5-RAP1 brought down FLAG-tagged POT1, TRF1, TRF2, TIN2, and GFP-tagged TPP1 (Fig. 1b, lane 1). With RAP1 as the anchor, we then assayed how the six-protein complex assembly might be affected when the remaining components were individually removed. As expected, in the absence of TRF2, the association of RAP1 with the other four telomeric proteins was disrupted (Fig. 1b, lane 3). Furthermore, POT1 appeared dispensable for the formation of the TRF2, RAP1, TIN2, TPP1, and TRF1 complex (lane 6). Similar results were also obtained for TRF1, because the remaining five proteins were able to complex without TRF1 (lane 2).
The most striking results were obtained when either TPP1 or TIN2 was omitted from the reaction. The complex completely failed to form when TIN2 was excluded (which added further proof that endogenous proteins were not affecting this assay) (Fig. 1b, lane 4). When TPP1 was absent (Fig. 1b, lane 5), TRF1 no longer coimmunoprecipitated with TRF2, and the levels of TIN2 and POT1 that associated with TRF2 were greatly reduced. The importance of TPP1 and TIN2 in six-protein complex formation was further confirmed by using V5-TRF2 as an alternative anchor (data not shown). We did observe a reduction of POT1 levels in the absence of TPP1. However, this lower level of POT1 could not account for the loss of TRF1 and decrease of TIN2 in the RAP1 complex, because the absence of POT1 did not disrupt high-order complex formation (Fig. 1b, lane 6). The above findings support the notion that TPP1 and TIN2 are critical for six-protein complex assembly and that the bridging between the TRF1 and TRF2 subcomplexes may be impaired in the absence of either TPP1 or TIN2.
Identification of Two Distinct POT1-Containing Complexes.
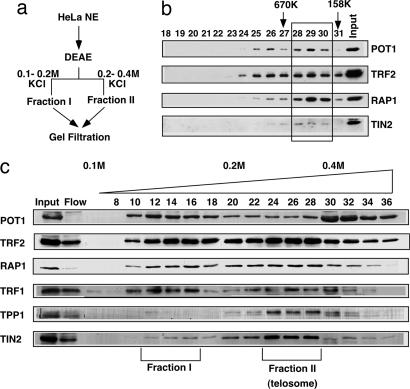
POT1 recruitment to the telomeres depends on TPP1 (9). When TPP1 was removed in the reconstitution experiments, interactions between POT1 and other components of the six-protein complex were indeed severely affected (Fig. 1b, lane 5). However, a small amount of POT1 was able to complex with RAP1, TRF2, and TIN2. To further investigate the physiological relevance of this finding, we analyzed endogenous POT1 protein complexes using HeLa cell nuclear extracts. Consistent with our reconstitution experiments, endogenous POT1 could be found in two major fractions on the DEAE ion-exchange column (Fig. 2a and b). The 0.2–0.4 M KCl fraction II contains POT1, TRF1, TRF2, TIN2, RAP1, and TPP1 (Fig. 2b). The majority of TIN2 and TPP1 were eluted in this fraction. Through gel filtration, we confirmed that fraction II corresponds to the high-molecular-mass mammalian telosome (≈1 MDa) described previously (9). Interestingly, fraction I (which eluted at 0.1–0.2 M KCl) contained barely detectable amounts of TPP1 (Fig. 2b), suggesting that this is a distinct POT1-containing complex. Indeed, upon further fractionation of fraction I on a gel filtration column, POT1 was found to elute in a smaller complex (≈500 kDa) (Fig. 2c). Notably, TRF2, RAP1, and a small amount of TIN2 were also coeluted with POT1 in this fraction, indicating that POT1, RAP1, TRF2, and TIN2 can indeed form a complex. Importantly, this POT1 complex found in fraction I of the DEAE column mirrors the four-protein complex formed in the 293T reconstitution experiments (Fig. 1b, lane 5). These findings indicate the existence of two POT1 complexes in mammalian cells: one that contains all six telomeric proteins and one that does not contain TPP1. Our results so far point to an essential role for TPP1 in the formation and function of higher-molecular-mass telomeric complexes. The possible functional differences of the complex without TPP1 and the six-protein complex are quite intriguing and merit further investigation.
