Abstract
A dynamic cycle of O-linked N-acetylglucosamine (O-GlcNAc) addition and removal acts on nuclear pore proteins, transcription factors, and kinases to modulate cellular signaling cascades. Two highly conserved enzymes (O-GlcNAc transferase and O-GlcNAcase) catalyze the final steps in this nutrient-driven “hexosamine-signaling pathway.” A single nucleotide polymorphism in the human O-GlcNAcase gene is linked to type 2 diabetes. Here, we show that Caenorhabditis elegans oga-1 encodes an active O-GlcNAcase. We also describe a knockout allele, oga-1(ok1207), that is viable and fertile yet accumulates O-GlcNAc on nuclear pores and other cellular proteins. Interfering with O-GlcNAc cycling with either oga-1(ok1207) or the O-GlcNAc transferase-null ogt-1(ok430) altered Ser- and Thr-phosphoprotein profiles and increased glycogen synthase kinase 3β (GSK-3β) levels. Both the oga-1(ok1207) and ogt-1(ok430) strains showed elevated stores of glycogen and trehalose, and decreased lipid storage. These striking metabolic changes prompted us to examine the insulin-like signaling pathway controlling nutrient storage, longevity, and dauer formation in the C. elegans O-GlcNAc cycling mutants. Indeed, we found that the oga-1(ok1207) knockout augmented dauer formation induced by a temperature sensitive insulin-like receptor (daf-2) mutant under conditions in which the ogt-1(ok430)-null diminished dauer formation. Our findings suggest that the enzymes of O-GlcNAc cycling “fine-tune” insulin-like signaling in response to nutrient flux. The knockout of O-GlcNAcase (oga-1) in C. elegans mimics many of the metabolic and signaling changes associated with human insulin resistance and provides a genetically amenable model of non-insulin-dependent diabetes.
Keywords: hexosamine, insulin signaling, nutrients, obesity
O-linked N-acetylglucosamine (O-GlcNAc) is a dynamic modification of nuclear pore complexes, transcription complexes, and kinases (1–4). Because of the diverse targets modified by O-GlcNAc, deciphering its role in cell physiology has proven challenging. Two enzymes regulate the cycling of O-GlcNAc: the O-linked GlcNAc transferase (OGT) and the glycosidase, O-GlcNAcase (OGA) (1–4). In mammals, the two enzymes of O-GlcNAc cycling are products of single genes; alternative splicing produces isoforms differing in subcellular location and substrate specificity (1, 5–9). The O-GlcNAc cycling enzymes act like kinases and phosphatases to modify Ser and Thr residues of target proteins. In addition, the hexosamine biosynthetic pathway giving rise to the O-GlcNAc donor, UDP-GlcNAc, is highly regulated and responsive to nutrient availability (4, 10–14).
OGA, originally identified as a meningioma auto antigen and termed MGEA5 (8), is a member of the family 84 glycoside hydrolase {Carbohydrate-Active Enzymes [CAZy] database [GH 84 (caz, a)]}. OGA is highly conserved in eukaryotic evolution from Drosophila melanogaster and Caenorhabditis elegans to rodents and man. In mammals, the MGEA5 gene produces at least two isoforms differing only in whether a histone acetyltransferase domain is present (9). OGA may be a bifunctional enzyme catalyzing both O-GlcNAc removal and histone acetylation (15). The N-terminal “hyaluronidase” domain has been proposed to be the catalytic site for OGA glycosidase activity (1) and is likely to conform to the TIM-barrel fold that typifies this class of glycosylhydrolases (4). The histone acetyltransferase (HAT) domain contains a zinc finger-like motif unique to the MYST family of HATs and interacts with histone tails (16). The gene encoding OGA has not been knocked out in mice, and no null alleles of OGA have been described.
The pathway terminating in O-GlcNAc addition and removal has features of a glycan-dependent signaling cascade, and we refer to it as the hexosamine-signaling pathway (1, 4, 17). The pathway is highly conserved in plants and animals and plays a key signaling function in Arabidopsis, where it impacts numerous pathways, including gibberellin signaling (18–24). We recently showed that the hexosamine-signaling pathway influences insulin-like signaling in C. elegans (25, 26) and mice (17). In man, the hexosamine-signaling pathway has been implicated in both diabetes mellitus and neurodegeneration (1, 2, 17, 27). One of the striking features of type 2 diabetes mellitus is insulin resistance. Resistance to insulin is associated with increased flux through the hexosamine biosynthetic pathway (28–33) as well as with inhibition of OGA (34–42). The link between hexosamine signaling and human diabetes mellitus was more directly shown by the finding that a single nucleotide polymorphism in the MGEA5 gene encoding OGA is associated with diabetes mellitus and age of onset in the Mexican-American population (43).
Development of appropriate mouse knockout models of the enzymes of hexosamine signaling has proven difficult because the pathway is essential in mammals. Knockouts of OGT are embryonic and stem cell lethal (44), and conditional knockouts of OGT in various tissues have not been fully completed (45). Overexpression of OGA induces a mitotic exit phenotype (46), suggesting that it too may perform essential functions. We originally identified C. elegans OGT (25) and have shown that knockouts of ogt-1 in the nematode, although viable and fertile, have effects on insulin-like signaling (25, 26). Because of the linkage of the human OGA to insulin resistance, we have now examined the impact of a knockout of the C. elegans OGA gene, oga-1. The knockout allele oga-1(ok1207) dramatically alters O-GlcNAc cycling, producing alterations in macronutrient storage, signaling, and dauer formation.
Results
The OGA gene, oga-1 (also known as T20B5.3), is an X-linked gene composed of 13 exons producing a transcript encoding an 854-aa protein (Fig. 1A, isoform b). We identified four major transcripts of oga-1 using RT-PCR that differ at the positions indicated in Fig. 1A by asterisks (for details, see the legend to Fig. 1). We failed to detect the partially confirmed cDNA assigned to oga-1 in WormBase release WS151 (oga-1a). All of our subsequent studies were performed on the cDNA corresponding to the most abundant transcript oga-1b.
