Abstract
Smad transcription factors are key signal transducers for the TGF-β/bone morphogenetic protein (BMP) family of cytokines and morphogens. C-terminal serine phosphorylation by TGF-β and BMP membrane receptors drives Smads into the nucleus as transcriptional regulators. Dephosphorylation and recycling of activated Smads is an integral part of this process, which is critical for agonist sensing by the cell. However, the nuclear phosphatases involved have remained unknown. Here we provide functional, biochemical, and embryological evidence identifying the SCP (small C-terminal domain phosphatase) family of nuclear phosphatases as mediators of Smad1 dephosphorylation in the BMP signaling pathway in vertebrates. Xenopus SCP2/Os4 inhibits BMP activity in the presumptive ectoderm and leads to neuralization. In Xenopus embryos, SCP2/Os4 and human SCP1, 2, and 3 cause selective dephosphorylation of Smad1 compared with Smad2, inhibiting BMP- and Smad1-dependent transcription and leading to the induction of the secondary dorsal axis. In human cells, RNAi-mediated depletion of SCP1 and SCP2 increases the extent and duration of Smad1 phosphorylation in response to BMP, the transcriptional action of Smad1, and the strength of endogenous BMP gene responses. The present identification of the SCP family as Smad C-terminal phosphatases sheds light on the events that attenuate Smad signaling and reveals unexpected links to the essential phosphatases that control RNA polymerase II in eukaryotes.
Keywords: osteosarcoma, Xenopus, signal transduction
TGF-β signals are initiated when ligands bind and activate receptor serine/threonine kinases at the cell surface (1–4). The resulting receptor complex propagates the signal through phosphorylation of cytoplasmic effectors, the receptor-regulated Smads (R-Smads). Once phosphorylated on two serine residues at the C-terminal sequence Ser-Xxx-Ser, the R-Smads translocate from the cytoplasm into the nucleus, where they modulate gene transcription. In the TGF-β superfamily, two groups of ligands can be distinguished: the TGF-β/activin/nodal subfamily and the bone morphogenetic protein (BMP) subfamily. They both activate specific pathways through different sets of receptors and cytoplasmic effectors and result in very different and sometimes antagonistic outcomes (5, 6). Smad2 and 3 respond to the TGF-β branch of the pathway, whereas Smad1, 5, and 8 mediate responses to the BMP branch. Given the prominent role of TGF-β signaling in metazoan biology, much attention is devoted to understanding how these pathways are regulated. Because the signal is propagated through phosphorylation, one might expect that protein phosphatases are directly involved in terminating the signal (7, 8). The critical dephosphorylation event appears to take place in the nucleus (7, 8). Yet, the nuclear phosphatases that are responsible for Smad1 dephosphorylation have not been identified.
In an unbiased expression cloning scheme to identify factors that affect dorsal/ventral polarity in Xenopus, we previously isolated a phosphatase, the Xenopus homologue of a human protein originally called Os4, for its ability to induce a secondary axis in early embryos (9). Secondary axis induction in frog embryos can occur by activation of the canonical Wnt pathway, by inhibition of the BMP pathway, or by activation of the activin/nodal pathway. The morphology of the secondary axis induced by Xenopus Os4 (i.e., lacking head structures such as cement gland and eyes) did not support activation of the Wnt pathway as a likely mechanism, leaving the other two possibilities open for investigation. However, our study using classical embryo explant assays left unresolved whether BMP inhibition or activation of activin was responsible for the secondary axis induction.
Human Os4, also called SCP2 [small C-terminal domain (CTD) phosphatase 2] or CTDSP2, the homolog of XOs4 that shares ≈90% similarity, belongs to a family of three closely related class C Ser/Thr phosphatases, the SCP family (10–13). SCP1–3 are related to the catalytic subunit of FCP1, which is the highly conserved, essential enzyme that dephosphorylates the CTD of RNA polymerase II (Pol II). Although SCPs can dephosphorylate the CTD of Pol II in vitro (13), they do not appear to be functionally redundant with FCP1, because they are also known to mediate silencing of neuronal-specific gene expression (12). To date, the identity of substrates that may explain the biological activities of Xenopus Os4 (XSCP2) has remained elusive.
Results
XSCP2 Acts as an Inhibitor of the BMP Pathway.
