Abstract
Considerable progress has been made in characterizing epidermal stem cells by microarray analysis of FACS-selected populations. One limitation of this approach is that the gene expression profiles represent the average of the cell population, potentially masking cellular heterogeneity of functional significance. To overcome this problem, we have performed single-cell expression profiling. We have generated cDNA libraries from single human epidermal cells, designated as stem or transit-amplifying cells on the basis of Delta1 and melanoma-associated chondroitin sulfate proteoglycan expression. Of the 14 putative stem cell markers identified, we selected one, the EGF receptor antagonist leucine-rich repeats and immunoglobulin-like domains 1 (Lrig1), for further study. Lrig1 was expressed in groups of basal cells in human interfollicular epidermis previously identified as enriched for stem cells. Overexpression of Lrig1 decreased keratinocyte proliferation but did not affect the proportion of stem and transit-amplifying cells, as judged by clonal growth characteristics. Down-regulation of Lrig1 using siRNA increased cell-surface EGF receptor levels, enhanced activation of downstream pathways, and stimulated proliferation. Lrig1 acted in part by negatively regulating the Myc promoter. We propose that Lrig1 maintains epidermal stem cells in a quiescent nondividing state, and that Lrig1 down-regulation triggers proliferation.
Keywords: gene profiling, keratinocyte, skin, EGF receptor
The epidermis is maintained by stem cells (SC) that reside in specific locations, or niches, and produce progeny that undergo differentiation along the distinct lineages of the hair follicle, sebaceous gland, and interfollicular epidermis (1, 2). The best-characterized reservoir of SC is in the bulge of the hair follicle (3–5). However, these cells are not essential for maintenance of the interfollicular epidermis, which instead depends on its own resident SC population (6–11).
Within the interfollicular epidermis, SC are not the only cells that are competent to proliferate (8). The progeny of a SC that are destined to terminally differentiate first undergo a few rounds of division, during which time they are known as transit-amplifying (TA) cells (12). Although SC can divide extensively to repair damaged epidermis, they tend not to be actively cycling in steady-state epidermis. Expression profiling of follicular bulge cells has identified a number of factors, including Wnt antagonists and bone morphogenetic proteins, that could maintain SC quiescence (3, 4). However, little is known about what regulates quiescence in interfollicular epidermis.
A range of markers of human interfollicular epidermal SC has been described, including high surface expression of β1 integrins (7, 13), high α6 integrin and low CD71 expression (14), high expression of Delta1 (15), low expression of desmoglein 3 (16), and low expression of the EGF receptor 1 (EGFR1; ref. 17). Gradients of marker expression would be consistent with gradients of “stemness” (12, 13, 18), as opposed to the existence of discrete SC and TA cell populations. However, melanoma-associated chondroitin sulfate proteoglycan (MCSP) is one SC marker that is undetectable in other basal cells of the human epidermis (19).
The criteria used to define cells as putative stem or TA cells differ somewhat among laboratories. In the case of β1, Delta1, and MCSP, the criteria were that cells expressing high levels of the markers were capable of sustained clonal growth in culture and lay within regions of the interfollicular epidermal basal layer that were relatively quiescent, with a low proportion of actively cycling cells (7, 13, 15, 19). Conversely, cells that expressed low levels of the markers formed small abortive clones of terminally differentiated cells in culture and lay in regions of the basal layer with a high proportion of actively cycling cells. The proportion of cells that express high levels of β1 integrins varies from 25% to 43%, depending on body site (7), whereas the proportion of SC is calculated as 10%, based on cell kinetic analysis (8). It is therefore likely that the cells that express MCSP and high levels of β1 and Delta1 are heterogeneous.
One way to characterize distinct cell populations is to generate global gene expression profiles. Such profiles have been described for the reservoir of SC in the hair follicle bulge of mouse (3, 4) and human epidermis (20). One limitation of the approach is that the data represent the average characteristics of a cell population, rather than the properties of individual cells. In addition, although bulge cells are well characterized, nonbulge populations are a heterogeneous mixture of SC and TA cells of a variety of cell lineages.
An alternative to conventional expression profiling methods is now emerging through the development of techniques that allow faithful amplification of mRNA from single cells (21–23). The cDNA generated can be used as a template to analyze specific genes (22) or can be fragmented and labeled for global expression profiling using DNA arrays (23). We now describe the generation of single-cell cDNA libraries from cultured human epidermal keratinocytes, identified as SC or TA cells on the basis of Delta1 and MCSP expression. Global expression profiling of these cDNA libraries enabled us to identify Kekkon/Lig1/leucine-rich repeats and Ig-like domains 1 (Lrig1; ref. 24), a negative regulator of EGFR/ErbB receptors, as a marker of putative SC. We present evidence that Lrig1 is a major determinant of SC quiescence.
Results
Generation of Single-Cell cDNA Libraries.
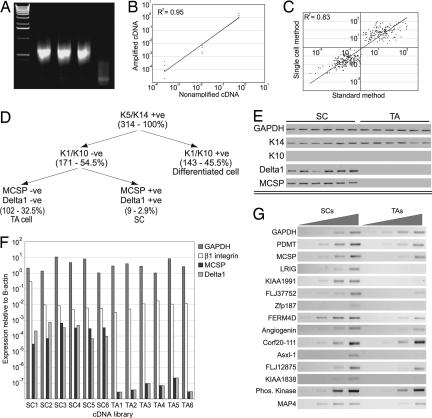
The single-cell expression profiling method (21–23) was validated by using purified total RNA from MCF7 and MCF10A cells. We could reproducibly generate cDNA from <50 pg of total RNA (Fig. 1A), the equivalent of the RNA content of one to two cultured cells. To determine whether the method distorted relative gene expression levels, we performed quantitative PCR for Gapdh, β-actin, β1-integrin subunit, and Dll1; relative expression levels were preserved when amplified samples were compared to traditional cDNA samples generated from the same starting material (Fig. 1B).