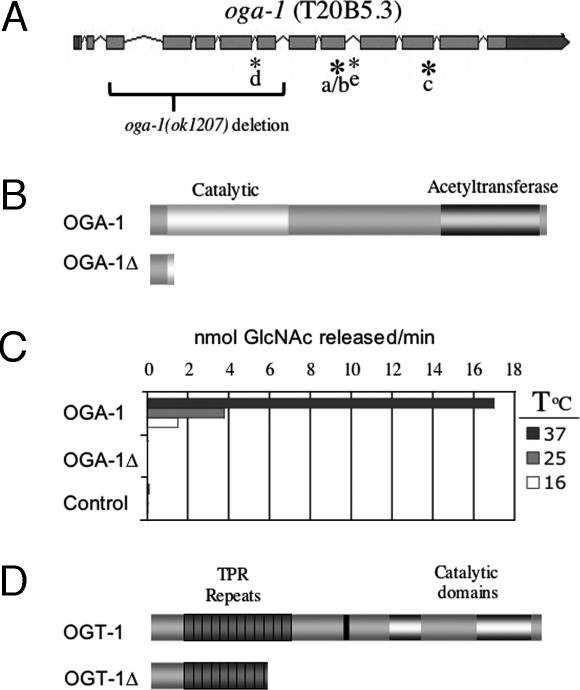
Fig. 1.
The C. elegans oga-1 gene. (A) A schematic of the oga-1 gene showing coding regions shaded light gray and noncoding regions shaded dark gray. We have confirmed four different splice variants (b–e). Novel sequences have been assigned the GenBank accession numbers: DQ407521, DQ407522, and DQ407523. Large bold asterisks indicate use of alternative 3′ splice sites, and small asterisks indicate in-frame utilization of the entire intron. The nine clones we isolated all have an insertion of 17 codons at the 3′ end of exon 9 in the coding sequence of presumptive isoform a. This insertion of 17 codons forms our canonic transcript, oga-1b (5/9 clones). Transcript oga-1c has two additional codons from exon 11 (2/9 clones). Two novel transcripts, d (1/9 clones) and e (1/9 clones), differ from the canonic oga-1b by inclusion of intron 5 or 9 sequences, respectively. Brackets indicate the oga-1(ok1207) deletion joining exon 3 with intron 7, producing a 41-aa protein with 13 novel amino acids. (B) The domain structure of OGA-1 reveals an OGA glycosidase catalytic domain and a region with homology to acetyltransferases. The truncated protein produced by the oga-1(ok1207) deletion allele, OGA-1Δ, lacks both catalytic domains. (C) Enzyme activity of recombinant C. elegans proteins was measured by using the OGA activity assay and normalized to equal amounts of epitope-tagged OGA-1 protein as described in Materials and Methods. The negative control is the PET43.1 expression vector, with human mOGT as insert. A positive control (data not shown in the graph) human OGA was found to release 120, 70, and 27 pmol of GlcNAc per min at 37°C, 25°C, and 16°C, respectively. (D) The ogt-1(ok430) deletion allele generates a truncated protein of 465 aa (OGT-1Δ) that lacks catalytic activity, as described (26).
The OGA-1 protein sequence encoded by oga-1b shows substantial primary sequence similarity (88%) to the human OGA MGEA5. It also shares the overall domain structure of mammalian OGA with a hyaluronidase domain and a C-terminal partial histone acetyltransferase (HAT) domain (Fig. 1B). To ensure that oga-1 encodes C. elegans OGA, we expressed the C. elegans OGA-1 cDNA in Escherichia coli and demonstrated the enzymatic hydrolysis of a synthetic OGA substrate (see Materials and Methods). C. elegans OGA-1 was active at all of the temperatures tested. (Fig. 1C)
The oga-1 deletion allele (ok1207) was obtained from the C. elegans Gene Knockout Consortium as a viable homozygote and was backcrossed before further study. The oga-1(ok1207) strain appears phenotypically normal upon simple inspection. Sequencing genomic DNA from oga-1(ok1207) animals revealed a 1,443-bp deletion beginning 10 bases into exon 3 and ending in intron 7 (Fig. 1A). This deletion allele is predicted to encode a truncated OGA-1 protein of 41 aa, the last 13 of which are unrelated to WT OGA-1 (Fig. 1B). The oga-1(ok1207) allele eliminates the entire “hyaluronidase-like” catalytic domain, and the resulting protein product is predicted to be a null for OGA-1 activity (see Fig. 1B). To test this prediction directly, the putative product encoded by the oga-1(ok1207) allele, which we term OGA-1Δ, was expressed in E. coli and was enzymatically inactive under conditions in which WT OGA was active (Fig. 1C). Several hexosaminidases exist in C. elegans, including hyaluronidases, lysosomal hexosaminidases, and OGA-1, rendering specific detection of OGA activity difficult in whole animal lysates. However, the total level of hexosaminidase activity measured by using a synthetic substrate (47, 48) was reduced by 30% in extracts of oga-1(ok1207) compared with the WT strain, consistent with total loss of OGA activity (data not shown).
Several methods were used to examine the effects of the oga-1(ok1207) mutant allele on O-GlcNAc abundance in the homozygous deletion strain. Levels of O-GlcNAc in oga-1(ok1207) animals were compared with WT and with ogt-1(ok430), encoding OGT-1Δ (Fig. 1D) (26). The levels of O-GlcNAc on nuclear pore proteins were assessed by immunofluorescence by using the O-GlcNAc-specific RL2 antibody (Fig. 2A). The ogt-1(ok430) mutant lacked any detectable level of O-GlcNAc on nuclear pores, whereas the oga-1(ok1207) mutant was slightly higher than WT. The monoclonal antibody CTD110 revealed more dramatic changes in the O-GlcNAc levels between WT and oga-1(ok1207) and recognized nothing in the ogt-1(ok430) mutants (Fig. 2B). Staining with CTD110, when detectable, was observed in a punctate pattern at the nuclear periphery in all tissues throughout development.
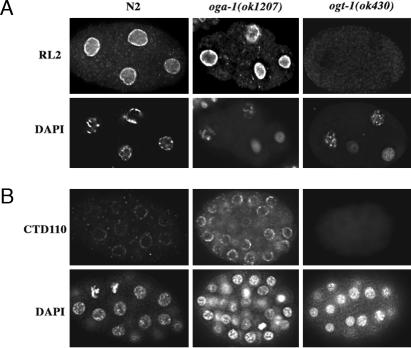
Fig. 2.
