Abstract
Fibrillin-1 is a 330-kDa multidomain extracellular matrix protein that polymerizes to form 57-nm periodic microfibrils, which are essential for all tissue elasticity. Fibrillin-1 is a member of the calcium-binding EGF repeat family and has served as a prototype for structural analyses. Nevertheless, both the detailed structure of fibrillin-1 and its organization within microfibrils are poorly understood because of the complexity of the molecule and the resistance of EGF arrays to crystallization. Here, we have used small-angle x-ray scattering and light scattering to analyze the solution structure of human fibrillin-1 and to produce ab initio structures of overlapping fragments covering 90% of the molecule. Rather than exhibiting a uniform rod shape as current models predict, the scattering data revealed a nonlinear conformation of calcium-binding EGF arrays in solution. This finding has major implications for the structures of the many other EGF-containing extracellular matrix and membrane proteins. The scattering data also highlighted a very compact, globular region of the fibrillin-1 molecule, which contains the integrin and heparan sulfate-binding sites. This finding was confirmed by calculating a 3D reconstruction of this region using electron microscopy and single-particle image analysis. Together, these data have enabled the generation of an improved model for microfibril organization and a previously undescribed mechanism for microfibril extensibility.
Keywords: elastic fibers, fibrillin microfibrils, solution x-ray scattering
Human fibrillin-1 is a 330-kDa modular glycoprotein, which assembles to form microfibrils in the extracellular matrix (1). Fibrillin microfibrils have a complex ultrastructure resembling “beads-on-a-string,” with an average periodicity of 57 nm and a diameter of 18 nm as revealed by electron microscopy (EM) (2, 3). They have a widespread distribution in nonelastin and elastin-containing tissues, such as the ciliary zonules, blood vessels, lung, and skin, and they act as a lattice for elastin deposition during elastic fiber formation. Mutations in the fibrillin-1 (FBN1) gene cause the connective-tissue disease Marfan syndrome, which is associated with severe cardiovascular, ocular, and skeletal defects (4).
The modular architecture of fibrillin-1 is dominated by epidermal growth factor-like (EGF) motifs, which are one of the most common motifs found in extracellular matrix macromolecules. More than 600 EGF motifs have been identified in >30 matrix and membrane proteins with wide-ranging functions (5). Forty-three of the 47 EGF domains in fibrillin-1 have a Ca2+ binding motif (cbEGF) and are arranged in contiguous arrays, separated in all but one case by an eight-cysteine-containing TB domain (Fig. 1) (6). Multiple tandem repeats of fibrillin-1 cbEGFs have been predicted to adopt rod-shaped structures in the presence of Ca2+ (7); because of this possibility, attention has been focused on the TB domain as a potential source of flexibility through its pairwise domain interactions. A recent study correlated Ca2+-binding affinity with the stability of the TB–cbEGF interface, providing a measure of flexibility (8). These data predicted that TB6–cbEGF would be the most flexible TB–cbEGF linker in the fibrillin-1 molecule.
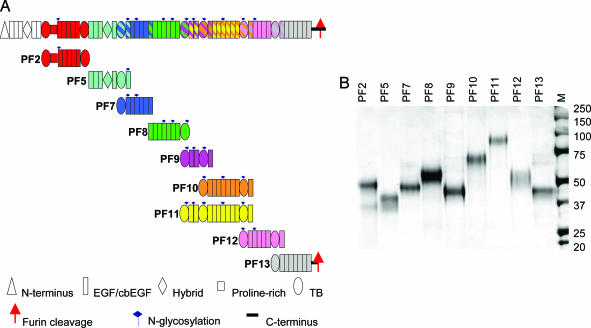
Fig. 1.
Schematic diagram of the domain structure of fibrillin-1 and the protein fragments used in this study. (A) The domain structure of fibrillin-1 is shown colored by protein fragment. All expressed fibrillin-1 fragments are shown below their position in the fibrillin-1 molecule. A striped representation on the fibrillin-1 molecule indicates overlapping fragments. A key indicating the different domains, N-linked glycosylation sites, and C-terminal furin cleavage site is shown. The protein fragments are shown in these corresponding colors throughout the figures. (B) SDS/PAGE showing fibrillin-1 fragments, PF2, -5, -7, -8, -9, -10, -11, -12, and -13, before HPLC. M shows standard molecular weight markers.
Microfibril elasticity is predicted to reside within reversible extension of the bead–interbead repeat (9), in turn proposed to arise by flexibility at the proline-rich region and some calcium-dependent cbEGF interfaces (8, 10). To establish the molecular basis of microfibril organization and mechanics, models of fibrillin-1 molecular alignment have been proposed, based on microfibril structural analysis and antibody (Ab) epitope mapping (10–12). One model proposed a parallel head-to-tail alignment, with molecules “compacted” in 57-nm periodic microfibrils but extended at higher periodicities (10, 11). A second model proposed a staggered alignment, with overlapping extended molecules (12).