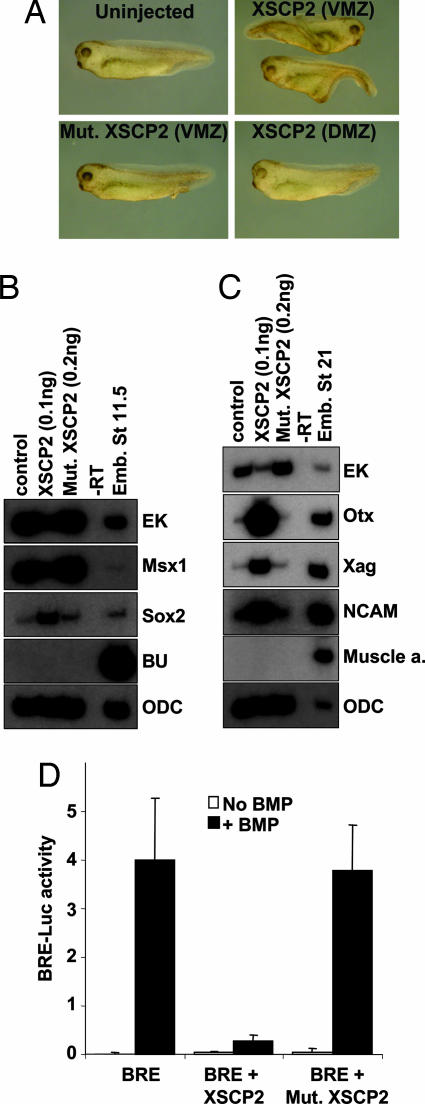
Microinjection of 100 pg of synthetic XSCP2 mRNA in the ventral marginal zone of the Xenopus embryo leads to the induction of a partial secondary axis (Fig. 1A). This secondary axis is elicited only if XSCP2 is microinjected in the ventral side of the embryo and requires the phosphatase function of XSCP2, as determined by the lack of effect of the catalytically inactive point-mutant form (Asp-107-Glu) (Fig. 1A). To understand the mechanism of action by which XSCP2 induces this secondary axis, we examined whether it mediates activation of the activin/nodal pathway or inhibition of the BMP pathway in Xenopus ectodermal explants. In intact uninjected animal caps, BMP signaling is active and induces epidermal fate, as can be observed by the induction of its immediate early response gene, Msx1 (14), and the epidermal keratin (EK) marker. In this ectodermal explant system, activation of the activin/nodal pathway leads to the induction of mesodermal fate, and inhibition of BMP signaling leads to the induction of neural genes (5). Analysis of intact ectodermal explants expressing 100 pg of XSCP2 with cell type-specific molecular markers reveals that the epidermal markers Msx1 and EK are strongly down-regulated, whereas both early (Fig. 1B) and late (Fig. 1C) neural-specific markers, Sox2, neural cell adhesion molecule, Xag, and Otx, are induced. In the meantime, the mesodermal genes Brachyury and muscle actin remain quiet.
Fig. 1.
XSCP2 acts as a BMP inhibitor. (A) Secondary axis phenotype. Embryos were injected at the four-cell stage in one blastomere with mRNAs encoding XSCP2 (100 pg) or a mutant form of XSCP2, Mut.XSCP2 (100 pg). Embryos were observed at tadpole stage. (B) Eight-cell-stage embryos were injected in four animal blastomeres with mRNAs encoding XSCP2 (100 pg) and Mut.XSCP2 (200 pg). Animal cap explants were assayed by RT-PCR for expression of the indicated molecular markers at gastrula (stage 11.5). Ornithine decarboxylase (ODC) is used as a loading control. (C) Same as B except that explants were assayed at neurula stages (stage 21). (D) Luciferase assay with BRE. Two-cell-stage embryos were injected with the BRE luciferase reporter construct (20 pg), the Renilla luciferase reporter construct (10 pg), and the indicated combinations of the following RNAs: BMP4 (200 pg), XSCP2 (100 pg), and Mut.XSCP2 (100 pg). Embryos were harvested at the onset of gastrulation (stage 10+) and assayed for luciferase activity. Results shown correspond to firefly luciferase values normalized with Renilla luciferase values.
We next investigated whether XSCP2 could prevent the transcriptional activation from a BMP/Smad1 responsive element (BRE) by BMP4 (15). The same dose of XSCP2 mRNA (100 pg) robustly abrogated the activation of BRE-driven transcription in response to BMP4, and the phosphatase activity of XSCP2 was essential for this effect (Fig. 1D). To eliminate the possibility of a general silencing effect of XSCP2 on transcription (13), we coinjected embryos with a Renilla luciferase reporter construct under the control of a noninducible promoter and normalized the results with the Renilla values. Taken together, these data strongly suggest that, at this dose, XSCP2 can act as a BMP inhibitor.
XSCP2 Can Directly Dephosphorylate Smad1 on C-Terminal Ser Residues.
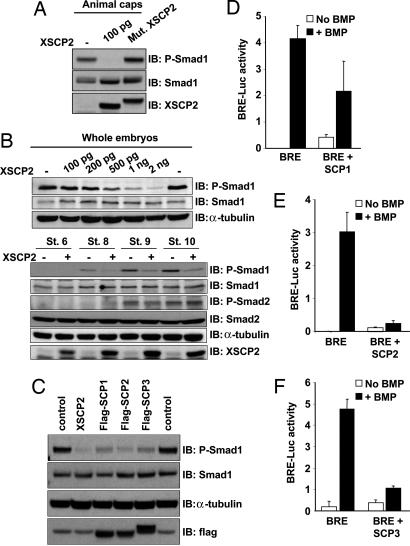
The phosphatase activity of XSCP2 being essential for secondary axis induction and neuralization of ectodermal explants prompted us to explore whether it was involved in Smad dephosphorylation. One hundred picograms of XSCP2 in animal caps significantly reduced the level of Smad1 C-terminal phosphorylation (Fig. 2A). As observed for the secondary axis induction and the neuralization of caps, the phosphatase function of XSCP2 is required for this effect, because the inactive XSCP2 does not affect Smad1 phosphorylation (Fig. 2A). We next tested whether this effect was restricted to the BMP branch of the pathway or whether XSCP2 was also involved in Smad2 dephosphorylation. However, because Smad2 is not phosphorylated in animal caps (16), we performed the experiment using whole embryos. The dose–response curve shown in Fig. 2B Upper demonstrates that the effect of XSCP2 is dose dependent but that 100 pg is not enough in the larger context of the whole embryo to elicit the same effect as in caps. Instead, 1 ng of XSCP2 induces a significant decrease in the level of phospho-Smad (P-Smad) 1, so this dose was used to perform experiments in whole embryos.