Fig. 2.
Two distinct POT1-containing protein complexes exist in human cells. (a) HeLa cell nuclear extracts (HeLa NE) were fractionated on a DEAE ion-exchange column. The bound proteins were eluted with increasing concentrations of KCl. The two main fractions (I and II) that contained telomeric proteins were then further fractionated on a gel filtration column. (b) Fractions from the DEAE column were collected and resolved by SDS/PAGE. Telomeric proteins were detected through Western blotting by using various antibodies as indicated. “Flow” indicates flow through. (c) Fraction I from the DEAE column was fractionated on a gel filtration column. Telomeric proteins at individual fractions (fractions 18–31) were detected through Western blotting with different antibodies as indicated. The majority of telomeric proteins were coeluted at ≈500 kDa (indicated by the box). Molecular masses of the gel-filtration standards are indicated.
TPP1 Promotes Higher-Order Complex Formation Through TIN2.
Within the six telomeric proteins, TIN2–TRF2 association establishes the TRF1–TIN2–TRF2 link and connects TRF1 to the TRF2 subcomplex (16, 33–35). Interestingly, in the absence of TPP1, TRF2 could no longer coimmunoprecipitate with TRF1, and its ability to coimmunoprecipitate with TIN2 (but not other telomere proteins) decreased by ≈70% (Fig. 1b). These findings imply that TIN2–TRF2 or TIN2–TRF1 interaction alone is insufficient to trigger six-protein complex formation. We therefore speculated that TPP1 might promote TIN2–TRF2 and TRF1–TIN2–TRF2 association, leading to proper high-order telomeric complex assembly.
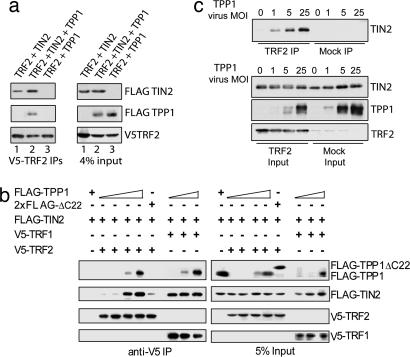
First, we investigated the function of TIN2 and TPP1 by examining the interactions of TRF2, TIN2, and TPP1 in transient transfection experiments. Consistent with our hypothesis, we could not detect direct interaction between TRF2 and TPP1 (Fig. 3a), but TPP1 coimmunoprecipitated together with TIN2 and TRF2, indicating the formation of a TPP1–TIN2–TRF2 complex. With increasing amounts of TPP1 (although the total amounts of TIN2 and TRF2 remained constant), the amount of TIN2 that associated with TRF2 increased as well (Fig. 3b). In contrast, when TRF2 was replaced with TRF1, binding of TRF1 to TIN2 was unaltered by changes in the amount of TPP1, confirming the specific role of TPP1 in promoting TRF2–TIN2 association (Fig. 3b). Furthermore, we demonstrated that TPP1 could promote TIN2–TRF2 binding by using recombinant TRF2, TIN2, and TPP1 proteins from insect cells. As shown in Fig. 3c, enhanced association of His-tagged TRF2 with TIN2 was observed with increased coexpression of TPP1, consistent with the notion that TPP1 directly regulates TIN2–TRF2 interaction. These data also ruled out the possible involvement of endogenous proteins in our experiments using 293T cells. In the absence of TPP1, we found little or no incorporation of TRF1 into the RAP1–TRF2 complex (Fig. 1b, lane 5), suggesting that TPP1 may stabilize the TRF1–TIN2–TRF2 three-protein complex through its regulation of TIN2–TRF2 interaction.
Fig. 3.