By using small-angle x-ray scattering (SAXS) and multiangle laser light scattering (MALLS) data collected for nine recombinant fibrillin-1 fragments, we have defined the previously undescribed shape and topography of large contiguous arrays of fibrillin-1 domains in physiological solution, bridging the information gap between the structural details of individual fibrillin-1 domains and microfibril organization. We also used single-particle image analysis to confirm our findings on the most compact region of the molecule, which contains integrin and heparan sulfate-binding sites (13, 14). These data provide exciting insights into the conformation of contiguous repeats of cbEGF domains in solution and the arrangement of fibrillin-1 in microfibrils.
Results
Solution Structure of Fibrillin-1 from Domain TB1 to the C Terminus.
The protein fragments used to produce a comprehensive study of the molecular shape of fibrillin-1 are shown in Fig. 1. Overall nine fragments are described, ranging in size from 5 to 13 contiguous domains. Constructs were designed aimed at studying specific areas that had previously been predicted to be folded or straight (10). The protein fragments were characterized by SDS/PAGE, gel filtration chromatography, and MALLS (see Figs. 6 and 7, which are published as supporting information on the PNAS web site) to confirm the fragments were the right size and monodisperse. The protein fragments also were found to have native conformations in binding studies with microfibril-associated proteins (15, 16), integrins (13), and heparan sulfate (14). Immediately before data collection, the protein fragments were chromatographed to ensure the sample was monomeric. Multiple attempts were made to collect data on the N-terminal region of fibrillin-1, but it was not possible because of sample aggregation at the concentrations required for data collection.
Ab Initio Simulated Structures.
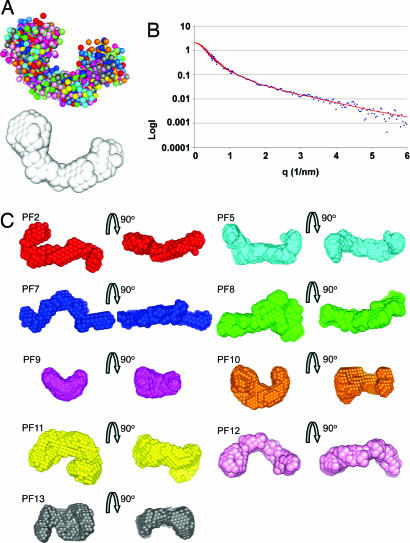
X-ray scattering measurements were made on protein solutions in the presence of Ca2+. The data quality was assessed by using Guinier plots, to check for aggregation in the sample (see Fig. 8, which is published as supporting information on the PNAS web site), and the radius of gyration (Rg) and maximum particle dimension (Dmax) were determined. The hydrodynamic radius of the monomeric protein (determined by MALLS) was compared with the Rg to ensure there was no aggregation in the sample (Table 1). To determine the 3D structure of fibrillin in solution, the ab initio programs GASBOR and DAMMIN (17, 18) were used. The modeling allowed us to fit the experimental data with discrepancy factors χ of between 0.6 and 1.2. A typical DAMMIN fit is displayed in Fig. 2B. For each program, at least 20 separate simulations were completed to determine the “uniqueness” of the solution (Fig. 2A). The mean normalized spatial discrepancy between the simulated structures was measured to establish the reliability of the solution (19). For each fragment, an averaged structure was created from the multiple runs to determine common structural features (19). These results are shown in Fig. 2C, where each structure is represented as an array of beads.
Table 1.
Comparison of results for each protein fragment
| Sample | MW, kDa | MALLS Rh, nm | Guinier Rg, nm | GNOM Rg, nm | Dmax, nm | Rod Dmax, nm | Rod, % | Rod Rg, nm | Rod χ |
|---|---|---|---|---|---|---|---|---|---|
| PF2 | 42.7 | 4.3 | 4.3 | 4.7 | 15.0 | N/D | N/D | N/D | N/D |
| PF5 | 37.8 | 3.8 | 3.6 | 3.9 | 11.5 | 14.6 | 79 | 4.1 | 2.65 |
| PF7 | 36.4 | 3.3 | 4.0 | 4.3 | 13.0 | 17.8 | 73 | 5.6 | 5.16 |
| PF8 | 39.1 | 3.8 | 4.2 | 4.5 | 14.0 | 20.5 | 68 | 6.3 | 6.77 |
| PF9 | 29.7 | 3.6 | 3.3 | 3.2 | 9.0 | 10.7 | 84 | 3.4 | 7.71 |
| PF10 | 52.6 | N/D | 3.6 | 3.8 | 11.0 | 23.4 | 47 | 7.7 | 9.42 |
| PF11 | 83.2 | 5.8 | 4.8 | 4.4 | 13.5 | 31.3 | 43 | 9.5 | 8.60 |
| PF12 | 42.7 | 3.9 | 3.8 | 4.1 | 12.5 | 17.2 | 73 | 5.6 | 5.52 |
| PF13 | 43.0 | N/D | 3.9 | 4.3 | 12.2 | 20.5 | 60 | 6.3 | 7.77 |
The following data are provided for each protein fragment: the predicted molecular mass (MW; calculated from the primary sequence); the hydrodynamic radius (Rh) from MALLS; the Rg from the Guinier approximation and GNOM; the Dmax; the measured maximum dimension of a rod-shaped model, i.e., a linear array of domains (Rod Dmax); the experimental Dmax/Rod Dmax expressed as a percentage (% Rod); the theoretical Rg of the rod model (Rod Rg); and the discrepancy factor χ between the rod model and experimental data (Rod χ). N/D, not determined.