Distribution of O-GlcNAc in WT and mutant embryos. The distribution of O-GlcNAc was assayed in fixed embryos by using antibodies RL2 and CTD110 that recognize O-GlcNAc modified epitopes. Fixation and antibody staining was performed as described (26). (A) Staining of 4-cell embryos with RL2 revealed heavy perinuclear staining in WT and oga-1(ok1207) mutant embryos; no staining was detected in ogt-1(ok430) embryos, as reported (26). (B) Staining of 16- to 48-cell embryos with CTD110 revealed punctate perinuclear staining that was faint in WT (N2) embryos and strong in oga-1(ok1207); no staining was detectable in ogt-1(ok430) or the ogt-1(ok430);oga-1(ok1207) double mutant. For all RL2 and CTD110 panels, the corresponding image for DNA staining with DAPI is shown immediately below.
To determine whether the altered levels of O-GlcNAc modifications in the mutant strains interfered with nuclear transport, we assayed the distribution of three transcription factors (HLH-1, HLH-2, and LIN-26) that have different temporal and spatial distribution patterns during embryogenesis. No difference in the nuclear accumulation of these transcription factors was observed between WT and oga-1(ok1207) or ogt-1(ok430) embryos (data not shown and ref. 26).
The levels of O-GlcNAc in each strain were quantified by immunoblotting by using a fluorescence-based detector as described in Materials and Methods. As shown in Fig. 3A Left, O-GlcNAc levels were significantly higher in the oga-1(ok1207) mutant strain than in the WT strain. The two major bands detected by RL2 were between 4.4- and 6.4-fold higher in the oga-1(ok1207) mutant strain as compared with WT. As previously reported, no O-GlcNAc was detectable in the ogt-1(ok430) strain (26). Quantitatively similar results were obtained with CTD110 (data not shown). As a loading control, blots were probed with RL1, an anti-nucleoporin antibody, and similar levels of nucleoporins were detected (Fig. 3A Right). The 82-kDa band detected by RL1 only in the ogt-1(ok430) mutant likely represents a nucleoporin whose mobility has shifted because of a lack of O-GlcNAc. The increased intensity of some of the lower molecular weight bands in the oga-1(ok1207) and ogt-1(ok430) mutants could be due to altered protein expression, stability or proteolysis (1, 2).
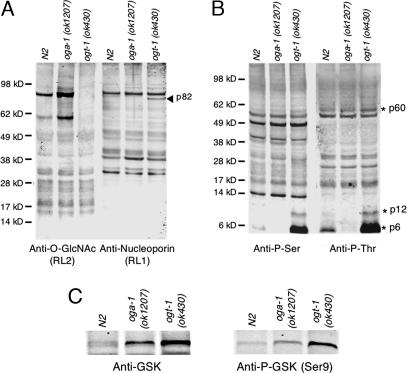
Fig. 3.
Altered levels of O-GlcNAc correlate with changes in phosphoprotein profiles. Shown is a comparison of lysates from WT (N2), oga-1(ok1207), and ogt-1(ok430) animals. Samples were resolved by SDS/PAGE and examined by Western blot analysis. (A) Blots were probed with a monoclonal antibody specific for O-GlcNAc-modified nucleoporins (RL2) and a monoclonal antibody specific for nucleoporins (RL1). Molecular mass standards are shown on the left. Altered migration because of the absence of O-GlcNAc on p82 is indicated by the arrowhead. (B) Phospho-serine (Anti-P-Ser) and phospho-threonine (Anti-P-Thr) antibodies were also used to detect differences in phosphorylation between the strains. Asterisks show migration of three proteins (p60, p12, and p6) with changes in apparent phosphorylation levels. In control experiments, phosphatase treatment reduced the observed levels of Ser- and Thr-phosphorylation by 70% and 66%, respectively. (C) Elevation of GSK protein (Left) and phospho-GSK (Right) levels in the mutant strains are shown. Data are representative of three separate experiments.
Many kinases are modified by O-GlcNAc, and we sought to determine the impact of blocked O-GlcNAc cycling on Ser- or Thr-phosphorylation profiles (1, 2). As shown by quantitative immunoblotting in Fig. 3B, the loss of either oga-1 or ogt-1 in C. elegans resulted in changes in Ser- and Thr-phosphorylation profiles. Phosphatase treatment reduced immunoreactivity, confirming the specificity of the phospho-antibodies. The apparent abundance of three phosphoprotein bands (p60, p12, and p6) was elevated between 1.4- and 9-fold with the loss of ogt-1. Interestingly, these bands were reduced and nearly undetectable in oga-1(ok1207). The observed changes in phosphoprotein profiles in the mutant strains may reflect different amounts of the target protein and/or their levels of phosphorylation. Intriguingly, these phosphoproteins do not correspond to RL2-positive proteins, suggesting that the altered phosphorylation is not simply explained by O-GlcNAc occupancy of available sites. We also observed a dramatic change in the levels of one of the key targets of insulin signaling, glycogen synthase kinase 3 (GSK-3) (Fig. 3C). With GSK-3, both protein and phosphoprotein levels were increased by 2.2-fold in ogt-1(ok430) and 1.3-fold in oga-1(ok1207).
We demonstrated an alteration in the levels of macronutrients stored in the ogt-1(ok430) knockout (26). As shown in Fig. 4, the OGA knockout oga-1(ok1207) also alters the levels of several macronutrients. Interestingly, the levels of both trehalose and glycogen were elevated in oga-1(ok1207) but were still lower than the very high levels observed in ogt-1(ok430) (Fig. 4A). The elevated storage of glycogen occurred in a subset of gut granules in N2, oga-1(ok1207), and ogt-1(ok430) that were readily detected by using carminic acid, a fluorescent reporter we have adapted for use in C. elegans (Fig. 4B) (26).
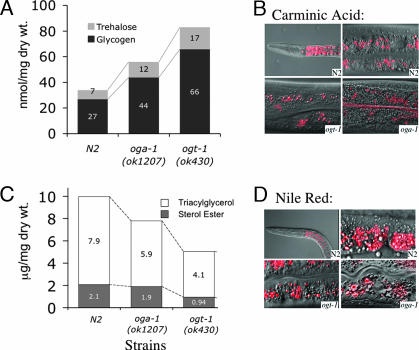
Fig. 4.