TPP1 promotes the interaction between TIN2 and TRF2. (a) TPP1 directly binds TIN2 but not TRF2. Expression vectors encoding V5-TRF2 were cotransfected into 293T cells, with expression vectors encoding FLAG-TIN2 (lane 1), both FLAG-TPP1 and FLAG-TIN2 (lane 2), or FLAG-TPP1 (lane 3). Approximately 3 μg of DNA was transfected for each construct. Whole-cell extracts were immunoprecipitated with anti-V5 antibodies and blotted with anti-FLAG-HRP antibodies. (b) TPP1 promotes interaction between TIN2 and TRF2. 293T cells were transiently transfected with expression vectors for V5-TRF2 (3.3 μg) or V5-TRF1 (4.5 μg) in combination with FLAG-TIN2 (1.5 μg) and increasing amounts of C-terminally tagged FLAG-TPP1 (0, 0.7, 2.4, and 5.6 μg for TRF2, and 0, 0.7, and 2.4 μg for TRF1). The TPP1 deletion mutant TPP1ΔC22 (2xFLAG-tagged at both termini) (5 μg) was also used as a control for TPP1. 2×FLAG-TPP1 and FLAG-TPP1 behaved similarly in these experiments (data not shown). Anti-V5 immunoprecipitates from lysates of these cells were resolved by SDS/PAGE and Western-blotted with anti-FLAG-HRP or anti-V5-HRP antibodies. (c) TPP1 promotes TIN2–TRF2 interaction in insect cells. Sf9 cells were coinfected with His-TRF2 and His-TIN2 baculoviruses along with increasing amounts of His-TPP1 viruses. Three days after infection, His-TRF2 was pulled down from insect cell extracts with anti-TRF2 antibodies. Extracts expressing TIN2 and TPP1 without His-TRF2 were used as controls (Mock IP). In the input whole-cell extracts there is a TRF2 antibody cross-reactive band. His-TIN2 or TPP1 proteins were detected with anti-TIN2 or anti-TPP1 antibodies. MOI, multiplicity of infection.
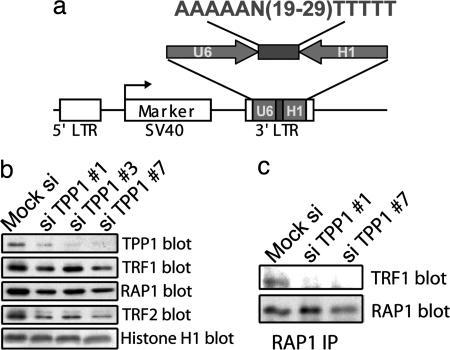
Our data so far support the model that TPP1 acts as an essential component of the six-protein complex by regulating TIN2–TRF2 interaction and the connectivity of TRF1 and TRF2 subcomplexes. Without TPP1, the formation of the TRF1–TIN2–TRF2 complex may be impaired. We previously showed altered telomeric localization of POT1 in TPP1 knockdown cells (9). However, the siRNA oligonucleotides (oligos) used previously only led to significant TPP1 knockdown in ≈50% of the cells, preventing us from performing biochemical analyses of TPP1 function in six-protein complex assembly. To achieve this goal, we generated and optimized a dual-promoter RNAi vector for screening oligos against TPP1 (Fig. 4a) (see Materials and Methods).
Fig. 4.
TPP1 regulates TRF1 association with the TRF2/RAP1 complex in vivo. (a) Design of the pRetro-dual RNAi vector for TPP1 knockdown. TPP1 RNAi oligos were cloned between the head-to-head-positioned U6 and H1 promoters. (b) Reducing endogenous TPP1 levels by RNAi decreases TRF1 recruitment to the TRF2/RAP1 complex. (Left) Nuclear extracts (420 mM KCl extracted) were prepared from HT1080 cells that were transfected with pRetro-dual control and TPP1 RNAi vectors. The extracts were resolved by SDS/PAGE and Western-blotted with the indicated antibodies. The anti-Histone H1 antibody was used as a loading control (note that the siTPP1 #7 lane was slightly underloaded). (Right) Nuclear extracts from HT1080 cells expressing pRetro-dual siTPP1 #1 and siTPP1 #7 were immunoprecipitated with anti-RAP1 antibodies, and Western-blotted with anti-RAP1 and anti-TRF1 antibodies.