Fig. 2.
XSCP2 and human SCPs cause Smad1 C-terminal dephosphorylation in vivo. (A) Overexpression of XSCP2 in animal caps decreases endogenous Smad1 C-terminal phosphorylation level. Two-cell-stage embryos were injected in the animal pole of both blastomeres with 100 pg of XSCP2 or Mut.XSCP2 mRNAs or left untreated. Animal cap explants were isolated at stage 9 and cultured until stage 10.5. Lysates were prepared and analyzed by immunoblotting with anti-P-Smad1, anti-Smad1, and anti-XSCP2 antibodies. The difference in electrophoretic mobility between Mut.XSCP2 and XSCP2 is due to the presence of a flag tag on the mutant form. (B) (Upper) XSCP2 effect on endogenous Smad1 phosphorylation level is dose dependent. Embryos were injected with increasing doses of XSCP2 mRNA and processed as in A except that the whole embryo was harvested to prepare the lysate. (Lower) XSCP2 overexpression decreases Smad1 phosphorylation levels without affecting Smad2. Two-cell-stage embryos were injected in both blastomeres with XSCP2 mRNA (1 ng) or left untreated and harvested at the indicated stages. Lysates were analyzed as in A by immunoblotting with the indicated antibodies. As a loading control, the anti-P-Smad1 membrane was stripped and reprobed with anti-α-tubulin. (C) Two-cell-stage embryos were injected with XSCP2 (1 ng), flag-SCP1, flag-SCP2, or flag-SCP3 mRNAs (500 pg), and whole embryos were harvested at stage 10.5. Samples were prepared and analyzed as described in B. (D–F) Luciferase assays with BRE and human SCP1 (D), SCP2 (E), and SCP3 (F). Embryos were injected with the indicated mRNAs and analyzed as in Fig. 1D.
Embryos injected with XSCP2 at two-cell stage and harvested at different time points up to gastrulation (stages 6, 8, 9, and 10) were subjected to immunoblotting using antibodies that specifically recognize the phosphorylated C-terminal tail of Smad1 or Smad2. Expression of XSCP2 strongly decreased the Smad1 phosphorylation without significantly affecting Smad2 phosphorylation (Fig. 2B Lower). This observation is consistent with our results in ectodermal explants, where XSCP2 affected the BMP pathway but not the activin/nodal pathway (Fig. 1 B and C). This result was not due to a nonselective dephosphorylation effect of XSCP2 overexpression, because it was not accompanied by extensive changes in phosphoprotein patterns when the immunoblots were probed with a panel of five different anti-P-Ser/Thr antibodies targeting major Ser/Thr protein kinase substrates (17–21) (Fig. 7, which is published as supporting information on the PNAS web site).
Human SCP1–3 were also able to mimic many of XSCP2’s effects. Expression of SCP1, 2, and 3 mRNAs in frog embryos resulted in a decrease in the endogenous Smad1 phosphorylation levels (Fig. 2C) and an inhibition of the transcriptional activation from a BMP responsive promoter (Fig. 2 D–F). Overexpression of the human SCPs also induced partial secondary axes at tadpole stages, albeit at lower frequency than was observed with XSCP2 (data not shown), suggesting that the biological activity is shared between XSCP2 and its mammalian counterparts.
SCPs Selectively Bind and Dephosphorylate Smad1.
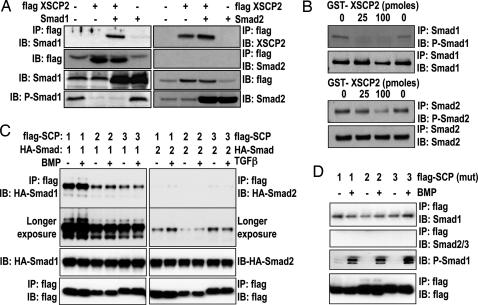
To investigate whether the effect of XSCP2 on Smad1 involves an interaction between these proteins, we coexpressed Smad1 and XSCP2 in embryos and performed immunoprecipitation (IP) experiments, which show that Smad1 and XSCP2 form a complex (Fig. 3A Left). No interaction could be detected between XSCP2 and Smad2 in a similar experimental setting (Fig. 3A Right). To further test our hypothesis that XSCP2’s effect on Smad1 could be direct, we asked whether bacterially produced recombinant GST-XSCP2 was able to dephosphorylate immunopurified P-Smad1 in vitro. Indeed, GST-XSCP2 can efficiently remove C-terminal phosphorylation from Smad1 in vitro (Fig. 3B Upper). Interestingly, the highest concentration of enzyme also leads to a small but reproducible decrease in Smad2 C-terminal phosphorylation (Fig. 3B Lower), suggesting that, under our assay conditions, XSCP2 acts as a robust Smad1 phosphatase and a weak Smad2 phosphatase.
Fig. 3.