Fig. 1.
Generation of single-cell cDNA libraries and marker validation. (A) Amplification of cDNA from 50 pg of total RNA from MCF7 cells. Triplicate samples are shown. (B) Real-time PCR demonstrating that relative expression levels of Gapdh, β1 Integrin, and Delta1 (in inverse order of abundance) compared with β-actin are preserved between amplified and nonamplified MCF7 cDNA samples. (C) Amplified, fragmented, and labeled cDNA from MCF7 and MCF10A cells was generated in hexaplicate and hybridized to HU133 Affymetrix gene arrays. The raw data were filtered as described in Materials and Methods. Fold values (y axis) for probes identified as differentially expressed using the single-cell method were truly differentially expressed when compared with fold values of the same samples analyzed using traditional methodology (x axis), because scatter is located in the upper-right and lower-left quadrants. (D) Markers used to identify cDNA libraries as originating from SC, TA cells, or terminally differentiated cells. Number of libraries of each type is shown in brackets. (E) Validation of marker expression by RT-PCR of cDNA libraries from individual SC and TA cells chosen for expression profiling. (F) Real-time PCR analysis of the picked cDNA libraries. (G) Validation of SC marker genes by semiquantitative PCR on total RNA from enriched populations of SC and TA cells by using a 4-fold dilution series of template.
As further validation, six independent cDNA libraries were each generated from 50 pg of MCF7 RNA and six from 50 pg of MCF10A RNA. Amplified material from the libraries was fragmented by RQ1 DNase and labeled with biotin-ddATP using terminal deoxynucleotidyltransferase (TdT) (23). Labeled cDNA from each library was analyzed on HU133A gene chip arrays. We compared the list of probes differentially expressed in each cell type with published array data generated by using traditional methodology (25). All but one of the probes on our list was truly differentially expressed (Fig. 1C). Therefore, the amplification method used to generate cDNA libraries from small amounts of starting material and the labeling technique can be applied to identify differentially expressed genes. Only differences in expression levels of ≥7-fold were reproducible; at lower fold differences, the number of false positives increased progressively.
We next generated single-cell cDNA libraries from cultured human epidermal keratinocytes. Small cells, chosen because keratinocytes enlarge during terminal differentiation (13), were picked from a diluted single-cell suspension and seeded into first-strand buffer. cDNA libraries were generated from >1,000 single cells. Qualitative PCR for Gapdh and β2 microglobulin was used to determine the quality of the cDNA libraries; on this basis, the number was reduced to 314 (Fig. 1D).
We screened the keratinocyte libraries by qualitative PCR to assign them into categories (Fig. 1D). All cells expressed K14, a marker of cells in the epidermal basal layer. One hundred forty-three cells also expressed K10, indicating they had initiated terminal differentiation. The K14+, K10− cells were further categorized on the basis of MCSP and Dll1 expression as putative SC (MCSP+, Dll1+) or TA cells (MCSP−, Dll1−; refs. 15 and 19). We identified nine libraries as derived from SC and 102 as derived from TA cells (Fig. 1D). The remaining 60 cDNA libraries were from Dll1+, MCSP− cells.
Expression Profiling of Single Epidermal SC and TA Cells.
Six cDNA libraries from MCSP+, Dll1+ cells (putative SC) and six from MCSP−, Dll1− cells (putative TA cells) were selected for further analysis (Fig. 1E). We verified expression of several marker genes by qualitative PCR (Fig. 1E) and of MCSP and Dll1 by quantitative PCR (Fig. 1F). Although high expression of β1 integrins is used to enrich for SC (7, 13), there was no significant difference in β1 integrin levels between the putative SC and TA cell libraries (Fig. 1F), which is to be expected, because integrin levels differ by only 2- to 3-fold between the cell populations (7, 13).
Some of the libraries were of higher quality than others, based on the number of present calls on the HU133A chip and pairwise correlation coefficients between samples. In high-quality samples, 30–40% of the genes were detected as present, and pairwise coefficient values >0.70 were observed (data not shown). We eliminated two cDNA libraries from each group on the basis of low numbers of present calls or poor correlation coefficients.
Based on a P value <0.05 as the statistical threshold, 14 genes were up-regulated at least 7-fold in the SC libraries compared with the TA cell libraries (Table 1). This was confirmed at the level of raw data for all 12 libraries. MCSP (19) has previously been reported as an epidermal SC marker, and the murine homologue of Lrig1, Lig1, is highly expressed in the hair-follicle bulge (26). The other genes have not been linked to epidermal SC previously.
Table 1.