Fig. 2.
Structures generated ab initio from the SAXS data. (A) Shapes were simulated ab initio by using the programs DAMMIN and GASBOR. An example of 20 independent DAMMIN simulations for PF12 are shown superimposed to highlight the uniqueness of the solution. These were used to calculate an average “most probable” shape, shown in white. (B) The experimental data for PF12 are plotted in blue as a function of q and compared with a typical theoretical fit obtained with DAMMIN (red). (C) For each protein fragment, the results of 20 independent simulations were averaged and filtered to give the most probable shape. The most probable shape is represented as arrays of beads and shown in two different orientations for each fragment.
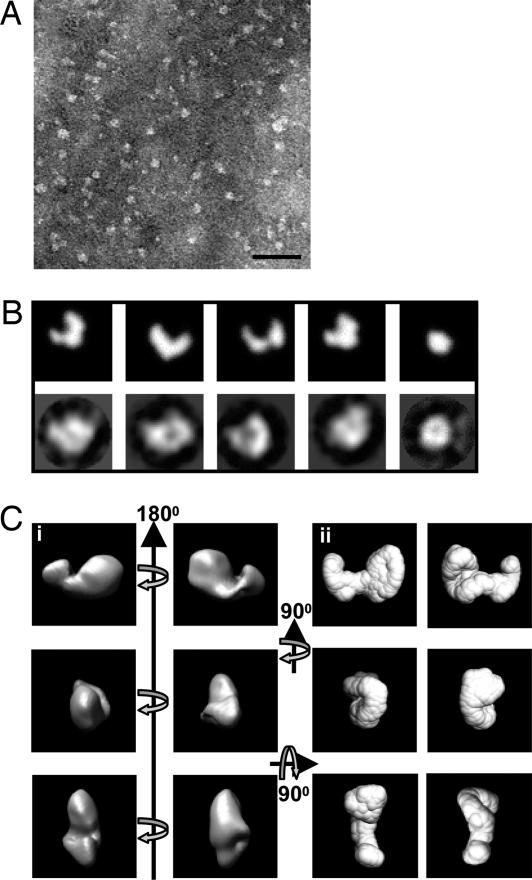
Single-Particle Image Analysis of PF11.
Negatively stained images were recorded of PF11 by using low-dose TEM (Fig. 3A). A 3D reconstruction of PF11 was calculated based on 1,168 particles; this reconstruction was performed without using a reference structure for alignment. The class averages generated from reference-free alignment were compared with the 2D projected images generated from the SAXS structure (Fig. 3B), and they compared very favorably. The 3D reconstruction was calculated by using angular reconstitution, and the structure was very similar to the solution structure generated with SAXS (Fig. 3C). The maximum dimension of the single-particle structure was ≈12 nm, which correlates well with the Dmax value of 13.5 nm from SAXS.
Fig. 3.
Comparison of the structures of PF11 determined by single-particle TEM and solution SAXS. (A) Negatively stained TEM image of fibrillin-1 fragment PF11 recorded on a Tecnai12 Twin at 120 keV. (Scale bar: 50 nm.) (B) (Upper) 2D projected images from the ab initio SAXS structure of PF11 were calculated by using IMAGIC. (Lower) These were compared with representative class averages from single-particle image processing of PF11. There are striking similarities in the size and shape of PF11 when compared by the two techniques. The class averages were generated without a reference and are completely independent of the SAXS structure. (Ci) The 3D reconstruction of PF11 was calculated by using angular reconstitution and is shown here in orthogonal views as a volume-rendered representation. (Cii) For comparison, the ab initio SAXS structure was drawn as a surface representation and shown in the same orientations. Again, there are very clear similarities between the single-particle EM and SAXS structures of PF11. For B and C, the boxes are 14 ×14-nm square.
Domain Path Modeling.
From the scattering data obtained, it is possible to test how appropriate predicted topologies of contiguous domain structures are. For example, the suitability of linear or more compact supradomain architectures can be examined. Linear models of contiguous domains were constructed for all protein fragments, with the exception of PF2, which was omitted because of the lack of structural information on the proline-rich region. The lengths of the linear models were measured in Quanta2000 and compared with the experimental Dmax value obtained for each protein fragment (Table 1). The predicted Rg of each linear model was calculated by using CRYSOL (20). The predicted scattering profile of each linear model was compared directly with the experimental scattering data again by using CRYSOL. The quality of the fit could be assessed by the discrepancy factor χ where a good fit would have a χ = 1.0 and a bad fit of >4.0. In general, a completely linear structure gave a poor fit to the experimental data, as indicated by the high χ values (Table 1). However, the linear model for PF5 had a reasonable fit to the data (χ = 2.65), indicating that this region may have a more linear structure.