We found three RNAi vectors to substantially (70–90%) knockdown TPP1 after their transfection into HT1080 cells (Fig. 4b). The TPP1 RNAi cells grew more slowly than did control cells (data not shown). The phenotypes of these cells with regard to cell cycle and telomere end capping currently require additional investigation. Interestingly, the level of RAP1-assocaited TRF1 was reduced by ≈80% in TPP1 knockdown cells (Fig. 4c), suggesting a key role of TPP1 in six-protein complex assembly and in regulating the connectivity between the TRF1 and TRF2 complexes in vivo.
TPP1 Promotes Six-Protein Complex Formation Through Its Interaction with TIN2.
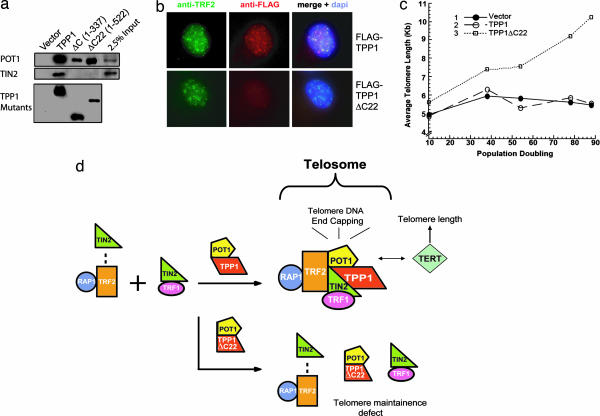
The data thus far indicate that both TPP1 and TIN2 are necessary for six-protein complex formation. Because TPP1 interacts with TIN2 directly (16, 33–35), the TPP1–TIN2 association may be coupled to six-protein complex assembly. Our deletional analyses in yeast indicated that the C-terminal half of TPP1 mediated its interaction with TIN2 (9). In fact, deletion of either the C terminus (TPP1ΔC) or just the last 22 residues of TPP1 (TPP1ΔC22) was sufficient to eliminate TIN2-binding (but not POT1-binding) in yeast (data not shown) (7–9) and in mammalian cells (Fig. 5a). Furthermore, TPP1ΔC22 no longer localized to telomeres (Fig. 5b), consistent with previous observations that demonstrated that TIN2 mediated the telomere targeting of TPP1 (16),
Fig. 5.
The extreme C terminus of TPP1 is required for direct TPP1–TIN2 interaction and enhanced TIN2–TRF2 association. (a) The last 22 aa of TPP1 are required for TPP1–TIN2 interaction in human cells. Whole-cell extracts from HT1080 cells expressing various FLAG-tagged TPP1 and TPP1 mutants (TPP1ΔC and ΔC22) were immunoprecipitated with anti-FLAG M2 antibodies. Endogenous POT1 or TIN2 was detected by Western blotting with anti-POT1 or anti-TIN2 antibodies. (b) TPP1ΔC22 no longer localizes to the telomeres. The telomere localization of FLAG-tagged wild-type or mutant TPP1 was analyzed by indirect immunofluorescence. The cells were doubly permeabilized and costained with anti-FLAG and anti-TRF2 antibodies. (c) TPP1–TIN2 mediated six-protein complex assembly is necessary for telomere length maintenance. Expression of TPP1ΔC22 but not TPP1 in HT1080 cells resulted in telomere elongation. Mean telomere restriction fragment length in control (vector alone) HT1080 cells and those expressing TPP1 or TPP1ΔC22 were plotted against population doublings. (d) A schematic representation of the six-protein complex assembly model. Overexpression of TPP1ΔC22 disrupts telosome assembly and favors the formation of subcomplexes containing TIN2–TRF2–RAP1, POT1–TPP1ΔC22, or TIN2–TRF1.