Stem cell markers identified by single-cell expression profiling
| Gene name | Unigene | Putative function |
|---|---|---|
| Putative dimethyladenosine transferase | AK057153 | Methylation of rRNA |
| MCSP | X96753 | Growth factor binding and cell adhesion |
| Lrig1 | BC071561 | EGFR degradation |
| KIAA1991 | XM_495886 | Protein ubiquitin ligase |
| Guanine nucleotide-binding protein-like 1 | NM_005275 | Signal transduction |
| Zinc-finger protein 187 | AL832741 | Transcription |
| FERM domain containing 4 | AB037715 | Cytoskeletal binding |
| Ribonuclease, RNase A family, 5 | NM_194430 | mRNA cleavage |
| C20-Orf-111 | AL133000 | |
| Additional sex combs like 1 | NM_015338 | Regulation of transcription |
| FLJ12875 | AK022937 | Protein ubiquitin ligase |
| KIAA1838 | AK056435 | Carbohydrate metabolism |
| Phosphorylase kinase, α2 | NM_000292 | Serine kinase |
| Microtubule-associated protein 4 | NM_002375 | Microtubule depolymerization |
To confirm that the genes were indeed differentially expressed, we isolated RNA from cells selected by differential adhesion to extracellular matrix (13). Primary human keratinocytes that adhere to collagen within 15 min have high β1 integrin levels and are capable of extensive clonal growth (putative SC). Cells that do not adhere to collagen within 45 min but subsequently adhere to fibronectin within 3 h are enriched for TA cells (low β1 integrin levels) and form abortive clones of terminally differentiated cells. Semiquantitative RT-PCR showed that the majority of genes identified as SC markers (Table 1) were indeed differentially expressed by the SC enriched population (Fig. 1G).
Validation of Lrig1 as a Marker of Putative SC.
We chose Lrig1 for further investigation because of its potential role in down-regulating EGF receptor responsiveness by increasing ubiquitinylation of the activated receptor (24, 27), and because Lig1-null mice exhibit epidermal hyperproliferation (26). These features suggested that Lrig1 might play a role in maintaining epidermal SC in a quiescent state.
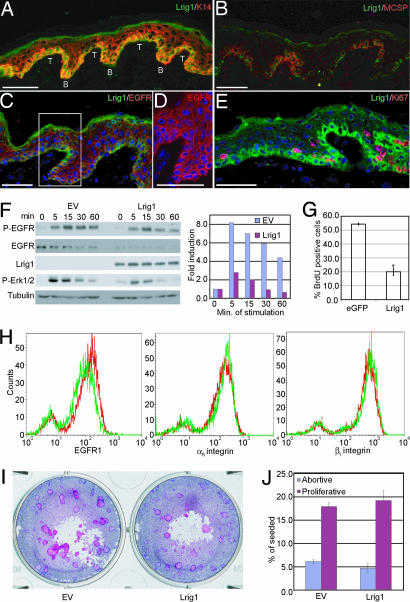
In human interfollicular epidermis, Lrig1 was expressed by K14+ cells in the basal layer (Fig. 2A). The basal layer is not flat but undulates, and the area where the epidermis projects furthest into the dermis is known as a rete ridge. Within the basal layer, Lrig1 was more abundant in groups of cells at the top of rete ridges (T in Fig. 2A), which also express MCSP (Fig. 2B), high levels of β1 integrins (7), and Delta1 (15). In keeping with Lrig1 negatively regulating activated EGFR1, Lrig1 bright cells expressed lower levels of EGFR1 than other basal keratinocytes (Fig. 2 C and D). In addition, the majority of Lrig1-expressing cells were negative for Ki67, a marker of proliferating cells (Fig. 2E; ref. 7). Lrig1 is thus expressed in the same cells previously identified as SC on the basis of proliferation and marker expression (7, 28).
Fig. 2.
Expression of Lrig1 in vivo and effects of overexpression in keratinocytes. (A–E) Immunofluorescence staining of human interfollicular epidermis with antibodies shown. (C–E) DAPI nuclear counterstain (blue). (A) Top (T) and bottom (B) of rete ridges are shown. The boxed area of C is shown at higher magnification in D. (Scale bars, 50 μm.) (F) Keratinocytes transduced with Lrig1 or empty retroviral vector (EV) were serum-starved for 24 h and stimulated with 10 ng/ml EGF for the number of minutes indicated. Western blots probed with antibodies are shown. Levels of P-ERK relative to tubulin were quantified by using NIH Image software, assigning the 0-min time point a value of 1. Data are representative of three independent experiments. (G) Percent BrdU-positive keratinocytes after transient transfection with eGFP or Lrig1. Mean ± SD of triplicate experiments (>100 cells per experiment). ∗P < 0.0001. (H) Flow cytometry of basal keratinocytes infected with empty retroviral vector (red) or Lrig1 (green). (I and J) Clonal growth assays of keratinocytes transduced with empty vector (EV) or Lrig1. (I) Representative dishes. (J) Mean ± SD of triplicate experiments.
High Levels of Lrig1 Reduce Keratinocyte Proliferation and EGF Responsiveness.
We examined the effects of overexpressing Flag-tagged Lrig1 by retroviral expression in primary human keratinocytes. Lrig1-expressing cells had lower total (Fig. 2F) and cell-surface (Fig. 2H) levels of EGFR1 than cells transduced with the empty vector, but surface expression of α6β4 and β1 integrins was unaffected (Fig. 2H). The level of EGFR1 phosphorylation in response to EGF was lower and less sustained in Lrig1-expressing cells, as was ERK1/2 phosphorylation (Fig. 2F and data not shown). This is consistent with the role of Lrig1 in reducing EGF responsiveness by mediating ubiquitinylation and degradation of activated EGFR1 (24, 27).
Overexpression of Lrig1 led to a reduction in proliferation, as evaluated by BrdU incorporation (Fig. 2G). However, Lrig1 did not compromise the clonal growth of primary keratinocytes (Fig. 2I), and there was no effect on the proportion of abortive colonies attributable to TA cell founders (Fig. 2J), which is in good agreement with the lack of an effect of Lrig1 on surface integrin levels (29). We conclude that Lrig1 may promote SC quiescence by reducing responsiveness to EGFR ligands.