Rigid-Body Domain Modeling.
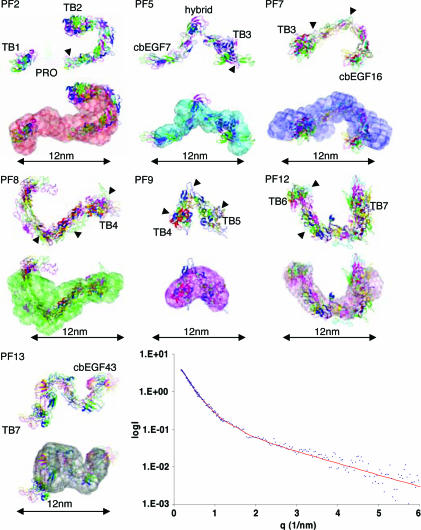
Before rigid-body modeling, homology models of all domains in fibrillin-1, where high-resolution structural information was not already available, were created by using SwissModel (21). This step was possible because of the high degree of sequence identity between the cbEGF domains in fibrillin-1. It was not possible to create a homology model for the proline-rich region, because there are no homologous structures in the Protein Data Bank. BUNCH and SASREF (22) were used for rigid-body modeling to the experimental scattering data, which was performed by using two- and three-domain subunits, with the conformation defined by structural studies (6, 7, 12, 23), or by using individual domains. The rigid-body modeling was restrained by defining linkers between domains to represent the connectivity of each protein fragment. BUNCH also fits missing loops between domains and was used to simulate the residues in the proline-rich region. For all fragments, multiple runs of BUNCH and SASREF were performed to check the reproducibility of the solution. The rigid-body modeling is completely independent of the ab initio simulation; therefore, when there is a good comparison between them, a high degree of confidence in the solution can be achieved. The structures generated are described in the following section (see Fig. 4 and Table 2; see also Table 3, which is published as supporting information on the PNAS web site).
Fig. 4.
Rigid-body modeling to experimental scattering data. Rigid-body modeling was performed with the programs SASREF and BUNCH to the experimental SAXS data. Multiple repetitions of modeling were performed, and resulting structures were superimposed to check for reproducibility. Structures are shown from repeated simulations (top of each image), and these are also superimposed onto the ab initio structure (bottom of each image) for fragments PF2, -5, -7, -8, -9, -12, and -13. Arrowheads indicate N-glycosylation sites. The experimental data for PF8 is plotted in blue as a function of q and compared with the theoretical fit for a modeled structure obtained with SASREF, shown in red.
Table 2.
Predicted Rg, Dmax, and discrepancy factor χ for the best-fit structure generated by rigid-body modeling
| Data set | Rg, nm | Dmax, nm | Discrepancy factor χ |
|---|---|---|---|
| PF2 | 4.3 | 14.4 | 2.56 |
| PF5 | 4.0 | 11.2 | 1.42 |
| PF7 | 3.6 | 11.0 | 2.13 |
| PF8 | 4.0 | 12.2 | 2.04 |
| PF9 | 2.5 | 7.6 | 2.89 |
| PF12 | 3.8 | 11.6 | 0.80 |
| PF13 | 4.0 | 11.2 | 2.66 |
PF2 comprises domains from TB1 to TB2 and contains the proline-rich region and one N-linked sugar. In all structures, domains cbEGF5–6 formed a curved shape, which compared well with the molecular envelope which had a hooked arrangement.
PF5 is from cbEGF7 to cbEGF11 and incorporates a hybrid domain; the structure of the hybrid domain is not known, but it contains features of both a TB domain and cbEGF motif. A partial model was created of the hybrid domain containing 49 of the 64 aa, but it was not possible to model the C-terminal region of this domain. BUNCH was used to model the missing 15 aa of the structure, and SASREF also was used, which ignored the missing residues in the simulation. In all simulations, the PF5 structure was V-shaped, with the bend occurring at the hybrid domain.
PF7 starts at TB3 and incorporates 6 cbEGF domains from the central 12 cbEGF region. In all simulations, the PF7 structure showed a kink in the stretch of cbEGF domains between cbEGF13 and cbEGF14. There are two N-linked sugars on domains cbEGF11 and cbEGF13; the latter of these may have some contribution to the kink.
PF8 overlaps with PF7 by one domain and continues to TB4. Rigid-body modeling showed that PF8 had a U-shaped structure with a curvature in the array of cbEGF domains between cbEGF19 and -20, which may be influenced by the N-linked glycosylation on domains cbEGF19 and -21.
The PF9 structure is a compact, almost globular structure with a bend between the cbEGF pair. There is an N-linked sugar attached to the second of these cbEGF domains, which may play a role in inducing this conformation.
Rigid-body modeling for PF10 and -11 yielded a number of different solutions rather than a single structure. These are both long fragments and therefore may have too many parameters to analyze in this way (data not shown).