Macronutrient storage in WT and O-GlcNAc cycling mutant strains. (A) Levels of the circulating disaccharide trehalose and glycogen were analyzed in WT and mutant strains as described in Materials and Methods. The results represent the averages of three separate experiments. For trehalose, the values (in nmol/mg dry weight) were as follows: 7 nmol/mg (SD = 1.6), 12 nmol/mg (SD = 2.1), and 17 nmol/mg (SD = 2.4) for N2, oga-1(ok1207), and ogt-1(ok430), respectively. For glycogen, the values (in nmol/mg dry weight) were: 27 nmol/mg (SD = 3.2), 44 nmol/mg (SD = 4.1), and 66 nmol/mg (SD = 4.8) for N2, oga-1(ok1207), and ogt-1(ok430), respectively. The ogt-1(ok430);oga-1(ok1207) double was nearly identical to ogt-1(ok430) in these assays. (B) Carminic acid was detected by fluorescence microscopy and was predominantly found in the intestine of the animal. The carminic acid-positive structures (red) are a distinct subset of granules in the intestinal cells in N2 (Upper Left, ×40 objective; Upper Right, ×100 objective), ogt-1(ok430) (Lower Left, ×100 objective), and oga-1(ok1207) (Lower Right, ×100 objective). Composites of fluorescence and Nomarski images are shown. (C) The levels of triacylglycerol and sterol ester in WT and mutant strains were assayed as described in Materials and Methods. Presented are average values from duplicate determinations that differed by no more than 4% of the total lipid mass. The ogt-1(ok430);oga-1(ok1207) double mutant was very similar to ogt-1(ok430). (D) Nile red was used to label lipid droplets of the intestinal cells. A composite of fluorescent and Nomarski image is shown for N2 (Upper Left, ×40 objective; Upper Right, ×100 objective), ogt-1(ok430) (Lower Left, ×100 objective), and oga-1(ok1207) (Lower Right, ×100 objective).
The storage of neutral lipids was also examined in strains with defects in O-GlcNAc cycling. Compared with N2, the levels of neutral lipids were decreased ≈40% in oga-1(ok1207) and 70% in ogt-1(ok430) (Fig. 4C). Although reduced in amount, compared with N2, the storage of neutral lipids by oga-1(ok1207) and ogt-1(ok430) was readily detected in a subset of gut granules by using Nile red, a fluorescent reporter of lipid droplets (Fig. 4D).
We examined the impact of inhibition of O-GlcNAc cycling on levels of UDP-GlcNAc. The amounts of UDP-HexNAc in the three strains were 0.5, 0.8, and 0.42 (nmol/mg dry weight) in WT, ogt-1(ok430), and oga-1(ok1207), respectively. The UDP-HexNAc pool consisted of a fixed ratio of UDP-GlcNAc to UDP-GalNAc of ≈2.1 in all strains. UDP-GlcNAc pools were elevated 30–40% in the ogt-1(ok430) mutant compared with WT and oga-1(ok1207), indicating that blocked UDP-GlcNAc utilization by ogt-1(ok430) produces a buildup of UDP-HexNAc.
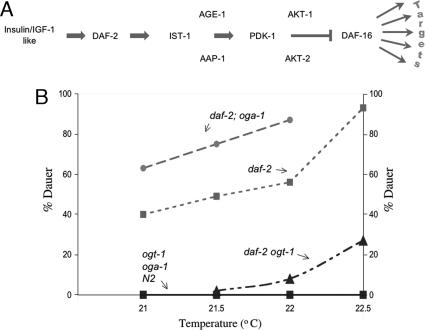
In C. elegans, insulin-like signaling modulates the storage of macronutrients in gut granules and other tissues (49–56) and regulates dauer larvae development (51, 54). This pathway is also linked to lifespan extension and stress responses (57–59). Insulin-like signaling functions through DAF-16, a homolog of the mammalian transcription factor Foxo1, which acts on many targets (Fig. 5A). Reduction in insulin-like signaling activates DAF-16, inducing dauer development and changes in macronutrient storage. The daf-2(e1370) temperature-sensitive constitutive dauer allele of the insulin-like receptor was used to modulate insulin-like signaling (60). We used the daf-2(e1370) allele, either alone or in combination with the oga-1 and ogt-1 mutant alleles, to provide a sensitive assay for perturbations of the signaling pathway regulating dauer formation. In the semipermissive range, the effects of disrupted O-GlcNAc cycling could be assayed by measuring the percentage of animals that became dauers (Fig. 5B). Over the range of temperatures tested, no dauers were formed in WT, oga-1(ok1207), and ogt-1(ok430) animals. However, the daf-2(e1370) strain produced an increasing percentage of dauer larvae at elevated temperatures, as expected (26). In the daf-2(e1370);oga-1(ok1207) double mutant background, dauer formation was significantly enhanced at each temperature compared with daf-2(e1370) alone. This result suggests that the oga-1(ok1207)-null allele blunts signaling pathways normally inhibiting DAF-16 activity. As we previously reported, the daf-2(e1370) ogt-1(ok430) double mutant significantly lowered dauer formation at each temperature compared with daf-2(e1370) alone (26). The ogt-1(ok430)-null is, therefore, hypersensitive to signaling that normally inhibits DAF-16 activity. These findings show that both of the enzymes of O-GlcNAc cycling contribute to the regulation of dauer formation in C. elegans.
Fig. 5.