SCPs selectively bind and dephosphorylate Smad1. (A) Two-cell-stage embryos were left untreated or injected with the indicated combinations of the following RNAs: Smad1 (2 ng), Smad2 (400 pg), and flag-XSCP2 (2 ng). Embryos were harvested at gastrula stage (stage 10.5). Lysates were prepared and immunoprecipitated with anti-flag M2 antibody and analyzed by immunoblot with anti-Smad1, anti-Smad2, and anti-XSCP2 antibodies. The expression of flag-XSCP2, Smad1, P-Smad1, and Smad2 was checked by immunoblotting the crude extracts used for the IP reaction. (B) In vitro XSCP2 phosphatase assay. Immunoprecipitated Smad1 (Upper) and Smad2 (Lower) from gastrula-stage embryos were incubated with increasing amounts of recombinant GST-XSCP2. Their phosphorylation status was monitored by immunoblotting with corresponding phospho-specific antibodies. To make sure that the same amounts of Smad1 and 2 were immunoprecipitated in the different conditions, we also probed with Smad1 and Smad2 antibodies after the IP. (C) HEK293 cells were transfected with flag-SCP1–3 and HA-Smad1 or HA-Smad2 as indicated, stimulated with BMP or TGF-β for 1 h, and lysed. Flag immunoprecipitates or lysates were immunoblotted with the indicated antibodies. The second blot from the top shows a longer exposure of the uppermost blot to demonstrate weak interaction of SCP1–3 with Smad2. (D) Same as C except that cells were transfected with catalytically inactive mutants of flag-SCP1–3 and treated with or without BMP for 1 h. P-Smad1 was used as a control for BMP stimulation efficiency.
Similarly, in HEK293 cells, we show that human flag-SCP1, 2, and 3 also interact efficiently with HA-Smad1 but poorly with HA-Smad2 (Fig. 3C). Furthermore, catalytically inactive mutants of flag-SCP1, 2, and 3 were able to pull down endogenous Smad1 but not Smad2/3 (Fig. 3D). Inactive SCP1–3 were used to ensure that endogenous Smad1 was not dephosphorylated upon BMP treatment. Under these conditions, treating cells with BMP or TGF-β did not enhance the binding of SCPs to Smad1 or Smad2, respectively, suggesting that the interaction between SCPs and Smad1 is BMP independent (Fig. 3 C and D). Taken together, these data suggest that BMP inhibition by SCPs could be mediated by a direct dephosphorylation of Smad1 C-terminal phosphorylation.
SCP1 and SCP2 Depletion Enhances BMP-Induced Smad1 Phosphorylation and Delays Smad1 Dephosphorylation.
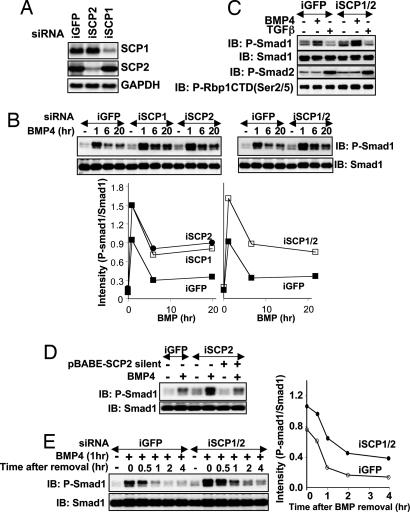
To definitely establish a role of endogenous SCPs in Smad1 dephosphorylation, we performed a loss-of-function analysis by introducing siRNA oligonucleotides targeting SCP1 (iSCP1), SCP2 (iSCP2), or both in human HaCaT keratinocytes. Transfection of iSCP1 and iSCP2 resulted in the reduction of the respective mRNA levels by >80%, whereas iGFP transfection, used as a control, had no effect on SCP1 or SCP2 mRNA levels (Fig. 4A). Transfection of iSCP1 and iSCP2 individually or together resulted in increased accumulation of P-Smad1 in response to BMP4 (Fig. 4B) compared with iGFP. Interestingly, TGF-β-induced Smad2 phosphorylation was unchanged in cells expressing iSCP1 and iSCP2 compared with iGFP, and the effect of SCP depletion on Smad1 phosphorylation was as pronounced as its effect on the phosphorylation level of Pol II CTD (Fig. 4C). The accumulation of P-Smad1 levels in BMP4-treated iSCP2-expressing cells was reversed to levels seen in iGFP-expressing cells in cells that expressed a SCP2 mutant that is resistant to iSCP2 silencing (Fig. 4D). These results indicate that endogenous SCP1 and/or SCP2 plays a role in regulating the levels of P-Smad1 in BMP4-treated cells.
Fig. 4.
RNAi inhibition of SCP1 and SCP2 enhances BMP-induced Smad1 phosphorylation and delays Smad1 dephosphorylation. (A) HaCaT cells were transfected with siRNA oligonucleotides targeted against GFP (iGFP), SCP1 (iSCP1), or SCP2 (iSCP2), and mRNA levels were checked by Northern blotting. GAPDH was used as a control. (B) HaCaT cells transfected with iGFP, iSCP1, iSCP2, or iSCP1/2 were left untreated (−) or treated with BMP4 for 1, 6, and 20 h. Lysates were analyzed by immunoblotting with P-Smad1 and Smad1 antibodies. In Lower, the intensities of the P-Smad1 and Smad1 bands were measured with NIH Image software, and the ratio of the intensities (P-Smad1/Smad1) was plotted against the time of BMP4 treatment for each transfection. (C) HaCaT cells transfected with iGFP or iSCP1/2 were left untreated (−) or treated with BMP4 or TGF-β for 20 h. Lysates were analyzed by immunoblotting using the indicated antibodies. CTD(P-Ser-2/5) recognizes the Pol II CTD phosphorylated at serines 2 and 5. (D) Same as C except that cells were infected with pBABE-puromycin vector (−) or pBABE-puromycin vector containing a SCP2 mutant that is resistant to iSCP2 silencing (+) and selected in 4 μg/ml puromycin before BMP treatment. (E) HaCaT cells transfected with iGFP or iSCP1/2 were left untreated (−) or treated with BMP4 for 1 h (+). After 1 h, BMP4 was removed from the media, and cells were lysed at the indicated time points after BMP4 removal. Smad1 dephosphorylation after BMP4 removal was assessed by immunoblotting lysates with P-Smad1 and Smad1 antibodies. In Right, the intensities of the P-Smad1 and Smad1 bands were measured and expressed as in B.