PF12 starts at TB6 and encompasses five contiguous cbEGF domains and TB7–cbEGF37. The structure of PF12 was U-shaped with a gradual curvature in the array of five cbEGF domains.
PF13 starts at TB7 and continues to the C terminus; however, the majority of PF13 is in the furin-processed form (15). The structures generated for PF13 had a curved, elongated S-shape.
Discussion
The functionality of fibrillin-1 within a tissue relies on a hierarchical assembly that spans from the molecule to the suprafibrillar architecture. The structure and organization of fibrillin-1 is only partly understood because of its complexity, and many details are still undefined, including the molecular structure and the organization within microfibrils. To address these fundamental questions, we have used solution SAXS, single-particle analysis, and MALLS to collect structural data on overlapping human fibrillin-1 fragments covering 2,400 aa (90% of the furin-processed molecule). Solution x-ray scattering is proving to be a powerful technique for obtaining information on shape and domain organization of molecules in physiological conditions (24, 25). Our data are a unique bridge between molecular and fibrillar levels of fibrillin-1 architecture. They provide previously undescribed evidence for the nonlinear structure of contiguous arrays of cbEGF domains, the surface-exposed arrangement of the hybrid domain, and the highly compact nature of the integrin-binding region of fibrillin-1.
NMR structures of pairs of cbEGF domains led to the suggestion that multiple cbEGF domains would have a rod-like structure (7). However, our data show clearly that long, contiguous arrays of cbEGF domains, in physiological solution, do not adopt a completely linear conformation in the presence of Ca2+ (Fig. 4). Rigid-body modeling was performed either by using domain linkages defined by structural studies or by allowing individual domains to have conformational variability relative to each other, while maintaining the correct connectivity and linker length. In all cases, the structures with the best fit to the scattering data corresponded to the structurally determined linkages for domain pairs (7, 23). However, the linkages between domain pairs did not maintain a linear conformation for the whole array, giving rise to more compact shapes. Of all of the protein fragments analyzed, PF5 had the most linear morphology, with an open V-shaped structure. The bend in the structure always occurred at the hybrid domain, which implies that this domain is accessible on the surface of fibrillin-1. The other hybrid domain, at the N terminus of fibrillin-1, has been implicated in lateral assembly via a free cysteine (15, 26), which suggests that the N-terminal hybrid domain is also surface exposed. Together, these data imply an interactive role on the surface of fibrillin-1 for both hybrid domains.
It is possible that recombinant protein fragments may not adopt the same conformation as the full-length fibrillin-1 molecule, because the protein fragments have untethered N and C termini, which may allow the fragments to adopt more compact arrangements. To investigate this possibility, data were collected on overlapping fragments PF9, -10, and -11. Theoretically, the combined shapes of PF9 and -10 will correspond to the shape of PF11. This theory was indeed the case. PF9 had a very compact shape, whereas PF10 was more elongated. PF11 contained the characteristics of both PF9 and -10, which indicates that these regions of fibrillin-1 do indeed retain their shape when produced as smaller fragments. PF11 was the most compact region of the molecule, which had a maximum dimension of only 43% of a completely rod-shaped particle (Table 1). This result was confirmed by using TEM and single-particle analysis, which revealed a 3D structure almost identical to the ab initio structure generated from the SAXS data. Significantly, this most compact region of the fibrillin-1 molecule is responsible for interacting with cells through integrins (13, 27) and heparan sulfate (14). From the structure of PF9, we can see that TB4 (integrin-binding) and TB5 (heparan sulfate-binding) are separated by a cbEGF pair in which there is a 60° kink; this configuration may enable binding of different substrates to the TB domains without steric hindrance.
The proline-rich region has been suggested to have a hinge-like structure, allowing the N terminus of the fibrillin-1 molecule to be folded back (10). The data presented here provide previously undescribed, direct structural information on the proline-rich region. The ab initio structure revealed a hook-shaped particle, and it was tempting to place the proline-rich region in the hook, allowing the TB1 domain to fold back over subsequent domains, consistent with the N terminus of fibrillin-1 folding back. However, the rigid-body modeling placed domains cbEGF5 and -6 in this hooked region. The data still indicate that this region has a turn within it, allowing the N terminus to be folded back.
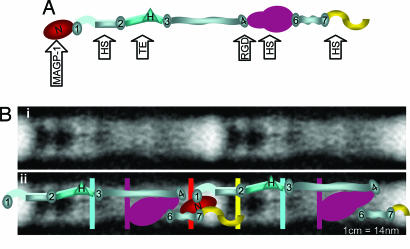
A putative structure for fibrillin-1 was generated by aligning the SAXS structures from TB1 to the C terminus (Fig. 5A). This structure had a length of 90 nm, which does not support a linear conformation of the fibrillin-1 molecule in solution. Even accounting for the absent N-terminal domains, our data imply that one fibrillin-1 molecule could not span more than one complete 57-nm microfibril repeat. Here, we propose a previously undescribed model for microfibril organization and extensibility, which supercedes the intramolecular folding model (10). We suggest that the N and C termini are overlapping and reside at the bead, and the fibrillin-1 molecule has a compact arrangement, particularly from domains TB4 to -6 (Fig. 5). This model is consistent with the previously mapped locations of monoclonal Ab epitopes (10, 11). The very globular structure of PF11 would allow this region to come in close proximity to downstream sequences and would account for the location of the polyclonal Ab PF2 epitope (10, 28). The locations of the globular TB and hybrid domains in the model correlate well with globular features of the microfibril (Fig. 5B). Domain–domain conformational rearrangements could allow some regions (e.g., from TB4 to -6) to become more linear upon imposed force, which would enable extension within the interbead region of the microfibril, providing a molecular basis for extensibility. In summary, we have determined the solution nanostructure of fibrillin-1, which has identified a highly compact region of the molecule; this finding has implications for microfibril organization and extensibility.