Knockdown of Lrig1 by siRNA Leads to Expansion of the SC Compartment.
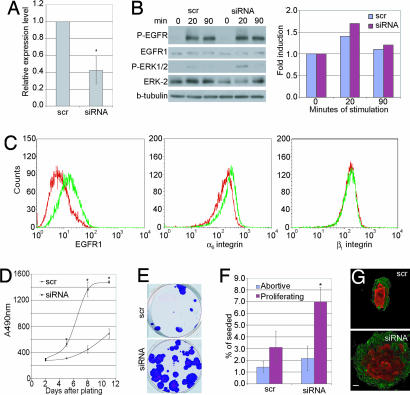
Using siRNA for nucleotides 1494–1512 of the coding sequence of human Lrig1 (27), it was possible to lower Lrig1 mRNA levels by on average 2.5-fold, as determined by quantitative PCR (Fig. 3A). Keratinocytes expressing Lrig1 siRNA had the same levels of total and phosphorylated EGFR1 as keratinocytes expressing a control scrambled siRNA (Fig. 3B). However, activation of ERK MAPK was increased (Fig. 3B). There was also a marked increase in the level of EGFR1 on the cell surface (Fig. 3C), reflecting the role of Lrig1 in endocytosis of activated receptor. Cell-surface β1 integrin levels were unaffected by Lrig1 knockdown, but there was a small increase in α6β4 (Fig. 3C), which might be attributable to increased EGFR1 signaling (30).
Fig. 3.
Consequences of reducing Lrig1 expression by siRNA. (A) Relative levels of Lrig1 mRNA, determined by quantitative PCR, in keratinocytes transduced with Lrig1 siRNA or scrambled siRNA (scr). Values are expressed relative to 18S ribosomal RNA. Mean ± SD of quadruplicate experiments. ∗, P = 0.0004. (B) Western blots of keratinocytes transduced with Lrig1 siRNA or scrambled (scr) siRNA treated with EGF for the times indicated. Levels of P-ERK relative to tubulin controls were quantified using NIH Image software, assigning the 0-min time point a value of 1. Data are representative of three independent experiments. (C) Flow cytometry of basal keratinocytes transduced with scrambled siRNA (red) or Lrig1 siRNA (green). (D) Quantitation of cell number using Celltiter96. Data are means ± SD of hexaplicate samples. ∗, P < 0.0001. (E and F) Clonal growth assays. (E) Representative dishes. (F) Mean ± SD of triplicate samples. ∗, P = 0.0004. (G) Individual clones stained with anti-α6 integrin (green) and involucrin (red) antibodies. (Scale bar, 100 μm.)
Keratinocytes expressing Lrig1 siRNA showed a marked increase in growth rate (Fig. 3D) and a dramatic increase in overall colony size (Fig. 3E). The proportion of nonabortive clones, which we attribute to SC founders, was increased, but there was no significant change in the proportion of abortive clones, attributable to TA cells (Fig. 3F; ref. 13). Keratinocytes transduced with Lrig1 siRNA were still capable of initiating terminal differentiation, as shown by the presence of suprabasal involucrin-positive keratinocytes in the clones (Fig. 3G). Thus, Lrig1 knockdown stimulated SC renewal without inhibiting terminal differentiation.
Lrig1 Regulates Myc Transcription.
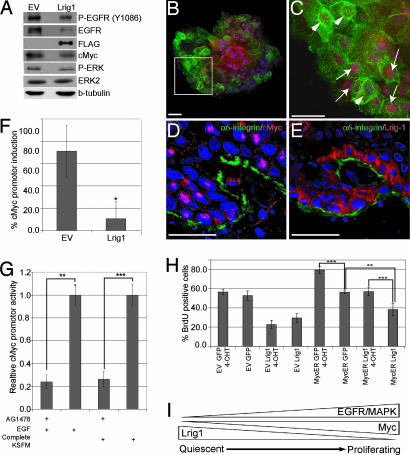
Exit from the epidermal SC compartment is linked to activation of cMyc in cultured keratinocytes (31) and transgenic mouse models (32, 33). Activation of EGFR1 and Myc levels could be linked, because Myc is a target gene of ERK2 stimulated transcription (34). Keratinocytes overexpressing Lrig1 not only had reduced EGFR1 phosphorylation and ERK activation (Figs. 2F and 4A) but also reduced levels of Myc protein (Fig. 4A). An inverse relationship between the level of Lrig1 and the level of Myc in individual cells was observed in cultured keratinocytes (Fig. 4 B and C) and in the basal layer of human interfollicular epidermis (Fig. 4 D and E). When keratinocytes were transiently transfected with a luciferase reporter driven by the Myc promoter, the level of luciferase activity was markedly reduced in cells expressing Lrig1 (Fig. 4F). This effect could be mimicked by Tyrphostin AG1478, a specific inhibitor of EGFR1 kinase activity, in the presence of 10 ng/ml EGF or complete keratinocyte serum-free medium (Fig. 4G). We conclude that Lrig1, EGFR1, and Myc levels in human keratinocytes are interrelated, both in culture and in interfollicular epidermis.
Fig. 4.