We next assessed the kinetics of Smad1 dephosphorylation in cells transfected with iGFP or iSCP1/2. Cells were treated with BMP4 for 1 h to yield maximal levels of P-Smad1. BMP4 was then removed from these cells, and P-Smad1 levels were assessed at various time points thereafter. In cells expressing iGFP, P-Smad1 levels reached to almost basal levels within 1 h of BMP4 removal, whereas P-Smad1 levels in cells expressing iSCP1/2 were significantly higher at all time points (Fig. 4E). Treatment of HaCaT cells with staurosporine, a broad-specificity protein kinase inhibitor that also inhibits BMP receptor type I kinase, did not enhance the rate of Smad1 dephosphorylation (Fig. 8, which is published as supporting information on the PNAS web site), indicating that the BMP receptor phosphorylation and activity had no effect on Smad1 dephosphorylation after BMP removal. The reduction in P-Smad1 levels in cells expressing iSCP1/2 (Fig. 4E) indicates that either the residual levels of SCP1 and/or SCP2 are mediating the remaining dephosphorylation of Smad1 or there are still other phosphatases that also play a role in dephosphorylating Smad1. These results strongly implicate SCP1 and SCP2 as prominent mediators of the dephosphorylation of BMP-activated Smad1.
RNAi Inhibition of SCP1/2 Enhances BMP-Induced Smad1 Phosphorylation and Depletes Nuclear SCP2 Immunofluorescence in Osteosarcoma Cells.
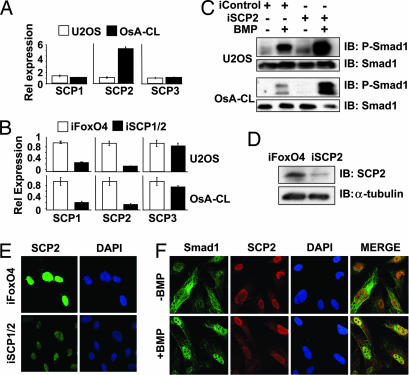
The chromosomal region containing the SCP2 gene is frequently amplified in a number of sarcomas and brain tumors (11). We assessed the levels of SCP2 in two osteosarcomas cell lines: OsA-CL, in which SCP2 is amplified, and U20S, in which SCP2 is not amplified (11) (Fig. 5A). Transfection of iSCP1/2 in both cells lines reduced the expression of SCP1 and SCP2 mRNAs by at least 80% as analyzed by quantitative real-time PCR (qRTPCR) (Fig. 5B). The BMP-induced phosphorylation of Smad1 was significantly enhanced in both U2OS and OsA-CL cells transfected with iSCP1/2 compared with iFoxO4 (Fig. 5C).
Fig. 5.
SCP2 is a nuclear phosphatase. (A) qRTPCR of SCPC1–3 was performed with cDNAs from U2OS and OsA-CL cells. Bars represent relative levels of expression. (B) qRTPCR of SCPC1–3 in cells transfected with the iSCP1 and iSCP2 oligonucleotides together or with iFoxO4 (siRNA against FoxO4) as a control. (C) U20S and OsA-CL cells were transfected as indicated and then treated with BMP or left untreated for 1 h. Protein lysates were subsequently analyzed by immunoblotting using the indicated antibodies. (D) Immunoblot analysis of endogenous SCP2 in OsA-CL cells in the presence of control iFoxO4 or iSCP2. (E) Immunofluorescence analysis of endogenously expressed SCP2 in OsA-CL cells in the presence of iSCP1/2 or control iFoxO4; a rabbit polyclonal SCP2 antibody was used. (F) Immunofluorescence analysis of endogenous Smad1 and SCP2 in OsA-CL cells in the presence or absence of BMP.
Using an antibody that recognizes SCP2 but not SCP1 or SCP3, we were able to detect endogenous SCP2 protein expression in OsA-CL cells and verified the decrease in protein expression in cells expressing iSCP2 (Fig. 5D) by immunoblotting. Using this antibody, we demonstrate that endogenous SCP2 is localized mainly to the nucleus in OsA-CL cells (Fig. 5E), which is consistent with previous studies (9, 13). This nuclear staining of SCP2 was greatly depleted when cells were transfected with iSCP2 (Fig. 5E), consistent with the reduction in SCP2 expression. When BMP is added to the cells, both Smad1 and SCP2 colocalize in the nucleus (Fig. 5F). As expected for a nuclear phosphatase, SCP2 did not prevent nuclear translocation of Smad1 upon BMP treatment.