Fig. 5.
Organization of the fibrillin-1 molecule in microfibrils. (A) A possible structure of the fibrillin-1 molecule from TB1 to the C terminus as predicted by SAXS. TB domains, ovals; the hybrid domain, triangle; contiguous cbEGF arrays, rectangles. The positions of Ab epitopes are indicated on the model by color (red, mAb2502; yellow, mAb2499; cyan, mAb11C1.3; purple, pAbPF2). The locations of heparan sulfate-binding sites (HS) (14), the RGD motif, and MAGP-1 (16) binding site are highlighted by arrows. (Bi) A negatively stained EM image of two microfibril repeats enhanced by image analysis. [Reproduced with permission from ref. 3 (Copyright 2005, Elsevier).] (Bii) A superimposition of Bi and a model of fibrillin-1 organization. The model predicts an overlap of the N and C termini and a molecule length of ≈90 nm and indicates how the fibrillin-1 molecule may be arranged within a microfibril. Only one molecule is shown for clarity, but we would expect there to be eight fibrillin-1 molecules in cross-section (10). The positions of Ab-binding sites on the microfibril are indicated by colored lines as described above. (For all images, the scale is ≈1 cm = 14 nm.) The predicted mass per unit length for the above model was calculated by using Mw and Dmax and multiplied by eight assuming there are eight molecules in cross-section (1, 10). The predicted mass is 68.0 and 25.8 kDa/nm for the bead and interbead. The experimental STEM mass of the bead and interbead of microfibrils from ciliary zonules is 55.0 and 28.3 kDa/nm (1).
Materials and Methods
Recombinant human fibrillin-1 fragments encoded by the following exons were expressed and purified by using 293-EBNA cells as described (13–16) (Fig. 1): PF2, exons 9–17; PF5, exons 18–25; PF7, exons 24–30; PF8, exons 30–38; PF9, exons 37–43; PF10, exons 41–52; PF11, exons 37–52; PF12, exons 50–58; and PF13, exons 57–64. The protein fragments were assessed by SDS/PAGE, gel-filtration chromatography, and MALLS (see Figs. 6 and 7).
SAXS of fibrillin-1 fragments (with the exception of PF13) was carried out on ID02 at the European Synchrotron Radiation Facility (Grenoble, France), by using either a 1- or 2-m (for PF5) sample to detector distance, which covers the momentum transfer interval 0.072 Å−1 < q < 0.69 Å−1 (1 m) or 0.016 Å−1 < q < 0.43 Å−1 (2 m). The modulus of the momentum transfer is defined as q = 4π sinθ/λ, where 2θ is the scattering angle, and λ is the wavelength. The q range was calibrated by using silver behenate powder based on diffraction spacings of 58.38 Å. Images were collected on a Thomson X-ray Intensifier (TH 49-427) lens coupled to a FReLoN CCD camera (2,048 × 2,048 pixels). This detector has an active area of size 180 mm and a frame rate of 14 images (1,024 × 1,024 pixels) per second with a 14-bit nominal dynamic range. The wavelength of x-rays was 0.1 nm. Samples containing 5% glycerol (to minimize beam damage) and in the concentration range 1.8–0 mg/ml were analyzed in a quartz capillary. Multiple images were obtained of each sample tested in time frames of 100 ms to check for radiation damage and protein aggregation between frames.
Low-angle scattering data were collected on station 2.1 of the Synchrotron Radiation Source at Daresbury Laboratory on fragments PF2, -7, -8, -11, and -12. Images were collected in frames of 60 s with a sample-to-detector distance of 5.75 m. During data collection the sample was maintained at 4°C. The scattering profiles were merged with the corresponding European Synchrotron Radiation Facility data to cover the momentum transfer interval 0.0091 Å−1 < q < 0.69 Å−1. Data on PF13 also was collected on station 2.1. Images were collected with sample-to-detector distances of 1 and 4 m. The corresponding profiles were merged to cover the momentum transfer interval 0.010 Å−1 < q < 0.76 Å−1.
