Abstract
Information processing pathways such as DNA replication are conserved in eukaryotes and archaea and are significantly different from those found in bacteria. Single-stranded DNA-binding (SSB) proteins (or replication protein A, RPA, in eukaryotes) play a central role in many of these pathways. However, whilst euryarchaea have a eukaryotic-type RPA homologue, crenarchaeal SSB proteins appear much more similar to the bacterial proteins, with a single OB fold for DNA binding and a flexible C-terminal tail that is implicated in protein–protein interactions. We have determined the crystal structure of the SSB protein from the crenarchaeote Sulfolobus solfataricus to 1.26 Å. The structure shows a striking and unexpected similarity to the DNA-binding domains of human RPA, providing confirmation of the close relationship between archaea and eukaryotes. The high resolution of the structure, together with thermodynamic and mutational studies of DNA binding, allow us to propose a molecular basis for DNA binding and define the features required for eukaryotic and archaeal OB folds.
Keywords: archaea/OB fold/protein structure/RPA/SSB protein
Introduction
The duplex structure of DNA confers enhanced protection and stability to its component nucleic acid strands. However, DNA duplexes are frequently distorted, melted or unwound, as a result of both DNA damage and programmed cellular pathways such as DNA replication and transcription. Whenever single-stranded DNA (ssDNA) is formed, ssDNA-binding (SSB) proteins function to protect and sequester the single-stranded regions until the double helix can be reformed (Chase, 1986; Wold, 1997).
SSB proteins are identified by the presence of an OB fold (oligonucleotide/oligosaccharide/oligopeptide binding fold) (Murzin, 1993), the principal DNA-binding unit, typically comprising 100 amino acids. The subunit composition of SSB proteins varies over different domains of life (Figure 1A). Bacterial SSB proteins have a single OB fold per polypeptide and form homotetramers, whereas eukaryotic SSB proteins (known as RPA, replication protein A) have a heterotrimeric structure. Archaea are split between the two major subdivisions: euryarchaea and crenarchaea. The former have a eukaryotic-type RPA (Chedin et al., 1998), and the latter resemble the bacterial SSB proteins in terms of domain organization (Wadsworth and White, 2001) (Figure 1A). The presence of the OB fold across all domains of life (and viruses) suggests that SSB proteins arose, through gene duplication events, from a common, ancestral SSB protein (Philipova et al., 1996; Suck, 1997).
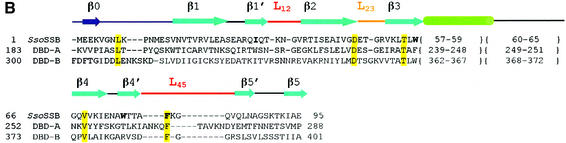
Fig. 1. (A) Domain organization of SSB proteins in eukaryotes, bacteria and archaea. Eukaryotes have a heterotrimeric SSB protein (RPA), with six OB folds (blue boxes), four of which participate in DNA binding. The third DNA-binding domain of RPA70 (DBD-C) has an insertion in the form of a zinc-binding domain (yellow box). In contrast, bacteria have a homotetrameric SSB protein with a single OB fold in each polypeptide. The C-terminal third of EcoSSB is a highly flexible ‘tail’ (green box) with an acidic terminus (red box) that does not participate in DNA binding but plays a role in mediating protein–protein interactions, essential for the recruitment of repair proteins. The domain organization of SsoSSB is strikingly similar to that of the bacterial SSB proteins, with a single OB fold, a flexible tail that is not required for DNA binding and an acidic patch at the extreme C-terminus (Wadsworth and White, 2001). (B) Structure-based sequence alignment of SsoSSB with the DBD-A and DBD-B subunits of hsRPA70. Important elements of structure are coloured as for Figure 2: β strands are in cyan, the L12 and L45 loops in red and the L23 loop is in orange. At the extreme N-terminus (blue), secondary structure is not conserved and therefore relates solely to SsoSSB. Residues conserved throughout all three homologues are shaded yellow. Important DNA-binding residues inferred from the structure and mutated in our analysis are in bold. The region connecting the β3 and β4 strands can be decomposed into a helical region (green in the alignment) and a coil. As this region differs in all structures, meaningful alignment of these residues is not possible. Visual inspection indicates that the structure-based sequence alignment extends over 84 residues with an r.m.s.d. of 1.4 Å, rather than the 87 residues with an r.m.s.d. of 1.6 Å calculated by LSQMAN. Automated alignment matches a slightly different subset of atoms.
Structural analysis of the SSB protein from Escherichia coli (EcoSSB) confirms the presence of four OB folds that are brought together to form the functional homotetramer (Raghunathan et al., 1997, 2000). In the chymotryptic fragment studied, ssDNA wraps around the tetramer in a complex pattern (Raghunathan et al., 2000). At low salt concentrations, EcoSSB saturates ssDNA with ‘unlimited’ inter-tetramer cooperativity, forming long clusters along the strand. Known as the (SSB)35-binding mode, this state is consistent with the occlusion of 35 nucleotides (nt) per tetramer and interaction with two of the four subunits of EcoSSB (Lohman and Overman, 1985; Bujalowski and Lohman, 1986). In the (SSB)65-binding mode, all four subunits interact with ssDNA to occlude 65 nt per tetramer, and at high salt concentrations ‘limited’ inter-tetramer cooperativity leading to the formation of beaded structures along the single DNA strand is observed (Lohman and Overman, 1985; Bujalowski and Lohman, 1986).