How does the disruption of TPP1-mediated six-protein complex formation affect telomere maintenance? We reasoned that the inability of TPP1ΔC22 to promote six-protein complex assembly might affect telomere length control. To examine this possibility, we determined the average telomere length in HT1080 cells expressing TPP1 and TPP1ΔC22. As shown in Fig. 5c, expression of TPP1ΔC22 but not TPP1 resulted in elongated telomeres. These data indicate that TPP1–TIN2 interaction regulates the formation of the six-protein complex necessary for telomere length control.
Collectively, our findings suggest possible cooperativity in TRF1–TIN2–TRF2–TPP1 complex formation and imply that the interaction between the TRF1 and TRF2 protein hubs, a key connection in the assembly and chromatin targeting of high-order telomeric complexes, may be stimulated by the heterodimerization of TIN2 and TPP1.
Discussion
Recent advances in elucidating the interaction and function of key telomeric proteins have allowed a better understanding of the intricacies of the mammalian telosome/shelterin that regulates telomere maintenance. At its core is the six-protein complex made up of TIN2, TRF1, TRF2, RAP1, TPP1, and POT1. Because of its ability to bind both TRF1 and TRF2, TIN2 is emerging as a key player in telomere chromatin formation (16, 33–35). Removal of TIN2, as done in our reconstitution experiments (Fig. 1), leads to the elimination of the bridge between the TRF1 and TRF2 subcomplexes. Surprisingly, TIN2–TRF2 interaction alone appeared to be insufficient to initiate TRF1 recruitment and six-protein complex formation. Given that TIN2 can bind either TRF1 or TRF2, TIN2–TRF1 and TIN2—TRF2 may in fact exist in separate subcomplexes. Our study has revealed that TPP1 also is a critical regulator of telomeric protein–protein interactions. By virtue of its interaction with TIN2, TPP1 stabilizes the TIN2–TRF2 and TRF1–TIN2–TRF2 interactions, stimulates the connection between the TRF1 and TRF2 subcomplexes, and promotes telosome assembly.
In budding yeast, telomeric proteins are packed into nuclease-resistant telosome structures (38). Whether the mammalian telomeric proteins and telomere DNA complex also can form into nuclease-resistant structures remains to be elucidated. The exact identities of the subunits of the mammalian telosome probably differ from the yeast telosome, because homologues of several mammalian telomeric proteins have yet to be found in budding yeast. Mammalian telosomes may very well act as key molecular machineries that regulate mammalian telomeres in a coordinated fashion. Our experiments here have highlighted the importance of TPP1–TIN2 interaction in six-protein complex function and represent a noteworthy step toward the understanding of this dynamic and complicated process.
How does TPP1–TIN2 interaction regulate TIN2–TRF2 binding? Our data are consistent with a direct role for TPP1 in regulating telosome stability. We speculate that the answer may at least partly lie in the structure of TIN2. The N-terminal half of TIN2 is predicted to have high helical contents and conserved throughout evolution (data not shown). This region interacts with TPP1 (D.L. and Z.S., unpublished data) and TRF2 (16, 33). C-terminal to this TIN2 region is the TRF1-binding domain (18). It is possible that TPP1 binding of TIN2 may stabilize or alter the conformation of TIN2, which in turn enhances TIN2–TRF2 interaction and enables TIN2 to simultaneously interact with both TRF2 and TRF1 (Fig. 5d).
What are the consequences of a stabilized link between TRF1 and TRF2? We think that the structural unit that contains TPP1, TIN2, TRF1, and TRF2 may have increased affinity for telomeric DNA, as observed for TRF1 (39). Several lines of evidence support this notion. First, multivalent interactions are known to enhance affinity, and in this case both TRF1 and TRF2 can bind telomeres. Second, recent studies suggest that the interaction of TRF1 and TRF2 with the telomere is in fact extremely dynamic. For example, Mattern et al. (40) demonstrated via fluorescence recovery after photobleaching that one fraction of GFP–TRF2 is bound more tightly to the telomeres, whereas GFP–TRF1 and the majority of GFP–TRF2 exhibited fast-off kinetics (<10 s) in live cells (40), a surprising result given the prevailing notion that TRF1 and TRF2 tightly bind to telomeres. This tightly bound TRF2 fraction may represent the six-protein complex, which may be stabilized by TPP1–TIN2 interaction. Finally, in TRF1-knockout mouse ES cells, TRF2 telomere localization was also impaired (37). This observation suggests that TRF2 requires TRF1 for stable telomere association and supports the model that the six-protein complex is one of the functional complexes that regulate telomere localization of multiple telomeric proteins.