Loss of O-GlcNAc cycling leads to altered dauer formation in O-GlcNAc cycling mutant strains. (A) Key components of the insulin-like signaling pathway regulating dauer formation in C. elegans are shown. Insulin-like ligands bind to DAF2 (insulin-like receptor) activating a series of kinases culminating in suppression of the DAF-16 transcription factor. DAF-16 regulates a variety of target genes including oga-1 and genes involved in the stress response, energy storage, longevity, and dauer formation. (B) Functional deletion of either of the O-GlcNAc cycling enzymes alters dauer formation in the temperature-sensitive daf-2(e1370) genetic background. The six strains, as indicated above each line, were examined at several temperatures that are semipermissive for the constitutive dauer mutant allele daf-2(e1370) (dotted line with squares). The daf-2(e1370);oga-1(ok1207) double mutation is shown to enhance dauer formation (dashed line with circles), and the daf-2(e1370) ogt-1(ok430) double mutation suppresses dauer formation (dashed–dotted line with triangles). Under conditions where daf-2(e1370) produced 82% dauer, both daf-2(e1370) ogt-1(ok430) and daf-2(e1370) ogt-1(ok430);oga-1(ok1207) produced ≈30% dauer (data not shown). Thus, ogt-1(ok430) appears epistatic to oga-1(1207) in its influence on dauer. Dauer formation was not detected in WT (N2), ogt-1(ok430), or oga-1(ok1207) alone (solid line with squares).
Discussion
OGA-1 Modulates Signaling and Macronutrient Storage.
The identification of a viable and fertile C. elegans OGA knockout provides a means of directly assessing the roles of this glycosidase in metabolism and animal physiology. The C. elegans oga-1(ok1207) mutant exhibits increased levels of O-GlcNAc on nuclear pores and other cellular targets and alterations in macronutrient storage and intracellular signaling. In mammals, reduction of O-GlcNAcase activity produces insulin resistance in tissue culture and isolated tissues (39, 40, 61). These findings, coupled with our previous findings that OGT overexpression produces insulin resistance in mice (17), demonstrate a role for O-GlcNAc cycling in metabolism and insulin signaling. Consistent with these findings, the human O-GlcNAcase gene, MGEA5, was shown to be a type 2 diabetes susceptibility locus in the Mexican-American population (43).
Many kinases are O-GlcNAc-modified, and an interrelationship between O-GlcNAc modification and phosphorylation of target proteins has been suggested (1, 62, 63). The C. elegans OGT-1 and OGA-1 knockouts allow a global analysis of changes in phosphorylation accompanying loss of O-GlcNAc turnover. We detected significant changes in only a few phosphoproteins that were not themselves heavily O-GlcNAc-modified. The subtle changes in phosphoprotein profiles accompanying the dramatic loss of O-GlcNAc cycling in ogt-1(ok430) and oga-1(ok1207) argue that only a small subset of kinases and/or their targets are normally modified by O-GlcNAc in C. elegans. One kinase that we have shown to be O-GlcNAc-modified is GSK-3 (64). Using anti-GSK-3 antibodies, we found that both the ogt-1(ok430) and oga-1(ok1207) mutations led to an elevation of the levels of GSK-3. In mammals, GSK-3β is both glycosylated (64) and phosphorylated. Insulin action stimulates the phosphatidylinositol 3-kinase (PI3-kinase) pathway, resulting in Akt activation that phosphorylates and inactivates GSK-3β (65, 66). Glycogen synthase, a key target of GSK-3β, is activated by dephosphorylation to increase glycogen synthesis (65–68). The observed increases in phosphorylated GSK-3 may be causally linked to the higher glycogen levels in the ogt-1(ok430) and oga-1(ok1207) mutant strains. Alternatively, the elevation in GSK-3 may be a compensatory response to the elevated glycogen in those strains. Intriguingly, GSK-3 has also been linked to the oxidative stress response in C. elegans (69).
The circulating disaccharide trehalose accumulated to significantly higher levels in both the ogt-1(ok430) and oga-1(ok1207) mutant strains compared with WT. In C. elegans, trehalose functions in sugar transport, energy storage, and protection from environmental stress (70). Transcriptional regulation through daf-16 is one of the mechanisms by which trehalose and glycogen synthesis are regulated in C. elegans (59, 71). However, both ogt-1(ok430) and oga-1(ok1207) strains modulate trehalose and glycogen levels under conditions where dauer larvae formation does not occur, suggesting a more complex regulation of trehalose levels in response to disruption of O-GlcNAc cycling.
Neutral lipid stores were significantly reduced in both ogt-1(ok430) and oga-1(ok1207) strains. Enzymes of lipid storage are transcriptionally responsive to insulin-like signaling in C. elegans, and fat stores increase in dauer larvae (72–76). Our findings suggest that active cycling of O-GlcNAc by both OGT-1 and OGA-1 are required to maintain normal fat stores in the worm. However, it is not yet clear whether the reduced storage seen in the mutant strains is due to decreased fat synthesis or increased lipolysis. In human type 2 diabetes, elevated plasma fatty acid levels result from enhanced lypolysis in adipocytes (77–79). The availability of C. elegans strains defective in O-GlcNAc cycling may allow a further examination of the complex metabolic changes associated with fat storage during normal and dauer development.
Enzymes of O-GlcNAc Cycling and Cellular Signaling Cascades Influencing Dauer.
The ability of both the oga-1(ok1207) and ogt-1(ok430) mutations to modulate dauer formation in C. elegans argues that active cycling of O-GlcNAc functions in the regulatory program leading to dauer formation. The oga-1(ok1207) mutation enhanced dauer formation in the sensitized daf-2 mutant background. This resistance to insulin-like signaling in the OGA-1 knockout is highly reminiscent of studies in mammalian cells where enhanced O-GlcNAc levels induced by OGT overexpression (17) or treatment with OGA inhibitors produce insulin resistance (17, 39, 40, 61, 80). In contrast, the ogt-1(ok430) mutation displayed hypersensitivity to insulin-like signaling in the dauer assay, suggesting that OGT-1 normally acts to blunt insulin-like signaling (26). OGT is the terminal step in the nutrient-sensing hexosamine-signaling pathway (4). Because UDP-GlcNAc synthesis is driven by nutrient availability, the pathway acts as a cellular detector of nutrient status. In the worm, food availability triggers insulin-like ligand release and activation of a signaling cascade producing normal lifespan and reproduction. With restricted food availability, insulin signaling is depressed, resulting in transcriptional activation of sets of genes involved in lifetime extension and inhibition of reproduction. Our findings suggest an intersection between the hexosamine- and insulin-mediated nutrient-sensing systems in the nematode. The enzymes of O-GlcNAc cycling act by “fine-tuning” the insulin-like signaling cascade, providing an additional level of metabolic control in response to nutrient availability. This fine-tuning mechanism is likely to apply to other proposed functions of O-GlcNAc cycling in the nematode, which include lifespan control, response to environmental stress, proteasomal regulation and transcription/translational control (4, 26). The C. elegans mutants in O-GlcNAc cycling may allow an examination of the potential role of the hexosamine-signaling pathway in mediating the well established link between nutrient deprivation and lifespan extension that has been observed from nematodes to mammals.