In contrast, structural analysis of the human RPA (hsRPA) has revealed a very different domain organization and mechanism of ssDNA binding. Typical of the RPA homologues, hsRPA is a heterotrimer composed of subunits of molecular weights 70, 32 and 14 kDa (RPA70, RPA32 and RPA14, respectively) (Wold, 1997). RPA70, which is thought to mediate initial binding of hsRPA to ssDNA (occluding 8–10 nt) (Blackwell and Borowiec, 1994), has four domains each with an OB fold: RPA70N, the N-terminal domain involved in protein– protein interactions (Jacobs et al., 1999); the principal DNA-binding domains DBD-A and DBD-B (Gomes and Wold, 1996; Bochkarev et al., 1997; Bochkareva et al., 2001); and DBD-C, which contains a ‘zinc-ribbon’ motif (Figure 1A). Although the precise role that zinc-binding plays in RPA is undefined, there is a suggestion it may modulate ssDNA binding by DBD-C (Bochkareva et al., 2000). RPA binds ssDNA through sequential interactions with DBD-A, DBD-B and DBD-C in tandem, followed by the OB fold of RPA32, DBD-D. It has been suggested that ssDNA is kinked around RPA to contact the last OB fold, allowing the RPA heterotrimer to occlude 30 nt in total (Bochkareva et al., 2002).
We report here the crystal structure of the SSB protein from Sulfolobus solfataricus (SsoSSB) determined to 1.26 Å, using selenomethionyl multiwavelength anomalous diffraction (MAD) methods. To date, this represents the highest-resolution structure of an SSB protein. We have crystallized a tryptic fragment (SsoSSB1–115) that lacks the C-terminal tail proposed to be involved in protein–protein interactions (Wadsworth and White, 2001). The structural data provide conclusive evidence that, although SsoSSB possesses a C-terminal tail similar to EcoSSB, its OB-fold domain is more similar to the eukaryotic RPAs (Figure 1B). The close relationship between the human and the crenarchaeal proteins is consistent with the closer phylogenetic relationship between the archaeal and eukaryotic domains (Keeling and Doolittle, 1995). It is this close relationship that has permitted studies of simpler archaeal systems to shed light on the more complex and less tractable human systems. In this study, we characterize the thermodynamics of interactions that control the recognition of DNA by SsoSSB. These results are directly relevant to the human systems.
Results
Overall structure
SsoSSB was expressed in E.coli and the purified protein truncated by trypsin digestion as described previously (Wadsworth and White, 2001). The crystallized protein contains residues 1–119. Our construct misses residues 120–148 of the full-length protein, which is predicted to be highly flexible and is involved in the mediation of protein–protein interactions. In the asymmetric unit, there are two copies of the SSB monomer, monomer A (residues 1–115) and monomer B (residues 1–114), related by a non-crystallographic 2-fold axis. The C-terminal 4–5 residues are apparently disordered. The two monomers superimpose with a root-mean-square deviation (r.m.s.d.) of 0.82 Å over 113 α carbons. This seemingly large difference is attributed to the N-terminal three residues and two flexible areas: residues 31–36 and 94–100. Excluding these regions leads to an r.m.s.d. of 0.42 Å over 98 α carbons. Residues 31–36 are found in a flexible loop on the surface that is intimately involved in crystal packing. The region between 94 and 100 is also on the surface in a crystal contact and, according to analysis of thermal factors, is more flexible than the rest of the structure. The N-terminal three residues adopt a different orientation between the subunits.
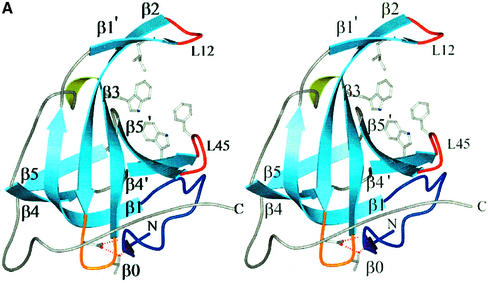
There is some debate as to whether SsoSSB exists as a monomer in solution (Wadsworth and White, 2001; Haseltine and Kowalczykoski, 2002). Although the asymmetric unit contains two monomers, this is an artefact of crystallization. The ‘dimer’ buries less than 1000 Å2, the interaction is confined to loops and there are no obvious protein–protein interactions suggesting specific recognition. Glutaraldehyde cross-linking of SsoSSB suggests a monomeric composition for full-length SSB protein (Wadsworth and White, 2001). On this basis, we define the biological unit as a monomer and refer in the Discussion to monomer A (Figure 2).
Fig. 2. SSB proteins from all three domains of life. Stereo views of (A) the SsoSSB monomer, (B) the DBD-B subunit of hsRPA70 with the loops oriented for DNA-binding and (C) the EcoSSB monomer. The SsoSSB monomer more closely resembles the overall fold and loop structure of DBD-B. In comparison, the EcoSSB monomer has loops that are far more extended, generating surfaces for the formation of the homotetramer (the biological unit). The important residues mentioned in the text are shown in ball-and-stick representation. The DNA-binding residues in SsoSSB are (clockwise from top) Ile30, Phe79, Trp75 and Trp56 and the protein secondary structure is labelled as described in the text, similar to the convention described by Murzin (1993). Residues at equivalent positions in DBD-B are shown. For EcoSSB, the four aromatic residues implicated in ssDNA binding are indicated (Raghunathan et al., 1997, 2000). In each picture, the L12 and L45 loops are coloured red, the L23 loop orange, the capping ‘helical’ region green and the N-terminus royal blue. The C-terminal helix in DBD-B, which may represent an interaction surface with RPA32, is also coloured green.