The scattering images obtained were all spherically averaged, and corrections for detector artifacts and normalization to absolute scattering intensities were performed by using in-house software. Buffer-scattering intensities were collected and subtracted from the sample image to remove background scattering using PRIMUS (29). The Rg, forward-scattering intensity, and 1D intraparticle distance distribution function p(r) in real space were evaluated with the indirect Fourier transform program GNOM (30). Particle shapes were restored from the experimental scattering profiles following ab initio procedures based on the simulated annealing algorithm using GASBOR (17) and DAMMIN (18). Many simulations (≈20) were performed for each protein fragment, facilitated by the distributed computational facilities afforded by CONDOR software, and these generated very similar but not identical shapes in each case. An averaged filtered structure was generated by using DAMAVER (19), and this structure was used as a fixed core in a final run of GASBOR and DAMMIN.
Because-high resolution structural information is available for the cbEGF32–33 (7), cbEGF12–13 (23), TB6 (6), and cbEGF22–TB4–cbEGF23 (12) domains, homology models were built by using the automated protein structure modeling server, SwissModel (21). SASREF and BUNCH performed rigid-body modeling of domains to the SAXS data set by using a simulated annealing protocol (22). The optimum position and orientation of contiguously linked subunits were found by minimizing the discrepancy between the experimental scattering data and the theoretical scattering curve calculated from the model. The program BUNCH can also find the probable conformations of missing loops by constructing dummy atoms. More than 10 BUNCH and SASREF simulations were computed for each protein fragment. These were performed using either single domains, pairs of domains, or three-domain subunits as input files. For each protein fragment, the structures generated from repeated runs were superimposed with the program SUPCOMB (31) and compared visually for similarity. Also using SUPCOMB, the structures with the best fit to the experimental data were superimposed with the ab initio structure. CRYSOL (20) was used for the simulation of scattering curves from the structural models; this analysis gave a discrepancy factor χ as a measure of how well the structural model fit the experimental data.
For TEM, 6 μl of PF11 (72 μg/ml) was allowed to absorb for 30 s onto a glow-discharged carbon-coated 400 mesh copper grid, after gel-filtration chromatography. The grid was washed three times with water, then negatively stained with 2% (wt/vol) uranyl acetate (pH 4.7). The grid was observed by using a FEI Tecnai Twin TEM equipped with a LaB6 filament operating at 120 kV. Negatives were recorded under low-dose conditions at 56,000× magnification, −700 nm defocus on a 2,048 × 2,048 CCD camera. The electron dose used for each image was ≈10 e−/Å2. Images were converted to Imagic5 format (Image Science), and the Imagic5 program suite was used for particle picking and image processing (32). The level of defocus of each individual image was checked by inspecting the position of the Thon rings in the power spectra and comparing this position with the calculated contrast transfer function. A total of 1,168 selected particles were windowed into 14 × 14-nm images. Band-pass filtering was used with a high-frequency cut-off of 20 Å and a low-frequency cut-off of 100 Å. The images were centered by cross-correlation to the total sum of the data set. Reference-free alignment and classification were used to generate the initial reference set. All of the particles then were aligned to the reference set by multireference alignment. Euler angles were assigned to the reference classes, and an initial 3D reconstruction was calculated. The 3D model then was subjected to refinement by multiple iterations. The first zero of the contrast transfer function at −700 nm defocus was calculated to be 20 Å; because a high-frequency cut-off was imposed, the contrast transfer function correction was deemed unnecessary. The Fourier shell correlation was calculated, and the resolution was determined to be 35 Å with a 3σ criteria.
Supplementary Material
Acknowledgments
We thank Hugh Beedie and James Osborne (Cardiff University Information Services) for implementation of distributed software; Drs. Dmitri Svergun and Maxim Petoukhov (European Molecular Biology Laboratory, Hamburg, Germany) for invaluable software support; the staff in the EM Facility and Dr. Thomas Jowitt in the Biomolecular Analysis Facility (University of Manchester) for their assistance; Amanda Morgan for excellent technical assistance; and Drs. Michael Sherratt and Bertrand Raynal (University of Manchester) for helpful discussions. C.B. has a Royal Society Olga Kennard fellowship. C.B. and M.-C.W. were supported by the Wellcome Trust. C.M.K. was supported by the Medical Research Council and is a Royal Society-Wolfson Merit Award holder. V.S., J.L.H., and T.J.W. were supported by Biotechnology and Biological Sciences Research Council Grant 98/S15326. This work also was supported by the European Synchrotron Radiation Facility and the Daresbury Synchrotron Radiation Source.