A Genetic Model of Insulin Resistance and Type 2 Diabetes Mellitus.
Recent findings suggest that the OGA locus (MGEA5) is a diabetes susceptibility locus in man. A single nucleotide polymorphism in intron 10 of the MGEA5 gene is associated with type 2 diabetes and age of onset in the Mexican-American population. This polymorphism may interfere with OGA activity in the susceptible population (43). This important finding links the human disease to a large body of literature implicating the hexosamine-signaling pathway and O-GlcNAc cycling to insulin resistance and diabetes mellitus (reviewed in refs. 1, 4, and 63). The knockout of OGA-1 in C. elegans produces a phenotype with striking parallels to human type 2 diabetes. The knockout has higher circulating levels of trehalose, higher levels of glycogen, and decreased fat stores. In addition, it exhibits resistance to the insulin-like signaling pathway. No other animal knockout of OGA has been reported, making C. elegans the first organism in which a knockout allele of OGA can be analyzed. The extensive genetic analysis previously carried out on the insulin-like signaling cascade in the nematode offers clear advantages for understanding how interference with O-GlcNAc cycling may impact cellular physiology.
Materials and Methods
Strains.
The following strains were used in this study: WT N2 Bristol; RB1169 [oga-1(ok1207)] and RB653 [ogt-1(ok430)] generated and kindly provided by the C. elegans Gene Knockout Consortium (Oklahoma Medical Research Foundation, Oklahoma City, OK); KM395 oga-1(ok1207) backcrossed five times [referred to as oga-1(ok1207)]; KM258 ogt-1(ok430) backcrossed four times [referred to as ogt-1(ok430)]; and KM396, a product of crossing KM395 to KM258 [referred to as ogt-1(ok430);oga-1(ok1207)]. KM396 was phenotypically similar to KM258 (see legends to Figs. 2, 4, and 5). KM258, KM395, and KM396 were each crossed to daf-2(e1370) and are referred to as daf-2(e1370) ogt-1(ok430), daf-2(e1370);oga-1(ok1207), and daf-2(e1370) ogt-1(ok430);oga-1(ok1207), respectively.
To generate these double and triple mutant strains, the presence of the oga-1(ok1207) and ogt-1(ok430) allele was verified by using nested PCR primers as described by the C. elegans Gene Knockout Consortium. Primers MF26 (ATCTCTACGCACCCAAAGATG) and MF14 (AACGTCGGAACCGCTCGTGAC) were used to identify animals that are heterozygous for the oga-1(ok1207) allele. The ogt-1(ok430) allele primer sets have been described (26) and MWK454 (GGGTGACACCTCGGCAGCAATCGCATG) was used to distinguish heterozygotes. Homozygosity of the daf-2(e1370) allele was confirmed by assaying dauers at restrictive temperature.
Characterization of oga-1 Transcripts.
Independent cDNA clones of oga-1 were generated by using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA), followed by amplification with primers MF01 (cgagatctaATGGAGCAAACAGAAAGC) and MF02 (ctagatcttCACTCTGAAGTAGAAG). Resulting cDNAs were subcloned into the pCR 2.1-TOPO vector (Invitrogen) and sequenced.
Bacterial Expression and O-GlcNAcase Assays.
Human OGA, C. elegans isoform b (See Fig. 1) of OGA-1, and the predicted deletion OGA-1Δ were each cloned into the PET43.1(EK/LIC) expression system by using the following PCR primers: sense, 5′-GACGACGACAAGATCGAGCAAACAGAAAGCTTCGATCGGCGAAAGGCTGTGCAAAAC-3′; antisense, 5′-GAGGAGAAGCCCGGTGCCTCAATTTGAATTTCTCTTTAACGGTTGAAAAAACAATCCTAGTCCGTAGAAGCCTTCAAC-3′. The C. elegans OGA-1, OGA-1Δ, and human OGA expression vectors were transformed into BL21(DE3) cells (Novagen, San Diego, CA). Bacterial extracts were prepared as described (7). The amounts of expressed protein used in the activity assay were normalized to the OGA-1 protein level measured by immunoblotting with anti-His-6 antibody (as described below). Assays were performed in 100 mM citrate phosphate (pH 6.5) and 10 μM FD-GlcNAc (fluorescein di-N-acetyl-β-d-glucosaminide) (47, 48) in a total volume of 100 μl. Samples were incubated at room temperature for 1 h and quenched with 900 μl of 0.5 M NaCO3. Fluorescence was measured on 200 μl (excitation, 485 nm; emission, 535 nm) by using a Victor 96 multiwell fluorescence reader (PerkinElmer, Wellesley, MA).
Immunoblotting and Immunofluorescence Microscopy.
Immunoblotting was carried out as described (26). Blots were imaged and quantitated by using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) according to the manufacturer’s instruction. Antibodies used were RL1 and RL2 (Affinity BioReagents, Golden, CO), CTD110 (CTD110.6; Covance Research Products, Denver, PA), rabbit polyclonal antibodies specific for phospho-serine and phospho-threonine (Research Diagnostics Inc., Flanders, NJ), and rabbit polyclonal antibodies directed against GSKβ and GSKβ (ser9) (Cell Signaling Technology, Danvers, MA). The 680-nm-conjugated secondary antibodies (Invitrogen) and the 800-nm-conjugated secondary antibodies (Rockland, Gilbertsville, PA) were diluted 1 in 10,000. Protein blots were dephosphorylated by treating with 1,000 units of calf intestinal phosphatase (New England Biolabs, Ipswich, MA) overnight with gentle shaking at 37°C.
Macronutrient Measurements.
Levels of trehalose and glycogen were measured as described (26). Carminic acid staining of glycogen and Nile red staining of neutral lipid containing gut-granules were carried out by feeding as described (26). Levels of neutral lipids were determined as described (26). Sugar nucleotide levels were measured after extraction of lyophilized worm pellets with 0.75 ml of cold 0.5 M perchloric acid followed by microcentrifugation at maximum speed for 10 min. After charcoal adsorption, eluted sugar nucleotides were made in 0.1 M HCl and heated to 95°C for 10 min. Released monosaccharides were assayed as described (26).