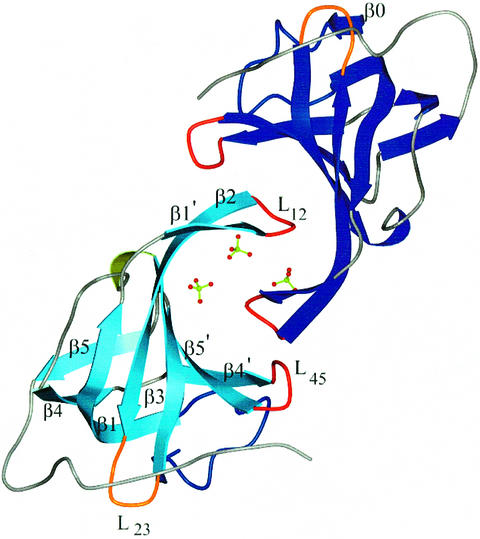
The crystal structure of SsoSSB1–115 shows that SsoSSB possesses a single OB fold as predicted (Wadsworth and White, 2001; Haseltine and Kowalczykoski, 2002). In comparison with the standard OB-fold template of five β strands and an α helix, SsoSSB shows some irregularities. Slight distortions centred at residues 26, 72–73 and 89 break up the regular β1, β4 and β5 strands, as is seen in RPA70 (Bochkarev et al., 1997). The β sheet still coils to form a closed β barrel, the hallmark of this fold. However, in SsoSSB, there is an extra β strand at the N-terminus (Figure 2). The most notable deviation from the OB-fold template is the absence of the α helix connecting the β3 and β4 strands. The helix caps the barrel and may be important for structural integrity; however, SsoSSB instead uses a distorted helical turn (residues 57–59) and a turn (residues 60–62) to accomplish the same effect. Two loops, L12 and L45, are known to be crucial in recognition and binding of the OB fold to its target molecule. These loops form β hairpins between β1′–β2 centred on residues 33–34 and β4′–β5′ centred on residue 80. The L12 loops from two monomers interact with each other via crystal contacts. This packing is strengthened by additional interactions with a network of three sulphate anions and water molecules (Figure 3).
Fig. 3. The crystallographic dimer. Sulphate ions are shown in ball-and-stick representation. The L12 loops from the two monomers interact with each other and sulphates, possibly mimicking the interaction with DNA. The important structural elements are labelled as discussed in the text. For clarity, the β0 strand is labelled in a different monomer.
SsoSSB resembles the eukaryotic homologues, not bacterial SSB proteins
In order to provide a rigorous analysis of the relationship of SsoSSB with other proteins containing OB folds, the coordinates were submitted to the DALI server to identify structural homologues (Holm and Sander, 1993). The DNA-complexed form of hsRPA70 (1JMC) was identified as the most similar protein (Z = 13.8). The α subunit of an ssDNA–telomere end binding protein complex from the protozoan Oxytricha nova (α-OnTEBP, 1OTC) (Horvath et al., 1998) was second on the list (Z = 9.0). In total, over 50 homologues were identified, reflecting the widespread occurrence of the OB fold.
In SsoSSB, we define the structural core of the OB fold as residues 15–25, 39–46, 50–56, 66–71, 74–77, 83–88 and 91–95 (47 residues in total). As we noted, the absence of the α helix from SsoSSB indicates that it is not a fundamental component of the OB fold. Remarkably, even this small core highlights that structurally the OB fold of SsoSSB is more like the DBD-B subunit of hsRPA70 (r.m.s.d. 0.9 Å, 1FGU) than EcoSSB (r.m.s.d. 2.1 Å, KAW) (Figure 2). The similarities between the structures were examined more thoroughly by adding matching atoms iteratively using the program LSQMAN (Kleywegt, 1996). Superposition of SsoSSB with EcoSSB matches in total 66 α carbons with an r.m.s.d. of 1.7 Å; for DBD-B of RPA70 with DNA bound, 87 atoms superimpose with an r.m.s.d. of 1.6 Å. The structural differences between EcoSSB and SsoSSB are reflected mainly in the loops (Figure 2). Of particular note are the L23 and L45 loops, which are longer in EcoSSB. These loops have aromatic residues that guide ssDNA around the tetramer through base stacking interactions. The L45 loop is proposed to be involved in cooperative assembly of tetramers along ssDNA (Raghunathan et al., 1997, 2000). In SsoSSB, the much shorter L23 and L45 loops preclude the possibility that SsoSSB binds DNA by wrapping it round a tetrameric arrangement of OB folds in the same way as EcoSSB.
We note the conservation of a DxT/S motif in the connection between β2 and β3 (Figure 1B). This occurs prior to an absolutely conserved key aromatic residue (Trp56 in SsoSSB) in archaea and eukaryotes. This motif is absent in EcoSSB and other bacterial SSB proteins but appears conserved in all eukaryotic sequences, with the exception of the mitochondrial protein (Yang et al., 1997), which has a bacterial origin. In SsoSSB, this region is remote from the DNA-binding site. The Asp and Thr residues (46 and 48, respectively) form a hydrogen bond network with the amide backbone at residues 5 and 6 (Figure 2). This results in the N-terminus of the protein folding into an initial turn of the β barrel. In DBD-B, Asp350 contacts the N-terminal amide backbone at residue 305 (Figure 2) and Ser352 makes a contact with the side chain of Asp306 (not shown in Figure 2), forcing this region of structure to adopt a helical turn. In the E.coli structure, the N-terminus is an elongated strand which makes a β sheet interaction to stabilize the dimer. This N-terminal strand appears to be stabilized by contacts with the L23 hairpin from a neighbouring monomer. SsoSSB and RPA70 may be unable to achieve the oligomeric states observed in EcoSSB because the N-terminus is folded back into the monomer through interactions with the L23 loop. This local region of structure is perhaps the key feature defining the eukaryotic and archaeal OB folds and differentiating them from the bacterial and mitochondrial SSB proteins.