What is the functional significance of TPP1–TIN2-mediated six-protein complex assembly? We would like to propose the following model (Fig. 5d). TRF2 and RAP1 form a stable subcomplex that weakly associates with TIN2. TRF1 and TIN2 are in a different stable subcomplex that does not interact with TRF2. Through direct interaction with TIN2, TPP1 promotes TIN2–TRF2 binding and stimulates TRF1–TIN2–TRF2 connectivity. This network of interactions ensures proper targeting of the business end of the six-protein complex, POT1, and allows the regulation of telomere length and end-capping. TPP1 may have additional function, such as modulating the stability of TRF2. It is clear, however, that the ability of TPP1 to bind TIN2 and promote TRF1–TIN2–TRF2 complex formation is an essential function of TPP1. The TPP1–TIN2 interaction has a two-pronged effect: telomeric targeting of POT1 and signaling for high-order telomeric complex assembly. This model predicts that perturbations in the six-protein complex and its components would result in the disruption of telomere maintenance. Indeed, knockdown or inactivation of any of the six telomeric proteins, including TPP1 (15, 30, 34, 41), led to misregulated telomere length and/or telomere end protection. The six-protein complex thus forms the basic platform on which layers of telomere signaling networks can be assembled into the telomere interactome (32) for the proper protection and maintenance of mammalian telomeres.
Materials and Methods
Expression Constructs and Antibodies.
For generating stable cell lines, singly or doubly FLAG-tagged full-length human TPP1 and its deletion mutants TPP1ΔC22 (residues 1–522) and TPP1ΔC (residues 1–337) were cloned into a pBabe-based retroviral vector. For expression in 293T cells, full-length POT1, TPP1, RAP1, TIN2, TRF1, or TRF2 were either cloned into the pCL vector (FLAG-tagged) or TOPO-cloned into pcDNA3.1 or pcDNA3.1-C-GFP (Invitrogen, Carlsbad, CA) to generate V5- or GFP-tagged fusion proteins. For insect cell expression, His-TPP1, His-TIN2, or His-TRF2 baculoviruses were produced by using the Blue-Bac baculovirus kit (Invitrogen).
The following antibodies were used: anti-V5 and anti-GFP (ChemiCON, Temecula, CA); anti-histone H1 (a generous gift from Estela Medrano, Baylor College of Medicine, Houston, TX); anti-FLAG M2 (Sigma, Lenexa, KS); anti-TRF2 (CalBiochem, San Diego, CA); anti-hRAP1 (30); anti-TIN2 and anti-TPP1 (9); goat anti-TRF1 (Bethyl Laboratories, Montgomery, TX); and anti-TRF1 (17) and POT1 (12) (gifts from Titia de Lange, The Rockefeller University, New York, NY). Anti-FLAG M2 antibody agarose beads (Sigma) and HRP-conjugated anti-V5 antibody (Bethyl Laboratories) also was used for immunoprecipitation and Western blotting, respectively.
Immunoprecipitation, Western Blot, and Immunofluorescence.
For six-protein complex reconstitution and interaction studies, 293T or HT1080 cells were cotransfected with plasmids encoding various telomeric proteins in different combinations. At 48 h after transfection, the cells were harvested and extracted with a high-salt-concentration buffer (20 mM Hepes, pH 7.9/420 mM KCl/0.1 mM EDTA/5 mM MgCl2/0.2%Nonidet P-40/1 mM DTT/0.2 mM PMSF/25% glycerol) (12). The extracts were then dialyzed in a low-concentration salt buffer (20 mM Hepes, pH 7.9/100 mM KCl/0.1 mM EDTA/1 mM DTT/0.5 mM PMSF/25% glycerol) (12). Subsequent immunoprecipitation and Western blotting with appropriate antibodies were carried out as previously described (9).