Loss of P-Smad1 Levels Is Due to SCP-Mediated Dephosphorylation and Proteasome-Mediated Degradation.
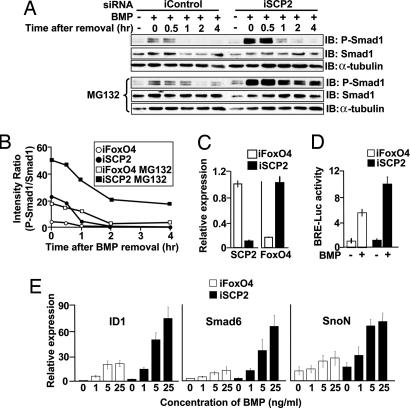
We assessed the kinetics of Smad1 dephosphorylation in OsA-CL cells treated with iSCP2 or control iFoxO4, similar to the data shown in Fig. 4E. In iFoxO4-treated cells, the levels of P-Smad1 reached to basal levels within 1 h of BMP4 removal (Fig. 6A), as seen in HaCaT (Fig. 4E) and U2OS (not shown) cells. On the other hand, in iSCP2-treated cells, which showed a reduction of SCP2 mRNA expression of >90% (Fig. 6C), the level of BMP-induced P-Smad1 remained elevated for a longer period (Fig. 6 A and B).
Fig. 6.
SCP2 depletion affects BMP responses in OsA-CL cells. (A) Time course of Smad1 dephosphorylation in OsA-CL cells performed as in Fig. 4E in the absence (Upper) or presence (Lower) of proteasome inhibitor MG132. (B) The intensities of P-Smad1 and Smad1 bands from A were measured as in Fig. 4, and the ratio of the intensities (P-Smad1/Smad1) is shown. (C) qRTPCR of SCP2 and FoxO4 in OsA-CL cells transfected as indicated. Bars represent relative levels of expression. (D) The BMP-induced reporter activity was tested in OsA-CL cells in the presence of iFoxO4 control or iSCP2 oligonucleotides by using a mammalian BMP-inducible luciferase reporter construct. Renilla luciferase was used for normalization purposes. (E) BMP-induced target gene expression in OsA-CL cells by qRTPCR. Cells were transfected with iFoxO4 or iSCP2 and, 48 h later, treated with different concentrations of BMP or left untreated for 3 h. Expression of BMP target genes ID1, Smad6, and SnoN was analyzed by qRTPCR.
Reduction of P-Smad levels after cell stimulation with TGF-β ligands can be mediated by protein dephosphorylation (7) as well as proteasome-dependent degradation (22). To investigate the relative contributions of each of these two possibilities, we used the proteasome inhibitor MG132. MG132 treatment in cells transfected with iFoxO4 delayed the clearance of P-Smad1 to basal levels by 1 h. In comparison, in cells transfected with iSCP2, MG132 addition resulted in maintenance of elevated levels of P-Smad1 for at least 4 h (Fig. 6 A and B), indicating that the clearance of P-Smad1 can result from a combination of protein dephosphorylation and degradation.
SCP2 Limits BMP Responsiveness in Mammalian Cells.
To test whether transcriptional activity of Smad1 was affected in the absence of SCP2, we used a luciferase reporter construct driven by an artificial Smad1 responsive promoter. Consistent with the increased levels of P-Smad1, transfection of iSCP2 led to an increase in the BMP-induced reporter activity in OsA-CL cells (Fig. 6D).
Similarly, treatment of OsA-CL cells expressing iFoxO4 with increasing amounts of BMP led to increased expression of the BMP target genes ID1 (23), Smad6 (2), and SnoN (24, 25), whereas treatment of cells expressing iSCP2 with the corresponding amounts of BMP led to a several-fold increase in the expression of these gene transcripts (Fig. 6E). Taken together, these experiments indicate that SCPs are essential in regulating BMP-induced gene responses.
Discussion
In the context of the Xenopus embryo, we propose that the secondary axis induction by XSCP2 is due to inhibition of BMP by means of Smad1 dephosphorylation. To draw this conclusion, we rely on the fact that the same dose that elicits the secondary axis can neuralize ectodermal explants, reduce Smad1 C-terminal phosphorylation in explants, and inhibit BMP-induced transcription in a reporter assay. Furthermore, our present data do not favor the alternative possibility that the secondary axis primarily results from activation of activin signaling. Indeed, the dose of XSCP2 that elicits the secondary axis does not induce mesodermal genes in explants, and XSCP2 only mildly dephosphorylates Smad2 compared with Smad1.
By addressing the biochemical roles of XSCP2 and its human counterparts, SCP1–3, we have identified nuclear phosphatases that reverse the receptor-mediated phosphorylation of Smad1, a mediator of BMP signals. Furthermore, loss-of-function studies in human cells by siRNA conclusively support our hypothesis that SCP family members are involved in Smad1 dephosphorylation in vivo. Collectively, the results suggest that the inhibitory effect of SCPs toward BMP responsive genes is rather selective. However, we cannot rule out that other phosphatase(s) are also involved in Smad1 dephosphorylation. Indeed, it was recently reported that pyruvate dehydrogenase phosphatase (PDP) can act as a phosphatase for MAD, the Drosophila homolog of Smad1 (26). PDP, a metabolic enzyme that dephosphorylates pyruvate dehydrogenase, is localized in the mitochondria, suggesting that Smad negative regulation by phosphatases may occur in several locations within the cell. Conversely, Smad1 may not be the only substrate of SCP phosphatases, as shown by its ability to also dephosphorylate the Pol II CTD (13) in vitro. It is likely that SCP family members, as is the case for many other catalytic subunits of phosphatases, can interact with different regulatory subunits that determine their substrate specificity and their subcellular localization (27, 28). Identification of such SCP partners will be the next crucial step in understanding the mechanism of action and specific functions of the members of this Smad phosphatase family.