A model for ssDNA binding by SsoSSB
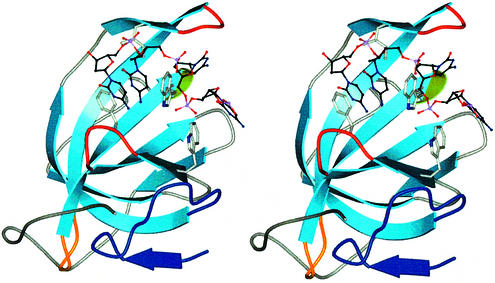
The crenarchaeal protein is structurally more similar to the DBD-B–DNA complex of RPA70 than it is to the apo-DBD-B structure. At first glance this is a surprising observation, as studies of OB folds have shown major conformational changes upon DNA binding (Bochkareva et al., 2001). However, we note that in SsoSSB the dimer, which arises from crystal packing, involves interlocking of the L12 loops with sulphate ions and water molecules, with the result that the L12 loop folds down over the barrel (Figure 3). Therefore, the closed form of the SsoSSB may have been trapped fortuitously due to crystal packing interactions and the presence of sulphate ions and water molecules in the binding cleft. The similarity between SsoSSB and DBD-B allows a facile superposition of the DNA-complexed DBD-B subunit of RPA70 onto apo-SsoSSB. Using this superposition, we generated a model of the SsoSSB–ssDNA complex (Figure 4). A close contact of 1.5 Å is observed between the backbone carbonyl of Gln31 and C5 on the corresponding nucleotide. All other protein–DNA contacts are 2.0 Å or over. The OB fold has a defined binding site that is located between the L12 loop, the L45 loop and the outside of the barrel, with the L12 loop closed for the reasons discussed above. It seems that the L45 loop is open, as in the DBD-B–ssDNA complex, to allow initial passage of DNA through the binding cleft. Although, in the absence of a ‘true’ apo-SsoSSB structure, we cannot categorically say that ssDNA binding would result in a large movement of the L12 and L45 loops as is seen in RPA70, we propose that ssDNA binding in SsoSSB follows the open/closed model suggested for RPA70 (Bochkarev et al., 1997).
Fig. 4. A model of the SsoSSB–DNA complex. Stereo view of SsoSSB complexed with ssDNA. ssDNA was modelled into the DNA-binding cleft by facile superposition of the DBD-B–DNA complex onto the SsoSSB structure. The model shows the similarity between the SsoSSB and RPA70 DNA-binding sites. The SsoSSB monomer is coloured as in Figure 2.
It also seems unlikely that SsoSSB wraps DNA around itself, as is seen in the EcoSSB–DNA complex. A single tetramer of EcoSSB wraps 65 nt of DNA, yielding a final stoichiometry of 16 nt of DNA per monomer. In contrast, SsoSSB forms complexes with a wide variety of lengths of ssDNA with a final stoichiometry of 4.5–5 nt per SSB monomer (Wadsworth and White, 2001; Haseltine and Kowalczykoski, 2002). This suggests a binding mode consisting of a tandem array of SSB monomers that multimerize along linear ssDNA.
Dissection of the thermodynamic contribution to DNA binding by hydrophobic residues
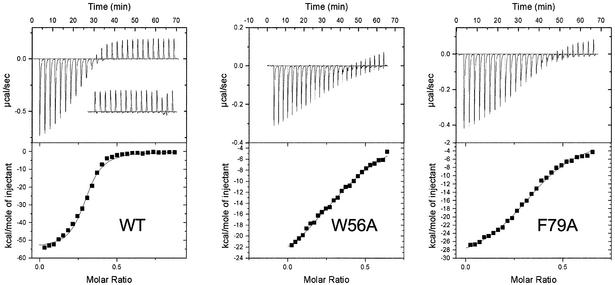
In order to quantify the individual contributions of key hydrophobic residues in the binding channel of SsoSSB, we created site-directed mutations, individually changing residues Ile30, Trp56, Trp75 and Phe79 to alanine. Using isothermal titration calorimetry (ITC), a thermodynamic analysis of the interaction of the wild-type and mutant proteins with a 21 nt oligonucleotide was undertaken (Table I). Wild-type SSB protein binds to the 21mer oligonucleotide with a final stoichiometry of approximately four SSB monomers bound per DNA molecule, confirming the binding density of 5 nt per monomer observed using fluorescence and gel-based assays (Wadsworth and White, 2001; Haseltine and Kowalczykoski, 2002). Accordingly, we have expressed the thermodynamic data in Table I both in terms of the concentration of 21 nt oligonucleotide and in terms of the concentration of 5 nt binding sites (in parentheses). Binding is highly exothermic (Figure 5), with ΔH equal to –56 kcal/mol (–230 kJ/mol) for the 21mer oligonucleotide (approximately –15 kcal/mol for a single monomer of SsoSSB binding). This is broadly similar to that seen for EcoSSB (–50 kcal/mol for a tetramer of EcoSSB) and reflects the enthalpically driven binding of SSB proteins thought to be due to extensive stacking interactions between the DNA bases and aromatic amino acids (Kozlov and Lohman, 1998). The binding affinity observed corresponds to an apparent dissociation constant of ∼90 nM for the 21mer oligonucleotide.
Table I. Thermodynamic data for wild-type and mutant SSB proteins binding to a 21mer oligonucleotide at 323K.