Indirect immunofluorescence was performed as described previously (30). FLAG-tagged proteins and endogenous TRF2 (an indicator of telomere localization) were detected with anti-FLAG and anti-TRF2 antibodies followed by secondary antibodies. Fluorescence microscopy was performed on a Nikon (Melville, NY) TE200 microscope equipped with a Coolsnap-fx charge-coupled device camera.
Fractionation of Telomere-Associated Complexes.
Chromatographic experiments were performed as previously described (9). Briefly, HeLa cell nuclear extracts were fractionated on an AKTA DEAE-Sepharose ion-exchange column (Amersham Pharmacia, Pittsburgh, PA) equilibrated with buffer A (50 mM Hepes, pH 7.5/0.2 mM EDTA/0.5 mM DTT/0.2 mM PMSF/5% glycerol) containing 100 mM KCl. Bound proteins were step-eluted with increasing concentrations of KCl (200–1,000 mM) in buffer A. The fractions from this column were then loaded on a Superose 6 HR 10/30 gel filtration column. The resulting fractions from the DEAE or gel filtration column were resolved by SDS/PAGE and probed with various antibodies as appropriate.
Binding Assays Using Insect Cell Expression Systems.
Sf9 cells (≈5 × 106) were coinfected with His-TRF2, His-TIN2, and increasing amounts of His-TPP1 baculoviruses. At 3 days after infection, the cells were collected, lysed in 1× NETN buffer (20 mM Tris, pH 8/1 mM EDTA/90 mM NaCl/0.5% NP-40), and incubated with anti-TRF2 antibody (ChemiCON) (3.5 μg) followed by protein G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at 4°C. Cells not infected with His-TRF2 were used controls. The eluted proteins were resolved by SDS/PAGE and blotted with anti-TIN2 antibodies.
RNAi Knockdown of TPP1.
To analyze the effects of TPP1 knockdown, we designed a vector system (pRetro-dual) based on the dual-promoter RNAi approach (42). Briefly, TPP1-specific RNAi oligos were cloned between the head-to-head positioned U6 and H1 promoters. This dual promoter RNAi cassette is then cloned into the 3′ ΔLTR (with the enhancer deleted) of the retroviral vector. After retroviral infection, two copies of the RNAi cassette will be integrated into the target genome, resulting in more efficient RNAi. The vector also contains a puromycin-resistance marker for the establishment of stable cells. These RNAi vectors also are suitable for transient transfection experiments.
The pRetro-dual vectors require shorter and fewer DNA oligo sequences. To identify the optimal knockdown sequences for TPP1, several vectors that cover different regions of the target gene were constructed. Of these constructs, we found three vectors (oligo 1, 5′-GACGTCAAAAACCAAGACTTAGATGTTCAGAATTTTTAGATCT-3′; oligo 3, 5′-GACGTCAAAAACTCTGAGAATGACCAGCTAATTTTTTAGATCT-3′, and oligo 7, 5′-AAAAAGTGGTACCAGCATCAGCCTTTTTTTAGATCT-3′) that significantly reduced TPP1 expression. HT1080 cells were transfected with the various siTPP1 constructs and assayed in immunoprecipitation experiments 72 h after transfection.
Telomere Restriction Fragment Assay.
HT1080 cells were infected with retroviruses encoding various TPP1 proteins. Puromycin was added to the culture media for 3 days. The surviving cells were subsequently maintained as a pool without clonal selection (at which point designated as P0), passaged, and collected at various time points. The telomere restriction fragment assay was performed as previously described (17). The data were then analyzed by using a PhosphorImager (Amersham Pharmacia) and the Telorun analysis tool (43).