Down-regulation of cMyc by Lrig1. (A) Western blot of keratinocytes transduced with Lrig1 or empty vector (EV) probed with antibodies to the proteins indicated. (B and C) Keratinocyte clone transduced with Lrig1 (green), stained for anti-cMyc (red) and DAPI (blue). (C) Higher magnification of boxed area in B. Cells with low Lrig1 and high Myc (arrows) or high Lrig1 and low Myc (arrowheads) are indicated. (Scale bars, 50 μm.) (D and E) Immunofluorescence staining of human epidermis with antibodies shown and DAPI nuclear counterstain (blue). Confocal images (Z stacks) of serial sections are shown. (Scale bar, 25 μm.) (F) Activity of Myc promoter (P1P2) determined by dual luciferase assays, showing percent induction by EGF in keratinocytes transduced with Lrig1 or empty vector (EV). Mean ± SD of quadruplicate experiments. ∗, P = 0.002. (G) Myc promoter activity in keratinocytes stimulated with EGF or keratinocyte serum-free medium in the presence or absence of AG1478. Mean ± SD of triplicate experiments. ∗∗, P = 0.0002; ∗∗∗, P < 0.0001. (H) Cells transduced with MycER or EV were transiently transfected with Lrig1 or GFP and either treated with 4OHT or untreated. Percent transfected cells that incorporated BrdU during a 24-h label is shown. Data represent mean ± SD of four or more samples with >100 transfected cells per sample. (∗∗∗, P < 0.0001; ∗∗, P = 0.0002). (I) Model for the role of Lrig1 in maintaining SC in a quiescent nondividing state.
To investigate whether the growth inhibitory effect of Lrig1 could be overcome by activation of Myc, keratinocytes stably expressing a 4-hydroxy-tamoxifen (4OHT)-inducible form of Myc (MycER; ref. 31) or the empty retroviral vector were transiently transfected with Lrig1 or a GFP control vector (Fig. 4H). Proliferation was measured as the fraction of transiently transfected cells that incorporated BrdU during a 24-h labeling period. As expected, Lrig1 suppressed proliferation of keratinocytes infected with the empty retroviral vector in the presence or absence of 4OHT. Lrig1 also decreased proliferation of cells transduced with MycER in the absence of 4OHT. 4OHT treatment of cells expressing MycER led to an increase in proliferation in the presence or absence of Lrig1, but the level of BrdU incorporation was lower when Lrig1 was present. We conclude that the inhibitory effect of Lrig1 on proliferation can partly be explained by reduced activation of Myc in response to EGF.
Discussion
We have generated a panel of cDNA libraries from single human epidermal MCSP+, Dll1+ cells and MCSP−, Dll1− cells, characteristics previously reported to distinguish SC and TA cells, respectively (15, 19). We used the libraries to identify previously undescribed markers of the putative SC compartment. The validation experiments performed with RNA from MCF7 and MCF10A cells showed that, although the chip-to-chip variation is greater and the number of detected probes is lower using single-cell libraries compared with traditional methods, genes that are truly differentially expressed could be identified. Because the signal-to-noise ratio increased progressively as the fold difference fell below 7-fold, markers reported to differ by 2- to 3-fold, such as the β1 integrins, could not be identified in the libraries.
Single-cell expression profiling is an important tool that complements SC profiling based on FACS-sorted populations (3, 4, 35). It can reveal heterogeneity at the single-cell level (23) and define cellular hierarchies that are masked in a general population. Dll1+, MCSP− cells (Fig. 1D) represent a new population, and it will be of interest to determine whether their characteristics place them in the putative stem or TA compartment.
Using differential cDNA hybridization and 2D protein gel electrophoresis of cultured keratinocytes fractionated according to β1 integrin expression, we found very few differentially expressed genes, none of which differed in abundance by >2-fold (35). In contrast, using single-cell expression profiling, we were able to detect 14 genes as up-regulated at least 7-fold in putative SC compared with TA cells. The new genes belonged to different ontological categories: rRNA modification (PDMT), ubiquitin ligases (KIAA1991 and FLJ12875), signal transduction (guanine nucleotide-binding protein-like 1, ZFP187, and Asxl-1), regulation of growth factor signaling (MCSP and Lrig1) cytoskeletal modulators (FERM4D and MAP4), and two housekeeping genes (phosphorylase protein kinase and KIAA1838). The SC profile demonstrates that, even when removed from their niche, SC are capable of retaining specific characteristics.
The identification of Lrig1 as a marker of epidermal SC fits well with the known importance of EGFR1 ligands for in vitro expansion of primary keratinocytes (36). The consequences of epidermal ablation of Lrig1 (26), Mig-6 (37), or the transcription factor AP2α (38) are all consistent with proliferation of the interfollicular SC compartment in response to EGFR ligand stimulation. Expression of Lrig1 ensures that SC are less responsive to growth-factor stimulation than their more differentiated progeny. Loss of Lrig1 is observed in psoriatic lesions and in squamous cell carcinomas (refs. 26 and 39 and data not shown), suggesting that, by triggering expansion of the SC compartment, it contributes to benign and neoplastic epidermal hyperproliferation. Our findings are in agreement with earlier studies showing that MAPK/ERK activation is required to maintain the epidermal SC compartment in culture (29). Rac1 is essential for epidermal homeostasis both in vivo and in vitro (40), and reduced expression of Lrig1 could potentiate EGF ligand-mediated activation of Rac1 by means of the exchange factor VAV (40, 41).
Our results lead us to propose a model in which Lrig1 expression maintains epidermal SC in a quiescent nonproliferative state (Fig. 4I), in part by negatively regulating Myc transcription. Down-regulation of Lrig1 triggers SC proliferation in response to EGF stimulation by causing a moderate increase in Myc that is not sufficient to stimulate terminal differentiation (32, 33, 42). This model is consistent with the finding that epidermal ablation of Myc (43) disrupts tissue homeostasis by impairing SC expansion. Furthermore, in psoriatic lesions where Lrig1 is lost, there is a prominent increase in the level of cMyc (ref. 44 and data not shown). One possibility that remains to be explored is that Myc-induced differentiation acts as a fail-safe device to prevent uncontrolled proliferation of SC when Lrig1 is down-regulated.