| Protein | Stoichiometry | Ka (app) | ΔH | ΔS | TΔS | ΔG |
|---|---|---|---|---|---|---|
| (n) | (×106 M–1) | (kcal/mol) | (cal/mol) | (kcal/mol) | (kcal/mol) | |
| Wild type | 0.28 ± 0.08 | 11.0 ± 1.9 (2.8 ± 0.5) | –56 ± 1.2 (–14 ± 0.3) | –140 ± 3 (–35 ± 0.7) | –45 ± 1.1 (–11 ± 0.3) | –10.5 ± 0.17 (–2.6 ± 0.04) |
| I30A | 0.29 ± 0.06 | 7.1 ± 1.1 (1.8 ± 0.3) | –44 ± 0.9 (–11 ± 0.2) | –110 ± 2 (–27 ± 0.5) | –34 ± 0.8 (–8.5 ± 0.2) | –10.1 ± 0.13 (–2.5 ± 0.03) |
| W56A | *0.41 ± 0.05 | 0.48 ± 0.04 (0.12 ± 0.01) | –32 ± 4.4 (–8.0 ± 1.1) | –74 ± 10 (–18 ± 2.5) | –24 ± 4.4 (–6.0 ± 1.1) | –8.3 ± 0.08 (–2.1 ± 0.02) |
| W75A | 0.30 ± 0.02 | 3.0 ± 0.55 (0.75 ± 0.14) | –41 ± 0.2 (–10 ± 0.05) | –97 ± 0.5 (–24 ± 0.1) | –31 ± 0.3 (7.8 ± 0.08) | –9.6 ± 0.22 (–2.2 ± 0.05) |
| F79A | *0.38 ± 0.01 | 1.24 ± 0.20 (0.31 ± 0.05) | –30 ± 2.0 (–7.5 ± 0.5) | –66 ± 4 (–17 ± 1) | –21 ± 2.2 (–5.2 ± 0.5) | –9.1 ± 0.13 (–1.8 ± 0.03) |
Data presented are mean values ± SD for three experiments. Thermodynamic values are based on the molar concentration of the oligonucleotide. As each oligonucleotide has four binding sites for SSB protein, we have divided these values by four to yield the thermodynamic parameters for a single SSB monomer binding to a 5 nt binding site, and these are shown in parentheses. *The weakest binding mutants, W56A and F79A, have apparent final stoichiometries lower than the wild-type protein; possible reasons for this are discussed in the Materials and methods.
Fig. 5. Thermodynamic analysis of DNA binding by wild-type and mutant forms of SsoSSB. (Upper panels) Representative raw titration data for wild-type SsoSSB and the mutant W56A and F79A SSB proteins during sequential injections of a 21mer oligonucleotide at 50°C. The inset on the wild-type panel shows the heat effects resulting from successive injections of buffer into the protein, which is dominated by the endothermic effect of mixing of the injectant at 20°C and the sample at 50°C. All binding isotherms were corrected for this effect before fitting. (Lower panels) Integrated heat responses fit to a single binding site model (continuous line). The derived thermodynamic parameters are summarized in Table I.
Slight systematic deviations from the fitted curve are observed for wild-type SSB protein in Figure 5, suggesting that a simple ‘one set of sites’ model does not describe the physical situation perfectly. Clearly, there is the possibility for some degree of cooperativity in the interaction of SSB monomers bound at adjacent sites on the oligonucleotide. This could include positive cooperative interactions between adjacent SSB monomers and negative cooperativity due to the presence of overlapping binding sites on the DNA. These caveats mean that absolute thermodynamic parameters stated here may be subject to some systematic error; further investigations of the presence of cooperativity will be required. It should also be noted that we cannot rule out the possibility that the point mutations made do not have more generalized, unforeseen consequences such as alterations in the protein conformation close to or more distant from the mutated residue or changes in the conformational flexibility of the SSB protein binding site. Therefore, the parameters presented in Table I do not necessarily reflect the individual thermodynamic contributions between individual mutated residues and the nucleic acid ligand.
The four mutant forms of SSB protein all have altered thermodynamic values for DNA binding, with a reduction in the favourable enthalpy change and a corresponding favourable change in entropy. The residue that contributes the greatest interaction energy to DNA binding is the central tryptophan, Trp56. Replacement of this residue with an alanine results in a loss of 2.2 kcal/mol (9.2 kJ/mol) in binding energy for the 21mer oligonucleotide, or 0.55 kcal/mol per SSB monomer, assuming a binding stoichiometry of four monomers per 21 nt oligonucleotide (Figure 5). Also of importance is Phe79, which contributes ∼1.5 kcal/mol to the binding interaction. The weaker binding of these two mutants resulted in weaker heat effects and less-well-defined end points for the calorimetric titration, and this may contribute to the uncertainties in the observed binding stoichiometries.
These two residues are conserved in equivalent positions in DBD-A (Phe238 and Phe269) and DBD-B (Trp361 and Phe386) of hsRPA, where they make stacking interactions with the bound ssDNA (Bochkarev et al., 1997). The other two residues mutated, Ile30 on the β hairpin involving the L12 loop, and Trp75, contribute ∼0.5 and 1 kcal/mol, respectively, to the binding energy per 21 nt oligonucleotide. Our model suggests that Trp75 recognizes the phosphate backbone via a hydrogen bond from the nitrogen of the indole ring, rather than forming a stacking interaction with the nucleotide bases. This is supported by the fact that there is no equivalent hydrophobic residue at the same position in either DBD-A or DBD-B (Figure 1B). Instead, DBD-B contacts the phosphate backbone of ssDNA via a hydrogen bond from Arg382. The thermodynamic analysis of DNA binding by the mutant proteins highlights the relatively small changes in ΔG that are the sum of much larger but compensatory changes in the enthalpy and entropy of binding
Discussion
All cellular life forms, and many prokaryotic and eukaryotic viruses, have an absolute requirement for a protein that can bind and protect exposed ssDNA during replication and repair. The 100 amino acid OB-fold domain appears to perform this role universally and has also been recruited as an ssDNA-binding module by other proteins such as BRCA2 (Yang et al., 2002) and TEBP (Horvath et al., 1998) in eukaryotes and RecG in bacteria (Singleton et al., 2001).