Acknowledgments
We thank Dr. Doug Chan and Hunter Richards, Andrew Laegeler, and Yiying Xie for technical help. This work was supported by the National Institutes of Health (Z.S.), the Department of Defense, the American Cancer Society, and the Welch Foundation.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Blackburn E. H. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Wright W. E., Shay J. W. Curr. Opin. Genet. Dev. 2001;11:98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 4.Kim S. H., Kaminker P., Campisi J. Oncogene. 2002;21:503–511. doi: 10.1038/sj.onc.1205077. [DOI] [PubMed] [Google Scholar]
- 5.Maser R. S., DePinho R. A. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 6.Wong J. M., Collins K. Lancet. 2003;362:983–988. doi: 10.1016/S0140-6736(03)14369-3. [DOI] [PubMed] [Google Scholar]
- 7.Baumann P., Cech T. R. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 8.Loayza D., Parsons H., Donigian J., Hoke K., de Lange T. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 9.Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. Nat. Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 10.Lei M., Baumann P., Cech T. R. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 11.Lei M., Podell E. R., Cech T. R. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 12.Loayza D., de Lange T. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 13.Colgin L. M., Baran K., Baumann P., Cech T. R., Reddel R. R. Curr. Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 14.Kelleher C., Kurth I., Lingner J. Mol. Cell. Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houghtaling B. R., Cuttonaro L., Chang W., Smith S. Curr. Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 17.van Steensel B., de Lange T. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 18.Kim S. H., Kaminker P., Campisi J. Nat. Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith S., de Lange T. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 20.Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M. R., Schnapp G., de Lange T. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith S., Giriat I., Schmitt A., de Lange T. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X. Z., Lu K. P. Cell. 2001;107:347–359. doi: 10.1016/s0092-8674(01)00538-4. [DOI] [PubMed] [Google Scholar]
- 23.Cong Y. S., Wright W. E., Shay J. W. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillard-Wetherell K., Machwe A., Langland G. T., Combs K. A., Behbehani G. K., Schonberg S. A., German J., Turchi J. J., Orren D. K., Groden J. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 25.Li B., Oestreich S., de Lange T. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X. D., Kuster B., Mann M., Petrini J. H., de Lange T. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 27.Stavropoulos D. J., Bradshaw P. S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M. S. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 28.Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., Bohr V. A. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., de Lange T. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor M. S., Safari A., Liu D., Qin J., Songyang Z. J. Biol. Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 31.Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M. M., Bohr V. A. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Songyang Z., Liu D. Crit. Rev. Eukaryotic Gene Expression. 2006;16:103–118. doi: 10.1615/critreveukargeneexpr.v16.i2.10. [DOI] [PubMed] [Google Scholar]
- 33.Kim S. H., Beausejour C., Davalos A. R., Kaminker P., Heo S. J., Campisi J. J. Biol. Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 34.Ye J. Z., Donigian J. R., Van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., De Lange T. J. Biol. Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 35.Liu D., O'Connor M. S., Qin J., Songyang Z. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 36.de Lange T. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 37.Iwano T., Tachibana M., Reth M., Shinkai Y. J. Biol. Chem. 2003;14:14. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 38.Wright J. H., Gottschling D. E., Zakian V. A. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 39.Kim S. H., Han S., You Y. H., Chen D. J., Campisi J. EMBO Rep. 2003;4:685–691. doi: 10.1038/sj.embor.embor872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattern K. A., Swiggers S. J., Nigg A. L., Lowenberg B., Houtsmuller A. B., Zijlmans J. M. Mol. Cell. Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J. Z., de Lange T. Nat. Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 42.Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P. G. Proc. Natl. Acad. Sci. USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouellette M. M., Liao M., Herbert B. S., Johnson M., Holt S. E., Liss H. S., Shay J. W., Wright W. E. J. Biol. Chem. 2000;275:10072–10076. doi: 10.1074/jbc.275.14.10072. [DOI] [PubMed] [Google Scholar]
- 44.Bianchi A., Smith S., Chong L., Elias P., de Lange T. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broccoli D., Smogorzewska A., Chong L., de Lange T. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 46.Yang Q., Zheng Y. L., Harris C. C. Mol. Cell. Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]