Materials and Methods
Antibodies and Chemicals.
The following antibodies were used: MCSP (9.2.27; PharMingen), Lrig-1 (gift from Satoshi Itami, University of Osaka, Osaka, Japan), phospho-EGFR1 (Y1086) (Zymed), EGFR1 (Ab-30; Abcam), EGFR (1005; Santa Cruz Biotechnology), Phospho-ERK (NEB), Ki67 (NCL-L-Ki67-MM1; Novocastra), β-tubulin (clone SAP.4G5; Sigma), K14 (LL002), cMyc (N-262; Santa Cruz Biotechnology), BrdU (ICR1; Oxford Biotechnology), ERK2 (K-23; Santa Cruz Biotechnology), Involucrin (SY-5), β1 integrin (P5D2), and α6 integrin (GoH3). BrdU, puromycin, 4OHT (all from Sigma-Aldrich), and AG1478 (Calbiochem) were used at 50 μM, 2 mg/ml, 100 ng/ml, and 10 μM, respectively.
Vectors.
The following vectors were used: pCMV-3xFLAG-Lrig1 [gift from Håkan Hedman, Umeå University, Umeå, Sweden (45)], pCMV-EGFP, pBabePuro-Lrig1Flag [gift from Yosef Yarden, Weizmann Institute, Rehovat, Israel (27)], pBabe Puro, pBabeMycER, pP2(-2489)-luc (46), pRL2, pSil5.1 Scr, and pRetroSuper-expressing siRNA to base pairs 1494–1512 of Lrig1.
Keratinocyte Culture and Retroviral Infection.
Primary human keratinocytes (strains kj, kt, and kv) were isolated from neonatal foreskin and cultured as described (15), unless otherwise stated. In some experiments, cells were starved of growth factors by overnight incubation in FAD + 0.5% FCS. Infection of human keratinocytes was carried out as described (15).
For transient transfections, keratinocytes were cultured in complete keratinocyte serum-free medium (Gibco) with EGF and bovine pituitary extract. Cells were transfected with a total of 5 μg of plasmid per well in six-well plates using Lipofectamine 2000 (Invitrogen). To measure proliferation, cells were allowed to recover for 1 day before incubation with BrdU for 24 h. To measure Myc promoter activity, plasmids pP2(-2489)-luc and pRL2 were transfected at a ratio of 4:1, lysed after 18 h, and assayed using the dual luciferase assay (Promega).
Single-Cell PCR.
Keratinocytes (strain kj) used to generate cDNA libraries had been cultured for no more than three passages, were subconfluent, and had received fresh medium 24 h before harvesting. Cells harvested in trypsin/EDTA were resuspended in FCS at 4°C and diluted in PBS. Single cells were collected by pipette and subjected to PCR essentially as described by Iscove et al. (21).
For the generation of cDNA, a single cell or 50 pg of purified total RNA was placed in 4.5 μl of first-strand buffer [1× Superscript III buffer (Invitrogen)/0.5% Nonidet P-40 (Pierce)/10 μM dNTP mixture (Roche)/3.4 nM oligo dT30 primer/1 mM DTT (Invitrogen)/5 units of SuperRNaseIN (Ambion)/7.5 units of PrimeRNase inhibitor (Eppendorf)], snap-frozen in liquid nitrogen, and lysed at 65°C for 5 min. Primer was allowed to anneal at 45°C for 2 min before addition of 0.5 μl of Superscript III and incubation at 45°C for 15 min. The reaction was inactivated at 65°C for 10 min. Five units of RNaseH (Invitrogen) were added in the presence of 5.77 mM MgCl2 in 6.5 μl for 15 min at 37°C before heat-inactivating at 65°C for 10 min. cDNA was polyadenylated at 37°C for 15 min with 6.5 μl of 2× terminal deoxynucleotidyltransferase buffer (Promega) supplemented with 1.5 mM dATP (Roche) and 30 units of terminal deoxynucleotidyltransferase (Promega) before heat-inactivating the enzyme at 65°C for 10 min.
Four microliters of polyadenylated cDNA was used as template for PCR amplification in 1× ExTaq buffer (TaKaRa)/0.65 mM dNTP (Roche)/8.25 μM oligo dT30 primer/5 units of ExTaq (TaKaRa) by incubating at 94°C for 1 min, 50°C for 2 min, and 72°C for 2 min to allow second-strand synthesis. Subsequently, 35 cycles of amplification were performed by incubating at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 2 min. A second round of amplification was performed as above by using 2 μl of the amplified cDNA as template according to the manufacturer's instructions.
Labeling of cDNA and Affymetrix Analysis.
Labeling was performed essentially as described (23). Forty micrograms of column-purified (Qiagen) cDNA was fragmented with 1 unit of RQ1 DNase (Promega) per 10 μg of cDNA in All-Phor-One buffer (Amersham Pharmacia) for 15 min at 37°C, and the enzyme was inactivated at 99°C for 10 min. Samples were labeled with 25 nmol of biotin-N6-dideoxyadenosine triphosphate (PerkinElmer) and 25 units of terminal deoxynucleotidyltransferase (Invitrogen) per 10 μg of cDNA for 1 h at 37°C. Fragmented and biotinylated cDNA was phenol/chloroform-precipitated and resuspended in distilled water (dH2O) at 1 μg/μl for Affymetrix analysis. Target cDNA generated from each sample was processed by using an Affymetrix GeneChip Instrument System. Spike controls were added to 10 μg of fragmented cDNA before overnight hybridization to GeneChip Human Genome 133 set oligonucleotide arrays (Affymetrix).