Interestingly, PPM1A (protein phosphatase 1A) was recently reported to act as a Smad2/3 phosphatase, resulting in attenuation of the TGF-β branch of the pathway, in mammalian cells as well as in zebrafish early embryogenesis (29). The SCP family of phosphatases is very distinct from PPM1A phosphatase. However, it remains possible that both families of phosphatases may act on the other Smad signaling branch under different conditions.
In sum, the present identification of SCPs as nuclear Smad1 phosphatases provides important insights into the physiological regulation of the central BMP signaling pathway, its alternate attenuation by Smad1 dephosphorylation and proteasome degradation routes, and its probable derangement in human osteosarcomas.
Materials and Methods
Xenopus Embryo Manipulations.
Xenopus embryo injections and dissections were performed as described in ref. 30. RNA synthesis and RT-PCR assays for molecular markers were performed as described in ref. 9. Indicated RNA doses injected are per embryo.
Plasmids.
The XSCP2 and flag-XSCP2 constructs are described in ref. 9. pGEX-XSCP2 was generated by PCR from pCs2-XSCP2. Recombinant GST-XSCP2 was expressed in BL21 cells as described in ref. 31. Human SCP1, 2, and 3 cDNAs were cloned from HaCaT RNA by using the SuperScriptIII One-Step RT-PCR System (Invitrogen, Carlsbad, CA) and subcloned into flag-pCMV5 vector. For the SCP2 silent mutant, C768T and A771T were introduced. pCs2-flag-SCP1, pCs2-flag-SCP2, and pCs2-flag-SCP3 were generated by PCR from pCMV5 constructs. All sequences were verified by DNA sequencing.
Luciferase Assays.
Luciferase assays in frog embryos and OsA-CL cells were performed as described in refs. 32 and 33. OsA-CL cells were transfected with siRNA oligonucleotides 24 h before transfection with the BRE-Luc and Renilla Luc reporter constructs.
Tissue Culture.
HaCaT and HEK293 cells were grown in DMEM supplemented with 10% FBS/penicillin-streptomycin/l-glutamine. OsA-CL and U20S cells were cultured as described in ref. 11. siRNA oligonucleotide transfection was performed by using Lipofectamine-2000 reagent (Invitrogen) (300 pmol of siRNA in 40 μl of Lipofectamine-2000). Thirty hours after transfection, the cells were starved in DMEM/2% FBS for 12 h and treated with BMP (25 ng/ml) or TGF-β (100 pM) for the indicated times before lysis. Transfection of pCMV5-HA or flag-tagged SCP1–3, Smad1, or Smad2 constructs (2 μg per 10-cm-diameter dish) was performed by using Lipofectamine-2000 as above. Flag IPs were performed with 0.5 mg of protein lysates by using flag-agarose beads (10 μl; Sigma, St. Louis, MO).
Northern Blots.
Total RNA from subconfluent HaCaT cells was harvested by using the RNeasy kit (Qiagen, Valencia, CA). Samples were electrophoresed in Mops buffer and transferred to a Hybond-N+ membrane (Amersham Pharmacia, Pittsburgh, PA). Radioactive probes for Northern blotting were derived from fragments of the relevant cDNA, and hybridization was performed at 68°C for 3 h. Detection was performed by exposure of membranes to a PhosphorImager screen.
Synthesis of siRNA Oligonucleotides.
The siRNA oligonucleotides were designed and synthesized by the High-Throughput Screening Core Facility in conjunction with the Organic Synthesis Core Facility at Memorial Sloan–Kettering Cancer Center. The siRNA sense strands (5′ → 3′) for the indicated targets were as follows: GFP (iGFP: CAAGCUGACCCUGAAGUUCTT), SCP1 (iSCP1: GCCGGUUGGGUCGAGACCUTT), and SCP2 (iSCP2: GCGGAGCAGAGGACGUCUATT). To knock down both SCP1 and SCP2, iSCP1 and iSCP2 were coexpressed in HaCaT cells.
Immunoblots.
Immunoblot analysis was performed as described in ref. 34 with minor modifications for frog embryo lysates: 70–80 μg of protein was loaded per lane, and for P-Smad2 Western blot analysis, the membrane was blocked for at least 10 h at room temperature in 5% polyvinyl pyrrolidone in TBST (Tris-buffered saline plus Tween 20) before being processed as described in ref. 34. Antibodies used are described in Supporting Text, which is published as supporting information on the PNAS web site.
Quantification of mRNA by Real-Time PCR Analysis.
cDNA used for the real-time PCR was synthesized from 1 μg of purified RNA from cells by using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), following the manufacturer’s protocols. Real-time PCR was performed by using the 7900HT Fast Real Time PCR System (Applied Biosystems, Foster City, CA). All reactions were performed in a volume of 10 μl containing 1 μl of cDNA template (20 ng), 0.1 μM primers, and 5 μl of the SYBR Green I Master Mix (Applied Biosystems). Each sample was analyzed in quadruplicate, and a no-template control was performed for each primer set used.