The structural data presented here suggest that the crenarchaeal SSB proteins represent the simplest form of SSB protein studied to date, with a single OB fold and a monomeric composition. Our mutagenesis and ITC represents the first attempt at a systematic dissection of the molecular basis for DNA binding by any SSB protein. Although uncertainties in the interpretation of the calorimetric data remain, the importance of two residues (Trp56 on the β3 strand and Phe79 on the L45 loop) has been highlighted. The aromatic stacking function performed by these residues is conserved in the two principal hsRPA DNA-binding domains, and an aromatic residue at the position corresponding to Trp56 is seen in the bacterial SSB proteins and in one of the OB folds of BRCA2 (Yang et al., 2002). The stoichiometry of 4.5–5 nt of DNA bound per monomer is preserved for all lengths of ssDNA studied (Wadsworth and White 2001), suggesting a high-density DNA-binding mode without significant wrapping or kinking of the DNA chain. This may reflect the hyperthermophilic lifestyle of S.solfataricus and most crenarchaea, as ssDNA will be particularly susceptible to DNA damage at elevated temperatures. This basic unit of DNA binding has evolved in eukaryotes into a complex heterotrimeric RPA, with four OB folds mediating interactions with ssDNA and other domains mediating protein–protein interactions. Initial DNA binding is carried out by the first two OB folds of the large subunit of RPA, which are organized in tandem, probably in a similar fashion to two consecutive SsoSSB monomers bound to ssDNA.
In contrast, bacteria have evolved a homotetrameric SSB protein that wraps a 65 bp segment of ssDNA in a highly structured fashion. The crenarchaeal and bacterial proteins share a remarkable flexible C-terminal tail that plays no part in DNA binding and is known in bacteria to be essential for the recruitment of repair proteins to sites of DNA damage. In all likelihood, the crenarchaeal tail performs the same function, and specific protein partners have been identified (D.Richard and M.F.White, unpublished data). However, the structure shows that the OB fold is actually distinct from that seen for EcoSSB and shares close structural similarity with the DNA-binding domains of hsRPA. The monomeric organization of the crenarchaeal OB fold is the simplest domain organization of the family and may reflect the structure of the ancestral SSB protein. The thermodynamic studies of SsoSSB mutants confirm the key residues, inferred from the structure, that control DNA recognition in both archaeal and eukaryotic systems. The evolutionary origins of the chimeric features of SsoSSB, with a eukaryotic-type DNA-binding domain coupled to a bacterial-like protein interaction domain, remain an intriguing puzzle at present.
Materials and methods
Purification and crystallization
Recombinant SsoSSB was prepared and purified as described previously and truncated by trypsin digestion (Wadsworth and White, 2001). Expression of the selenomethionyl derivative was achieved using E.coli Rosetta cells (Novagen), following the protocol described by VanDuyne et al. (1993). Protein with selenomethionine incorporated was purified as described for native recombinant SSB protein, except that all purification buffers contained 4 mM DDT to prevent selenomethionine oxidation. Both native and selenomethionine protein were treated with porcine trypsin to determine the flexibility of the C-terminus. We have been unable to crystallize full-length protein. Details of the crystallization have already been reported (Kerr et al., 2001). The presence of sulphate appears essential. The selenomethionyl derivative effectively crystallizes identically to the native protein.
Data collection
A 1.26 Å native data set was collected on Daresbury station PX 9.6 (Table II). Data were obtained from a single, frozen, bipyramidal crystal suspended in mother liquor and paraffin oil. Data collection was achieved in four passes, to account for the presence of ‘overloaded’ reflections in the low-resolution shells and to ensure high completeness and redundancy in all shells. Data were integrated in MOSFLM (Leslie, 1992) and merged in SCALA (Evans, 1997). The space group was determined to be either P61 or P65 from the systematic absences, with cell dimensions a = b = 75.81, c = 70.12, α = β = 90°, γ = 120°. Assuming two molecules in the asymmetric unit, the Matthews coefficient was 2.25 Å3/Da, giving a solvent content of ∼45%.
Table II. Data collection and refinement statistics.
| Data collection | Native | λ1 | λ2 | λ3 |
|---|---|---|---|---|
| Wavelength (Å) | 0.87 | 0.9780 | 0.9786 | 0.9600 |
| Beamline | SRS PX 9.6 | SRS PX 14.2 | ||
| Resolution (Å) | 47.92–1.26 (1.29–1.26) | 37.32–1.69 (1.73–1.69) | 37.26–1.69 (1.73–1.69) | 37.27–1.66 (1.70–1.66) |
| Space group | P61 | P61 | ||
| Cell constants | a = b = 75.81 Å, c = 70.12 Å, α = β = 90°, γ = 120° | a = b = 74.69 Å, c = 69.06 Å, α = β = 90°, γ = 120° | ||
| VM | 2.25 | 2.14 | ||
| Total measurements | 463924 | 408408 | 407283 | 432531 |
| No. of unique reflections | 60733 | 24609 | 24521 | 25989 |
| Average multiplicity | 7.5 (3.6) | 16.6 (16.3) | 16.6 (16.3) | 16.6 (16.3) |
| I/σ | 4.5 (2.0) | 8.1 (1.6) | 8.3 (1.4) | 8.1 (1.4) |
| Completeness (%) | 99.9 (99.9) | 100 (100) | 100 (100) | 100 (100) |
| Rmergea | 8.1 (35.2) | 5.4 (45.6) | 5.6 (54.6) | 5.5 (53.5) |
| Wilson B-factor (Å2) | 15 | 21 | 21 | 21 |
| Anomalous completeness (%)b | N/A | 100 (100) | 100 (100) | 100 (100) |
| f′/f″ (refined) |
N/A |
–5.71/4.05 |
–4.69/4.64 |
–1.75/3.53 |
| Refinement |
|
|
|
|
| Resolution (Å) | 65–1.26 | |||
| R-factor | 18.3 (25.1) | |||
| Rfree | 21.0 (28.1) | |||
| r.m.s.d. bonds (Å)/angles (°) | 0.015/1.6 | |||
| B-factor deviation bonds (Å2)/angles (°) | ||||
| Main chain | 1.4/2.2 | |||
| Side chains | 2.7/4.0 | |||
| Residues in Ramachandran core (%)c | 95.9 | |||
| Protein atoms | 1731 | |||
| Water atoms | 273 | |||
| Average B-factor (Å) | 11 | |||
| PDB accession code | 1o7i |
aRmerge = ΣΣI(h)i – <I(h)>/ΣΣI(h)i, where I(h)i is the measured diffraction intensity and the summation includes all observations.
bAnomalous completeness corresponds to the fraction of possible acentric reflections for which an anomalous difference has been measured.
cRamachandran core refers to the most favoured region in the Φ/Ψ Ramachandran plot as defined by Laskowski et al. (1993).