Array images produced by the Affymetrix GeneChip scanner were converted into raw files by using Microarray Suite 5.0 (Affymetrix). Unscaled raw files were analyzed by using Genespring 6.0 (Agilent Technologies). Measurements <0.01 were set to 0.01, chips were normalized to the 50th percentile, and genes were normalized to the median of the measurements for that gene. Pairwise correlation coefficients and the number of present probes were used to assess the quality of the samples.
Gene lists were from samples SC2, SC3, SC4, SC6, TA1, TA2, TA3, and TA5. Probes carrying a present call in four of four SC arrays, having average raw expression values >200, a SD of <1.4, and a fold difference of >7 were filtered on confidence for P values <0.05.
Adhesion Fractionation of Cultured Epidermal Keratinocytes.
Keratinocytes (5 × 106) were enriched for SC and TA cells by differential adhesion to 50 μg/ml type IV collagen (Sigma) and 50 μg/ml fibronectin, as described (13). RNA was isolated from adhesion-selected cells after lysis in TRIzol (Helena Bioscience).
Analysis of Genes by PCR, RT-PCR, and Real-Time PCR.
Screening of single-cell cDNA libraries by PCR was carried out with ExTaq (TaKaRa), by using 0.2 μl of the amplified samples as template. For RT-PCR from purified total RNA, 1 μg was used as template. Initially, samples were incubated with 1 unit of RQ1 DNase per microgram of total RNA to remove any genomic contamination. Subsequently, reverse transcription was carried out by using Superscript II (Invitrogen) and an oligo dT20 primer. Semiquantitative PCR was performed with gene-specific primers by using a serial dilution of template. All primer sequences are included in Supporting Text, which is published as supporting information on the PNAS web site.
Quantitative PCR was performed as described (42) for genes Actin, Gapdh, β1 Integrin, Delta1, Mcsp, 18S ribosomal rRNA, and Lrig1. Real-time PCRs and analysis were performed with an ABI Prism 7700 Sequence Detection System (Applied Biosystems). Relative quantification of each gene was determined using the standard curve method. The relative amount of each mRNA was normalized to the level of β-Actin or 18S ribosomal rRNA, as indicated.
Immunostaining and Flow Cytometry.
Frozen tissue sections and cultured cells were fixed with 2% paraformaldehyde for 10 min and, if necessary, permeabilized for 5 min with 0.4% Triton X-100. After blocking with 10% FCS in PBS, sections and cells were incubated for 1 h with antibodies diluted in 10% FCS in PBS. Secondary antibodies were conjugated with Alexa Fluor 488 or 594 (Molecular Probes).
For flow cytometry, primary human keratinocytes were labeled with antibodies to EGFR1, α6 integrin, or β1 integrin for 30 min at 4°C. Cells were labeled with anti-mouse or -rat secondary Alexa Fluor 488-conjugated antibody (Molecular Probes) and analyzed by using a FACscaliburII sorter and Cell Quest FACS analysis system (BD Biosciences).
Protein Lysates and Western Blotting.
Keratinocytes were scraped into RIPA buffer and lysed on ice for 30 min. Equal amounts of protein were loaded for analysis using SDS/PAGE and Western blotting as described (40).
Supplementary Material
Acknowledgments
We thank Osawa Masatake and Shin-Ichi Nishikawa (RIKEN Institute, Kobe, Japan) for teaching us how to make single-cell cDNA libraries; Håkan Hedman, Yosef Yarden, and Satoshi Itami for reagents; and the Cancer Research UK Microarray Facility for expert technical assistance. K.B.J. is the recipient of fellowships from the Carlsberg Foundation and the Danish Medical Research Council. This work was supported by Cancer Research UK.
Abbreviations
- MCSP
melanoma-associated chondroitin sulfate proteoglycan
- 4OHT
4-hydroxy-tamoxifen
- SC
stem cell(s)
- TA
transit amplifying
- EGFR1
EGF receptor 1
- Lrig1
leucine-rich repeats and immunoglobulin-like domains 1
- MycER
4OHT-inducible form of Myc.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The raw data from the expression profiling experiments reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE4858).