Abbreviations
- cbEGF
calcium-binding EGF
- MALLS
multiangle laser light scattering
- SAXS
small-angle x-ray scattering.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kielty C. M, Sherratt M. J., Marson A., Baldock C. Adv. Protein Chem. 2005;70:405–436. doi: 10.1016/S0065-3233(05)70012-7. [DOI] [PubMed] [Google Scholar]
- 2.Kielty C. M., Cummings C., Whittaker S. P., Shuttleworth C. A., Grant M. E. J. Cell Sci. 1991;99:797–807. doi: 10.1242/jcs.99.4.797. [DOI] [PubMed] [Google Scholar]
- 3.Lu Y., Holmes D. F., Baldock C. J. Mol. Biol. 2005;349:73–85. doi: 10.1016/j.jmb.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 4.Robinson P. N., Booms P., Katzke S., Ladewig M., Neumann L., Palz M., Pregla R., Tiecke F., Rosenberg T. Hum. Mutat. 2002;20:153–161. doi: 10.1002/humu.10113. [DOI] [PubMed] [Google Scholar]
- 5.Campbell I. D., Bork P. Curr. Opin. Struct. Biol. 1993;3:385–392. [Google Scholar]
- 6.Yuan X., Downing A. K., Knott V., Handford P. A. EMBO J. 1997;16:6659–6666. doi: 10.1093/emboj/16.22.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downing A. K., Knott V., Werner J. M., Cardy C. M., Campbell I. D., Handford P. A. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 8.Jensen S. A., Corbett A. R., Knott V., Redfield C., Handford P. A. J. Biol. Chem. 2005;280:14076–14084. doi: 10.1074/jbc.M412832200. [DOI] [PubMed] [Google Scholar]
- 9.Wess T. J., Purslow P. P., Kielty C. M. J. Struct. Biol. 1998;122:123–127. doi: 10.1006/jsbi.1998.3992. [DOI] [PubMed] [Google Scholar]
- 10.Baldock C., Koster A. J., Ziese U., Rock M. J., Sherratt M. J., Kadler K. E., Shuttleworth C. A., Kielty C. M. J. Cell Biol. 2001;152:1045–1056. doi: 10.1083/jcb.152.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. J. Mol. Biol. 1996;258:104–116. doi: 10.1006/jmbi.1996.0237. [DOI] [PubMed] [Google Scholar]
- 12.Lee S. S., Knott V., Jovanovic J., Harlos K., Grimes J. M., Choulier L., Mardon H. J., Stuart D. I., Handford P. A. Structure (London) 2004;12:717–729. doi: 10.1016/j.str.2004.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bax D. V., Bernard S. E., Lomas A., Morgan A., Humphries J., Shuttleworth C. A., Humphries M. J., Kielty C. M. J. Biol. Chem. 2003;278:34605–34616. doi: 10.1074/jbc.M303159200. [DOI] [PubMed] [Google Scholar]
- 14.Cain S. A., Baldock C., Gallagher J., Morgan A., Bax D. V., Weiss A. S., Shuttleworth C. A., Kielty C. M. J. Biol. Chem. 2005;280:30526–30537. doi: 10.1074/jbc.M501390200. [DOI] [PubMed] [Google Scholar]
- 15.Marson A., Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Shuttleworth C. A., Baldock C., Kielty C. M. J. Biol. Chem. 2005;280:5013–5021. doi: 10.1074/jbc.M409029200. [DOI] [PubMed] [Google Scholar]
- 16.Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K. T., Marson A., Shuttleworth C. A., Weiss A. S., Kielty C. M. J. Biol. Chem. 2004;279:23748–23758. doi: 10.1074/jbc.M400212200. [DOI] [PubMed] [Google Scholar]
- 17.Svergun D. I., Petoukhov M. V., Koch M. H. J. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svergun D. I. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volkov V. V., Svergun D. I. J. Appl. Cryst. 2003;36:860–864. [Google Scholar]
- 20.Svergun D. I., Barberato C., Koch M. H. J. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 21.Schwede T., Kopp J., Guex N., Peitsch M. C. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petoukhov M. V., Svergun D. I. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smallridge R. S., Whiteman P., Werner J. M., Campbell I. D., Handford P. A., Downing A. K. J. Biol. Chem. 2003;278:12199–12206. doi: 10.1074/jbc.M208266200. [DOI] [PubMed] [Google Scholar]
- 24.Mould A. P., Symonds E. J., Buckley P. A., Grossmann J. G., McEwan P. A., Barton S. J., Askari J. A., Craig S. E., Bella J., Humphries M. J. J. Biol. Chem. 2003;278:39993–39999. doi: 10.1074/jbc.M304627200. [DOI] [PubMed] [Google Scholar]
- 25.Vestergaard B., Sanyal S., Roessle M., Mora L., Buckingham R. H., Kastrup J. S., Gajhede M., Svergun D. I., Ehrenberg M. Mol. Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 26.Reinhardt D. P., Gambee J. E., Ono R. N., Bachinger H. P., Sakai L. Y. J. Biol. Chem. 2000;275:2205–2210. doi: 10.1074/jbc.275.3.2205. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto H., Broekelmann T., Cheresh D. A., Ramirez F., Rosenbloom J., Mecham R. P. J. Biol. Chem. 1996;271:4916–4922. [PubMed] [Google Scholar]
- 28.Maddox B. K., Sakai L. Y., Keene D. R., Glanville R. W. J. Biol. Chem. 1989;264:21381–21385. [PubMed] [Google Scholar]
- 29.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 30.Semenyuk A. V., Svergun D. I. J. Appl. Cryst. 1991;24:537–540. [Google Scholar]
- 31.Kozin M. B., Svergun D. I. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 32.van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.