Dauer Assay.
Embryos from 20 to 30 gravid adults laid at 16°C overnight were incubated at the indicated temperatures and scored for dauer phenotype up to 5 days later. Incubation and accurate temperature monitoring was as described (26).
Acknowledgments
We acknowledge Dr. Tetsu Fukushige for assistance with dauer assays. This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Abbreviations
- O-GlcNAc
O-linked N-acetylglucosamine
- OGT
O-linked GlcNAc transferase
- OGA
O-GlcNAcase
- GSK-3
glycogen synthase kinase 3.
Footnotes
References
- 1.Hanover J. A. FASEB J. 2001;15:1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- 2.Wells L., Hart G. W. FEBS Lett. 2003;546:154–158. doi: 10.1016/s0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- 3.Wells L., Whalen S. A., Hart G. W. Biochem. Biophys. Res. Commun. 2003;302:435–441. doi: 10.1016/s0006-291x(03)00175-x. [DOI] [PubMed] [Google Scholar]
- 4.Love D. C., Hanover J. A. Sci. STKE. 2005 doi: 10.1126/stke.3122005re13. 2005, re13. [DOI] [PubMed] [Google Scholar]
- 5.Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A. J. Cell Sci. 2003;116:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 6.Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., Love D. C. Arch. Biochem. Biophys. 2003;409:287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 7.Lazarus B. D., Roos M. D., Hanover J. A. J. Biol. Chem. 2005;280:35537–35544. doi: 10.1074/jbc.M504948200. [DOI] [PubMed] [Google Scholar]
- 8.Heckel D., Comtesse N., Brass N., Blin N., Zang K. D., Meese E. Hum. Mol. Genet. 1998;7:1859–1872. doi: 10.1093/hmg/7.12.1859. [DOI] [PubMed] [Google Scholar]
- 9.Comtesse N., Maldener E., Meese E. Biochem. Biophys. Res. Commun. 2001;283:634–640. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- 10.Marshall S., Bacote V., Traxinger R. R. J. Biol. Chem. 1991;266:10155–10161. [PubMed] [Google Scholar]
- 11.Marshall S., Bacote V., Traxinger R. R. J. Biol. Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 12.Marshall S., Garvey W. T., Traxinger R. R. FASEB J. 1991;5:3031–3036. doi: 10.1096/fasebj.5.15.1743436. [DOI] [PubMed] [Google Scholar]
- 13.Traxinger R. R., Marshall S. J. Biol. Chem. 1991;266:10148–10154. [PubMed] [Google Scholar]
- 14.Traxinger R. R., Marshall S. J. Biol. Chem. 1992;267:9718–9723. [PubMed] [Google Scholar]
- 15.Toleman C., Paterson A. J., Whisenhunt T. R., Kudlow J. E. J. Biol. Chem. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- 16.Toleman C. A., Paterson A. J., Kudlow J. E. J. Biol. Chem. 2005;281:3918–3925. doi: 10.1074/jbc.M510485200. [DOI] [PubMed] [Google Scholar]
- 17.McClain D. A., Lubas W. A., Cooksey R. C., Hazel M., Parker G. J., Love D. C., Hanover J. A. Proc. Natl. Acad. Sci. USA. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D., Juarez S., Hartweck L., Alamillo J. M., Simon-Mateo C., Perez J. J., Fernandez-Fernandez M. R., Olszewski N. E., Garcia J. A. J. Virol. 2005;79:9381–9387. doi: 10.1128/JVI.79.15.9381-9387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenboim-Wainberg Y., Maymon I., Borochov R., Alvarez J., Olszewski N., Ori N., Eshed Y., Weiss D. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartweck L. M., Scott C. L., Olszewski N. E. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izhaki A., Swain S. M., Tseng T. S., Borochov A., Olszewski N. E., Weiss D. Plant J. 2001;28:181–190. doi: 10.1046/j.1365-313x.2001.01144.x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen S. E., Binkowski K. A., Olszewski N. E. Proc. Natl. Acad. Sci. USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olszewski N., Sun T. P., Gubler F. Plant Cell. 2002;14(Suppl):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swain S. M., Tseng T. S., Thornton T. M., Gopalraj M., Olszewski N. E. Plant Physiol. 2002;129:605–615. doi: 10.1104/pp.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lubas W. A., Frank D. W., Krause M., Hanover J. A. J. Biol. Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- 26.Hanover J. A., Forsythe M. E., Hennessey P. T., Brodigan T. M., Love D. C., Ashwell G., Krause M. Proc. Natl. Acad. Sci. USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells L., Vosseller K., Hart G. W. Cell. Mol. Life Sci. 2003;60:222–228. doi: 10.1007/s000180300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buse M. G., Robinson K. A., Gettys T. W., McMahon E. G., Gulve E. A. Am. J. Physiol. 1997;272:E1080–E1088. doi: 10.1152/ajpendo.1997.272.6.E1080. [DOI] [PubMed] [Google Scholar]
- 29.Buse M. G., Robinson K. A., Marshall B. A., Mueckler M. J. Biol. Chem. 1996;271:23197–23202. doi: 10.1074/jbc.271.38.23197. [DOI] [PubMed] [Google Scholar]
- 30.Hawkins M., Barzilai N., Liu R., Hu M., Chen W., Rossetti L. J. Clin. Invest. 1997;99:2173–2182. doi: 10.1172/JCI119390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebert L. F. J., Daniels M. C., Zhou J., Crook E. D., Turner R. L., Simmons S. T., Neidigh J. L., Zhu J. S., Baron A. D., McClain D. A. J. Clin. Invest. 1996;98:930–936. doi: 10.1172/JCI118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossetti L., Hawkins M., Chen W., Gindi J., Barzilai N. J. Clin. Invest. 1995;96:132–140. doi: 10.1172/JCI118013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Liu R., Hawkins M., Barzilai N., Rossetti L. Nature. 1998;393:684–688. doi: 10.1038/31474. [DOI] [PubMed] [Google Scholar]
- 34.Roos M. D., Xie W., Su K., Clark J. A., Yang X., Chin E., Paterson A. J., Kudlow J. E. Proc. Assoc. Am. Physicians; 1998. pp. 422–432. [PubMed] [Google Scholar]