Collection of data from previous SSB protein crystals had alerted us to the presence of merohedral twinning in the crystal lattice (Kerr et al., 2001), highlighted by the fact that the data merged in the higher symmetry Laue groups and the anomalous behaviour of the cumulative intensity distribution in TRUNCATE (French and Wilson, 1978). The twinning fraction of the 1.26 Å data was determined to be ∼0.30 using the Yeates Merohedral Crystal Twinning Server (Yeates, 1997). The data were successfully detwinned using the CCP4 program DETWIN (Taylor and Leslie, 1998). We were unable to discern any trend in crystal morphology, resolution limit or structure that correlated with the twinning fraction.
A three-wavelength MAD data set was collected from selenomethionyl crystals at Daresbury station PX 14.2 (Table II). Data were collected from a single, frozen, bipyramidal crystal. Analysis of the data was used to select a non-twinned crystal. Cryoprotection was achieved by pulling the crystal through the mother liquor and then through a single drop of 50% PEG 600. Highly redundant data were collected to ensure accurate measurement of the anomalous signal. Two hundred and seventy images were recorded for each MAD wavelength, as 5 s, 1°, non-overlapping oscillations per frame using a crystal to detector distance of 140 mm. The data were integrated in MOSFLM and merged in SCALA.
MAD phasing and refinement
Using all the data to 1.7 Å, the scaled intensities from the three-wavelength data were used to locate selenium sites in SOLVE (Terwilliger and Berendzen, 1999). Four out of an expected six were found, with a figure of merit of 0.51 and Z-score of 11.46. The raw MAD phases were used to calculate electron density maps showing a clear protein–solvent boundary and regions of secondary structure. Successive rounds of maximum-likelihood density modification in RESOLVE (Terwilliger, 2000) gave an improved figure of merit of 0.61. These phases were used to build the initial main chain of the crystallographic dimer in ARP/wARP using the ‘warpNtrace’ mode (Perrakis et al., 1999). After some manual intervention to build less-well-ordered regions, the model refined smoothly using REFMAC5 (Murshudov et al., 1997) against the detwinned native data (Table II). Where necessary, the model was manually adjusted in ‘O’ (Jones et al., 1991). Once the refinement had extended to 1.5 Å, thermal factors were refined anisotropically. Hydrogen atoms were included in their riding positions but were not included in the final model, as they could not be seen in the electron density.
The use of NCS restraints was explored; however, the Rfree decreased when they were removed. The final model was refined to an Rfree of 21.0% and an R-factor of 18.3%, with good geometry as judged by PROCHECK (Laskowski et al., 1993) and WHATIF (Vriend, 1990). The slightly higher R-factors in the higher shells are probably due to the inevitable errors incorporated by the detwinning process.
Site-directed mutagenesis
Isogenic SsoSSB mutants were generated with the Stratagene QuickChange mutagenesis protocol (Stratagene, La Jolla, CA) using the pET19b–SSB construct as the template. Single amino acid changes to alanine were made at Ile30, Trp56, Trp75 and Phe79. All mutations were confirmed by nucleotide sequencing using the Sequenase 2.0 protocol (Amersham Pharmacia Biotech) and an ABI 377 automated DNA sequencer (Applied Biosystems). The mutant proteins were expressed and purified as described for wild-type SSB protein. The mutant proteins are sufficiently stable to survive a 30 min heat step at 70°C during the course of purification, suggesting that their stability has not been seriously compromised compared with the wild-type protein.
ITC of SsoSSB mutants
ITC experiments were carried out using a VP-ITC device (MicroCal, Northampton, MA). All solutions were degassed. SSB protein samples were dialysed extensively against 20 mM MES buffer, pH 6.5, 100 mM potassium glutamate and 1 mM MgCl2. The binding experiments were performed in triplicate at 50°C. A 370 µl syringe with stirring at 400 r.p.m. was used to titrate the 21mer oligonucleotide (sequence 5′-CCGAGTACCAGCATGAACTTA) into a cell containing ∼1.4 ml of SSB protein (15 mM). Each titration consisted of a preliminary 1 µl injection followed by up to 30 subsequent 10 µl injections of 50 mM oligonucleotide. Calorimetric data were analysed using MicroCal ORIGIN software using a ‘single binding site’ model. The residual endothermic effect observed in Figure 5 for the titration of DNA into wild-type SSB protein was due to the difference in temperature between the injectant in the syringe (20°C) and the sample chamber (50°C) and was observed in control titrations where buffer was injected into the protein sample and for buffer into buffer titrations. These effects were corrected before analysis of the binding curves.