References
- 1.Owens D. M., Watt F. M. Nat. Rev. Cancer. 2003;3:444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 2.Alonso L., Fuchs E. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl. 1):11830–11835. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris R. J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J. S., Sawicki J. A., Cotsarelis G. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 5.Cotsarelis G., Sun T. T., Lavker R. M. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 6.Levy V., Lindon C., Harfe B. D., Morgan B. A. Dev. Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Jones P. H., Harper S., Watt F. M. Cell. 1995;80:83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 8.Potten C. S., Morris R. J. J. Cell Sci. 1988;10(Suppl.):45–62. doi: 10.1242/jcs.1988.supplement_10.4. [DOI] [PubMed] [Google Scholar]
- 9.Ghazizadeh S., Taichman L. B. EMBO J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claudinot S., Nicolas M., Oshima H., Rochat A., Barrandon Y. Proc. Natl. Acad. Sci. USA. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito M., Liu Y., Yang Z., Nguyen J., Liang F., Morris R. J., Cotsarelis G. Nat. Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 12.Niemann C., Watt F. M. Trends Cell Biol. 2002;12:185–192. doi: 10.1016/s0962-8924(02)02263-8. [DOI] [PubMed] [Google Scholar]
- 13.Jones P. H., Watt F. M. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 14.Li A., Simmons P. J., Kaur P. Proc. Natl. Acad. Sci. USA. 1998;95:3902–3907. doi: 10.1073/pnas.95.7.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowell S., Jones P., Le Roux I., Dunne J., Watt F. M. Curr. Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 16.Wan H., Stone M. G., Simpson C., Reynolds L. E., Marshall J. F., Hart I. R., Hodivala-Dilke K. M., Eady R. A. J. Cell Sci. 2003;116:4239–4248. doi: 10.1242/jcs.00701. [DOI] [PubMed] [Google Scholar]
- 17.Fortunel N. O., Hatzfeld J. A., Rosemary P. A., Ferraris C., Monier M. N., Haydont V., Longuet J., Brethon B., Lim B., Castiel I., et al. J. Cell Sci. 2003;116:4043–4052. doi: 10.1242/jcs.00702. [DOI] [PubMed] [Google Scholar]
- 18.Potten C. S., Loeffler M. Development (Cambridge, U.K.) 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 19.Legg J., Jensen U. B., Broad S., Leigh I., Watt F. M. Development (Cambridge, U.K.) 2003;130:6049–6063. doi: 10.1242/dev.00837. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama M., Terunuma A., Tock C. L., Radonovich M. F., Pise-Masison C. A., Hopping S. B., Brady J. N., Udey M. C., Vogel J. C. J. Clin. Invest. 2006;116:249–260. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iscove N. N., Barbara M., Gu M., Gibson M., Modi C., Winegarden N. Nat. Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- 22.Osawa M., Egawa G., Mak S. S., Moriyama M., Freter R., Yonetani S., Beermann F., Nishikawa S. Development (Cambridge, U.K.) 2005;132:5589–5599. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- 23.Tietjen I., Rihel J. M., Cao Y., Koentges G., Zakhary L., Dulac C. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 24.Ghiglione C., Carraway K. L., 3rd, Amundadottir L. T., Boswell R. E., Perrimon N., Duffy J. B. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 25.Wilson C. L., Pepper S. D., Hey Y., Miller C. J. BioTechniques. 2004;36:498–506. doi: 10.2144/04363RN05. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y., Miura H., Tanemura A., Kobayashi K., Kondoh G., Sano S., Ozawa K., Inui S., Nakata A., Takagi T., et al. FEBS Lett. 2002;521:67–71. doi: 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- 27.Gur G., Rubin C., Katz M., Amit I., Citri A., Nilsson J., Amariglio N., Henriksson R., Rechavi G., Hedman H., Wides R., Yarden Y. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen U. B., Lowell S., Watt F. M. Development (Cambridge, U.K.) 1999;126:2409–2418. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- 29.Zhu A. J., Haase I., Watt F. M. Proc. Natl. Acad. Sci. USA. 1999;96:6728–6733. doi: 10.1073/pnas.96.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashimata M., Gresik E. W. Dev. Dyn. 1997;208:149–161. doi: 10.1002/(SICI)1097-0177(199702)208:2<149::AID-AJA2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Gandarillas A., Watt F. M. Genes Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waikel R. L., Kawachi Y., Waikel P. A., Wang X. J., Roop D. R. Nat. Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 33.Arnold I., Watt F. M. Curr. Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 34.Chuang C. F., Ng S. Y. FEBS Lett. 1994;346:229–234. doi: 10.1016/0014-5793(94)00480-3. [DOI] [PubMed] [Google Scholar]
- 35.O'Shaughnessy R. F., Seery J. P., Celis J. E., Frischauf A., Watt F. M. FEBS Lett. 2000;486:149–154. doi: 10.1016/s0014-5793(00)02252-3. [DOI] [PubMed] [Google Scholar]
- 36.Rheinwald J. G., Green H. Nature. 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- 37.Ferby I., Reschke M., Kudlacek O., Knyazev P., Pante G., Amann K., Sommergruber W., Kraut N., Ullrich A., Fassler R., Klein R. Nat. Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Bolotin D., Chu D. H., Polak L., Williams T., Fuchs E. J. Cell Biol. 2006;172:409–421. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanemura A., Nagasawa T., Inui S., Itami S. Dermatol. Surg. 2005;31:423–430. doi: 10.1111/j.1524-4725.2005.31108. [DOI] [PubMed] [Google Scholar]
- 40.Benitah S. A., Frye M., Glogauer M., Watt F. M. Science. 2005;309:933–935. doi: 10.1126/science.1113579. [DOI] [PubMed] [Google Scholar]
- 41.Fernandez-Zapico M. E., Gonzalez-Paz N. C., Weiss E., Savoy D. N., Molina J. R., Fonseca R., Smyrk T. C., Chari S. T., Urrutia R., Billadeau D. D. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 42.Frye M., Gardner C., Li E. R., Arnold I., Watt F. M. Development (Cambridge, U.K.) 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- 43.Zanet J., Pibre S., Jacquet C., Ramirez A., de Alboran I. M., Gandarillas A. J. Cell Sci. 2005;118:1693–1704. doi: 10.1242/jcs.02298. [DOI] [PubMed] [Google Scholar]
- 44.Honma M., Benitah S. A., Watt F. M. Mol. Biol. Cell. 2006;17:1888–1896. doi: 10.1091/mbc.E05-12-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson J., Starefeldt A., Henriksson R., Hedman H. Cell Tissue Res. 2003;312:65–71. doi: 10.1007/s00441-003-0697-1. [DOI] [PubMed] [Google Scholar]
- 46.Lee T. C., Ziff E. B. J. Biol. Chem. 1999;274:595–606. doi: 10.1074/jbc.274.2.595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.