- 35.Hanover J. A., Lai Z., Lee G., Lubas W. A., Sato S. M. Arch. Biochem. Biophys. 1999;362:38–45. doi: 10.1006/abbi.1998.1016. [DOI] [PubMed] [Google Scholar]
- 36.Gao Y., Parker G. J., Hart G. W. Arch. Biochem. Biophys. 2000;383:296–302. doi: 10.1006/abbi.2000.2094. [DOI] [PubMed] [Google Scholar]
- 37.Akimoto Y., Kreppel L. K., Hirano H., Hart G. W. Diabetologia. 2000;43:1239–1247. doi: 10.1007/s001250051519. [DOI] [PubMed] [Google Scholar]
- 38.Liu K., Paterson A., Konrad R., Parlow A., Jimi S., Roh M., Chin E., Kudlow J. Mol. Cell. Endocrinol. 2002;194:135. doi: 10.1016/s0303-7207(02)00155-7. [DOI] [PubMed] [Google Scholar]
- 39.Arias E. B., Kim J., Cartee G. D. Diabetes. 2004;53:921–930. doi: 10.2337/diabetes.53.4.921. [DOI] [PubMed] [Google Scholar]
- 40.Park S. Y., Ryu J., Lee W. Exp. Mol. Med. 2005;37:220–229. doi: 10.1038/emm.2005.30. [DOI] [PubMed] [Google Scholar]
- 41.Perreira M., Kim E. J., Thomas C. J., Hanover J. A. Bioorg. Med. Chem. 2005;14:837–846. doi: 10.1016/j.bmc.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Goldberg H. J., Whiteside C. I., Hart G. W., Fantus I. G. Endocrinology. 2006;147:222–231. doi: 10.1210/en.2005-0523. [DOI] [PubMed] [Google Scholar]
- 43.Lehman D. M., Fu D. J., Freeman A. B., Hunt K. J., Leach R. J., Johnson-Pais T., Hamlington J., Dyer T. D., Arya R., Abboud H., et al. Diabetes. 2005;54:1214–1221. doi: 10.2337/diabetes.54.4.1214. [DOI] [PubMed] [Google Scholar]
- 44.Shafi R., Iyer S. P., Ellies L. G., O’Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. Proc. Natl. Acad. Sci. USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donnell N., Zachara N. E., Hart G. W., Marth J. D. Mol. Cell. Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. J. Biol. Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 47.Kim E. J., Kang D. O., Love D. C., Hanover J. A. Carbohydr. Res. 2006;341:971–982. doi: 10.1016/j.carres.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim E. J., Perreira M., Thomas C. J., Hanover J. A. J. Am. Chem. Soc. 2006;128:4234–4235. doi: 10.1021/ja0582915. [DOI] [PubMed] [Google Scholar]
- 49.Cypser J. R., Johnson T. E. Biogerontology. 2003;4:203–214. doi: 10.1023/a:1025138800672. [DOI] [PubMed] [Google Scholar]
- 50.Dorman J. B., Albinder B., Shroyer T., Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gottlieb S., Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holt S. J., Riddle D. L. Mech. Ageing Dev. 2003;124:779–800. doi: 10.1016/s0047-6374(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 53.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 54.Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 55.Rea S., Johnson T. E. Dev. Cell. 2003;5:197–203. doi: 10.1016/s1534-5807(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 56.Tissenbaum H. A., Ruvkun G. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Libina N., Berman J. R., Kenyon C. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 58.Lin K., Hsin H., Libina N., Kenyon C. Nat. Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 59.Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 60.Riddle D. L. Stadler Genet. Symp. 1977;9:101–120. [Google Scholar]
- 61.Vosseller K., Wells L., Lane M. D., Hart G. W. Proc. Natl. Acad. Sci. USA. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hart G. W., Greis K. D., Dong L. Y., Blomberg M. A., Chou T. Y., Jiang M. S., Roquemore E. P., Snow D. M., Kreppel L. K., Cole R. N., et al. Adv. Exp. Med. Biol. 1995;376:115–123. [PubMed] [Google Scholar]
- 63.Wells L., Vosseller K., Hart G. W. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 64.Lubas W. A., Hanover J. A. J. Biol. Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 65.Cohen P., Frame S. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 66.Frame S., Cohen P. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole A., Frame S., Cohen P. Biochem. J. 2004;377:249–255. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harwood A., Braga V. M. Nat. Cell Biol. 2003;5:275–277. doi: 10.1038/ncb0403-275. [DOI] [PubMed] [Google Scholar]
- 69.An J. H., Vranas K., Lucke M., Inoue H., Hisamoto N., Matsumoto K., Blackwell T. K. Proc. Natl. Acad. Sci. USA. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pellerone F. I., Archer S. K., Behm C. A., Grant W. N., Lacey M. J., Somerville A. C. Int. J. Parasitol. 2003;33:1195–1206. doi: 10.1016/s0020-7519(03)00173-5. [DOI] [PubMed] [Google Scholar]
- 71.Lamitina S. T., Strange K. Am. J. Physiol. 2005;288:C467–C474. doi: 10.1152/ajpcell.00451.2004. [DOI] [PubMed] [Google Scholar]
- 72.Ambros V. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 73.Chiang S. H., MacDougald O. A. Trends Genet. 2003;19:523–525. doi: 10.1016/j.tig.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 75.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 76.Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- 77.DeFronzo R. A. Int. J. Clin. Pract. 2004;58(Suppl. 143):9–21. doi: 10.1111/j.1368-504x.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 78.Nawrocki A. R., Scherer P. E. Curr. Opin. Pharmacol. 2004;4:281–289. doi: 10.1016/j.coph.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 79.Wyne K. L. Am. J. Med. 2003;115(Suppl. 8A):29S–36S. doi: 10.1016/j.amjmed.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 80.Buse M. G., Robinson K. A., Marshall B. A., Hresko R. C., Mueckler M. M. Am. J. Physiol. 2002;283:E241–E250. doi: 10.1152/ajpendo.00060.2002. [DOI] [PubMed] [Google Scholar]