For these experiments, the protein concentration for each mutant was estimated using the molar extinction coefficient computed from the amino acid composition of the protein. As three of the mutations resulted in changes in aromatic residues, the mutant proteins had quite different calculated extinction coefficients. In the case of the wild-type and the I30A and W75A mutants, the final stoichiometry observed during ITC was ∼0.28; within error this corresponds to four SSB monomers bound per 21mer oligonucleotide, commensurate with the calculated binding site size of 5 nt per SSB monomer. For the W56A and F79A mutants, the stoichiometry was observed to approach 0.4, or 2–3 monomers per oligonucleotide (asterisk in Table I). Cross-linking studies suggest that the W56A mutant retains the stoichiometry observed for the wild-type protein (data not shown), though we cannot absolutely rule out changes in binding density in these mutants. It is also possible that this discrepancy arose from the variation in extinction coefficient for these mutants. Correction of the protein concentrations of these mutants to bring the binding stoichiometry into line with the wild-type protein results in higher calculated association constants (W56A Ka = 2.2 × 106 M–1, F79A Ka = 5.1 × 106 M–1), but does not change the overall ranking of the effect of each mutation on binding. A further possibility is that the titration of the two weaker binding proteins gave smaller exothermic signals and did not reach defined end points (Figure 5), resulting in corresponding uncertainties in the calculated binding stoichiometries. Although the absolute thermodynamic parameters for these two mutants contain some degree of uncertainty, the primary importance of these residues for ssDNA binding is clear.
Acknowledgments
Acknowledgements
Thanks to John Ladbury, Sham Haq and Alan Cooper for helpful discussions regarding ITC. Figures 2, 3 and 4 were generated with BOBSCRIPT (Esnouf, 1997) through the GL Render interface (L.Esser, personal communication) and rendered using POV-Ray. This work was funded by the BBSRC. M.F.W. is a Royal Society University Research Fellow; J.H.N. is BBSRC Career Development Fellow.
References
- Blackwell L.J. and Borowiec,J.A. (1994) Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol., 14, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A., Pfuetzner,R.A., Edwards,M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- Bochkareva E., Korolev,S. and Bochkarev,A. (2000) The role for zinc in replication protein A. J. Biol. Chem., 275, 27332–27338. [DOI] [PubMed] [Google Scholar]
- Bochkareva E., Belegu,V., Korolev,S. and Bochkarev,A. (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J., 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E., Korolev,S., Lees-Miller,S.P. and Bochkarev,A. (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J., 21, 1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalowski W. and Lohman,T.M. (1986) Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry, 25, 7799–7802. [DOI] [PubMed] [Google Scholar]
- Chase J.W. (1986) Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem., 55, 103–136. [DOI] [PubMed] [Google Scholar]
- Chedin F., Seitz,E.M. and Kowalczykowski,S.C. (1998) Novel homologs of replication protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends Biochem. Sci., 23, 273–277. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capablilities. J. Mol. Graph. Model., 15, 112–113, 132–134. [DOI] [PubMed] [Google Scholar]
- Evans P.R. (1997) SCALA. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography, no. 33, pp. 22–24. [Google Scholar]
- French G.S. and Wilson,K.S. (1978) On the treatment of negative intensity observations. Acta Crystallogr. A, 34, 517–525. [Google Scholar]
- Gomes X.V. and Wold,M.S. (1996) Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry, 35, 10558–10568. [DOI] [PubMed] [Google Scholar]
- Haseltine C.A. and Kowalczykoski,S.C. (2002) A distinctive single-strand DNA-binding protein from the Archaeon Sulfolobus solfataricus. Mol. Microbiol., 43, 1505–1515. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- Jacobs D.M., Lipton,A.S., Isern,N.G., Daughdrill,G.W., Lowry,D.F., Gomes,X. and Wold,M.S. (1999) Human replication protein A: global fold of the N-terminal RPA 70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR, 14, 321–331. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. and Doolittle,W.F. (1995) Archaea: narrowing the gap between prokaryotes and eukaryotes. Proc. Natl Acad. Sci. USA, 92, 5761–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr I.D., Wadsworth,R.I., Blankenfeldt,W., Staines,A.G., White,M.F. and Naismith,J.H. (2001) Overexpression, purification, crystallization and data collection of a single-stranded DNA-binding protein from Sulfolobus solfataricus. Acta Crystallogr. D Biol. Crystallogr., 57, 1290–1292. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr., 52, 842–857. [DOI] [PubMed] [Google Scholar]
- Kozlov A.G. and Lohman,T.M. (1998) Calorimetric studies of E.coli SSB protein–single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J. Mol. Biol., 278, 999–1014. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography, no. 26. [Google Scholar]
- Lohman T.M. and Overman,L.B. (1985) Two binding modes in Escherichia coli single strand binding protein–single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem., 260, 3594–3603. [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr., 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Philipova D., Mullen,J.R., Maniar,H.S., Lu,J., Gu,C. and Brill,S.J. (1996) A-hierarchy of SSB protomers in replication protein A. Genes Dev., 10, 2222–2233. [DOI] [PubMed] [Google Scholar]
- Raghunathan S., Ricard,C.S., Lohman,T.M. and Waksman,G. (1997) Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl Acad. Sci. USA, 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan S., Kozlov,A.G., Lohman,T.M. and Waksman,G. (2000) Structure of the DNA binding domain of E.coli SSB bound to ssDNA. Nat. Struct. Biol., 7, 648–652. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Scaife,S. and Wigley,D.B. (2001) Structural analysis of DNA replication fork reversal by RecG. Cell, 107, 79–89. [DOI] [PubMed] [Google Scholar]
- Suck D. (1997) Common fold, common function, common origin? Nat. Struct. Biol., 4, 161–165. [DOI] [PubMed] [Google Scholar]
- Taylor H.O. and Leslie,A.G.W. (1998) A program to detwin merohedrally twinned data. CCP4 Newsletter on Protein Crystallography, no. 35, p. 9. [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr., 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr., 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDuyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Wadsworth R.I.M. and White,M.F. (2001) Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res., 29, 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Yang C., Curth,U., Urbanke,C. and Kang,C. (1997) Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol., 4, 153–157. [DOI] [PubMed] [Google Scholar]
- Yang H. et al. (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science, 297, 1837–1848. [DOI] [PubMed] [Google Scholar]
- Yeates T.O. (1997) Detecting and overcoming crystal twinning. Methods Enzymol., 276, 344–358. [PubMed] [Google Scholar]