Supplementary Material
Acknowledgments
We thank all members of A.H.B.’s and J.M.’s laboratories for helpful discussions. This work was supported by The Rockefeller University’s Women and Science Fellowship Program (M.K.) and National Institutes of Health Grants HD32105 (to A.H.B.) and CA34610 (to J.M.). G.S. is a Laura Hartenbaum Breast Cancer Postdoctoral Fellow of the Damon Runyon Cancer Research Foundation. J.M. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- BMP
bone morphogenetic protein
- CTD
C-terminal domain
- SCP
small CTD phosphatase
- IP
immunoprecipitation
- P-Smad
phospho-Smad
- Pol II
RNA polymerase II
- BRE
BMP/Smad1 responsive element
- qRTPCR
quantitative real-time PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Feng X. H., Derynck R. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J., Seoane J., Wotton D. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 3.Schier A. F., Talbot W. S. Annu. Rev. Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y., Massagué J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Sanjuan I., Brivanlou A. H. Nat. Rev. Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 6.Wilson P. A., Lagna G., Suzuki A., Hemmati-Brivanlou A. Development (Cambridge, U.K.) 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- 7.Inman G. J., Nicolas F. J., Hill C. S. Mol. Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 8.Xu L., Kang Y., Col S., Massagué J. Mol. Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- 9.Zohn I. E., Brivanlou A. H. Dev. Biol. 2001;239:118–131. doi: 10.1006/dbio.2001.0420. [DOI] [PubMed] [Google Scholar]
- 10.Kamenski T., Heilmeier S., Meinhart A., Cramer P. Mol.Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 11.Su Y. A., Lee M. M., Hutter C. M., Meltzer P. S. Oncogene. 1997;15:1289–1294. doi: 10.1038/sj.onc.1201294. [DOI] [PubMed] [Google Scholar]
- 12.Yeo M., Lee S. K., Lee B., Ruiz E. C., Pfaff S. L., Gill G. N. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 13.Yeo M., Lin P. S., Dahmus M. E., Gill G. N. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A., Ueno N., Hemmati-Brivanlou A. Development (Cambridge, U.K.) 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- 15.Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A., Massagué J. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 16.Faure S., Lee M. A., Keller T., ten Dijke P., Whitman M. Development (Cambridge, U.K.) 2000;127:2917–2931. doi: 10.1242/dev.127.13.2917. [DOI] [PubMed] [Google Scholar]
- 17.Demonacos C., Krstic-Demonacos M., Smith L., Xu D., O’Connor D. P., Jansson M., La Thangue N. B. Nat. Cell Biol. 2004;6:968–976. doi: 10.1038/ncb1170. [DOI] [PubMed] [Google Scholar]
- 18.James D., Levine A. J., Besser D., Hemmati-Brivanlou A. Development (Cambridge, U.K.) 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 19.Kohn E. A., Yoo C. J., Eastman A. Cancer Res. 2003;63:31–35. [PubMed] [Google Scholar]
- 20.Lynch D. K., Daly R. J. EMBO J. 2002;21:72–82. doi: 10.1093/emboj/21.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt J. M., Stork P. J. Mol. Cell. 2002;9:85–94. doi: 10.1016/s1097-2765(01)00432-4. [DOI] [PubMed] [Google Scholar]
- 22.Lo R. S., Massagué J. Nat. Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 23.Ogata T., Wozney J. M., Benezra R., Noda M. Proc. Natl. Acad. Sci. USA. 1993;90:9219–9222. doi: 10.1073/pnas.90.19.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuide M., Hara T., Furuya T., Takeda M., Kusanagi K., Inada Y., Mori M., Imamura T., Miyazawa K., Miyazono K. J. Biol. Chem. 2003;278:531–536. doi: 10.1074/jbc.C200596200. [DOI] [PubMed] [Google Scholar]
- 25.Takeda M., Mizuide M., Oka M., Watabe T., Inoue H., Suzuki H., Fujita T., Imamura T., Miyazono K., Miyazawa K. Mol. Biol. Cell. 2004;15:963–972. doi: 10.1091/mbc.E03-07-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H. B., Shen J., Ip Y. T., Xu L. Genes Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barford D., Das A. K., Egloff M. P. Annu. Rev. Biophys. Biomol. Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 28.Bollen M., Beullens M. Trends Cell Biol. 2002;12:138–145. doi: 10.1016/s0962-8924(01)02247-4. [DOI] [PubMed] [Google Scholar]
- 29.Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., et al. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brivanlou A. H., Harland R. M. Development (Cambridge, U.K.) 1989;106:611–617. doi: 10.1242/dev.106.3.611. [DOI] [PubMed] [Google Scholar]
- 31.Baratte B., Meijer L., Galaktionov K., Beach D. Anticancer Res. 1992;12:873–880. [PubMed] [Google Scholar]
- 32.Korchynskyi O., ten Dijke P. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 33.Vonica A., Weng W., Gumbiner B. M., Venuti J. M. Dev. Biol. 2000;217:230–243. doi: 10.1006/dbio.1999.9551. [DOI] [PubMed] [Google Scholar]
- 34.Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. J. Biol. Chem. 2001;276:19469–19482. doi: 10.1074/jbc.M009953200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.