Abstract
The phylogenetic enigma of snail hemoglobin, its isolated occurrence in a single gastropod family, the Planorbidae, and the lack of sequence data, stimulated the present study. We present here the complete cDNA and predicted amino acid sequence of two hemoglobin polypeptides from the planorbid Biomphalaria glabrata (intermediate host snail for the human parasite Schistosoma mansoni). Both isoforms contain 13 different, cysteine-free globin domains, plus a small N-terminal nonglobin “plug” domain with three cysteines for subunit dimerization (total Mr ≈ 238 kDa). We also identified the native hemoglobin molecule and present here a preliminary 3D reconstruction from electron microscopical images (3 nm resolution); it suggests a 3 × 2-mer quaternary structure (Mr ≈ 1.43 MDa). Moreover, we identified a previously undescribed rosette-like hemolymph protein that has been mistaken for hemoglobin. We also detected expression of an incomplete hemocyanin as trace component. The combined data show that B. glabrata hemoglobin evolved from pulmonate myoglobin, possibly to replace a less-efficient hemocyanin, and reveals a surprisingly simple evolutionary mechanism to create a high molecular mass respiratory protein from 78 similar globin domains.
Keywords: evolution, hemocyanin, Mollusca
Among the plethora of gastropods using copper-containing hemocyanin for O2 transport (1–4), a single family of freshwater snails, the Planorbidae (trumpet snails), contains a high molecular mass extracellular hemoglobin (5–10). Planorbid hemoglobin has been analyzed in many details biochemically (5–10), and some peculiar features have been revealed. In contrast to the more usual hemoglobin polypeptides of Mr ≈ 17 kDa, planorbid hemoglobin is composed (according to SDS/PAGE) of polypeptides with an apparent Mr of 175–200 kDa, encompassing at least 10 globin domains. It has been proposed that two such subunits dimerize via several disulfide bridges and that four or five such dimers constitute the native protein (5–10). In electron micrographs, rosette-like rings and rather irregular particles, each of ≈20 nm in diameter, have been observed, and it has been suggested that both structures represent different views, or different stability forms, of the hemoglobin molecule (6–8). Light scattering and scanning transmission electron microscopy indicated, for the native protein, a molecular mass of ≈1.8 MDa in the case of Helisoma (Planorbella) trivolis (8), whereas analytical ultracentrifugation suggested 1.65 MDa in the case of Planorbis corneus (6). On the basis of small angle x-ray scattering, a 4 × 2-mer quarternary structure with pointgroup D2 symmetry has been proposed for Biomphalaria glabrata hemoglobin (BgHb) (10). However, because of the complete lack of amino acid sequence data, and the lack of more detailed electron microscopical data, not only the subunit structure but also the quaternary structure of planorbid hemoglobin remained unclear. On the other hand, the planorbid B. glabrata is a major intermediate host species for the human parasite Schistosoma mansoni (11), and as the major hemolymph protein, hemoglobin may contribute to parasite–host interaction, thereby probably influencing parasite survival (12). This interesting connection, and the insular occurrence of planorbid hemoglobin within the usually blue-blood gastropods, stimulated us to trace its phylogenetic origin, solve its structure, and unravel whether it really once replaced a hemocyanin.
Results and Discussion
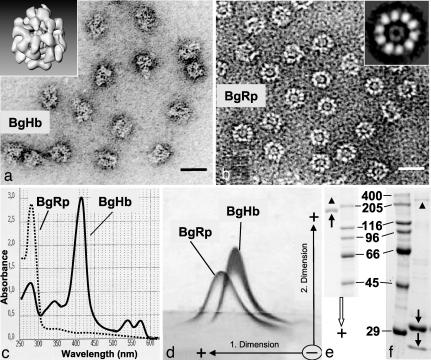
Electron microscopy of B. glabrata hemolymph proteins revealed ≈95% of asymmetric particles and ≈5% of symmetrical double rings (rosettes) of ≈20 nm in diameter; this result corresponds to observations in other planorbids (6, 7), and both structures have been interpreted as hemoglobin (7). After their HPLC separation (Fig. 1a and b), we identified the asymmetric molecules as BgHb by correlation with their characteristic UV-visible spectrum (Fig. 1c). However, the rosettes (BgRp, from B. glabrata “rosette protein”) are neither hemoglobin nor hemocyanin (Fig. 1 c–f): They possessed the typical UV-visible spectrum of a protein lacking a pigment, did not fuse with BgHb in 2D immunoelectrophoresis, and showed two polypeptides of 31 and 25 kDa instead of a single 180-kDa band as obtained from BgHb. Further characterization of this protein is warranted, also in view of other annular proteins found in gastropods (13) and growing evidence that B. glabrata hemolymph proteins contribute to parasite–host interaction (e.g., ref. 12).
Fig. 1.
Identification of purified BgHb and BgRp. (a) Electron microscopy of HPLC-purified BgHb molecules. (a Inset) Three-dimensional reconstruction (see Fig. 5). (b) Electron microscopy of HPLC-enriched BgRp molecules. (b Inset) Class sum image from 10 top views aligned by IMAGIC. (Scale bars: 25 nm.) (c) UV-visible spectra of BgHb and BgRp. (d) Tandem-crossed immunoelectrophoresis of purified BgHb and BgRp against rabbit antibodies versus B. glabrata hemolymph proteins. Note the separate precipitation of the two protein peaks indicating nonidentity. (e) SDS/PAGE of BgHb (arrow) showing an apparent Mr of 180 kDa. A trace of the disulfide-bridged dimer of BgHb is also visible (arrowhead). Marker protein masses are indicated in kilodaltons. (f) SDS/PAGE of BgRP, showing two polypeptides (arrows) with 31 and 25 kDa, respectively. Traces of the hemocyanin subunit (arrowhead; see also Fig. 2) are also visible.
In the hemolymph, we also detected trace quantities of molecules that morphologically resembled molluscan hemocyanin (Fig. 2). However, in contrast to the semihollow didecameric cylinders of a typical gastropod hemocyanin (3), the molecules in B. glabrata appear as decamers and hollow rings because of the absence of the internal collar complex (see Fig. 2). Indeed, the polypeptide chain that most probably represents B. glabrata hemocyanin (BgHc) has an apparent molecular mass of only 300 kDa compared with 400 kDa as usual for gastropod hemocyanins (refs. 1 and 4; see Fig. 1f, the largest marker polypeptide is keyhole limpet hemocyanin). We considered that the trace expression of a mutated hemocyanin in B. glabrata would have little or no significance for oxygen transport but it might still function as a phenoloxidase that, according to data from crustaceans, is required only in minute quantities (14). However, a staining assay of protein bands in native PAGE for phenoloxidase activity carried out in the laboratory of H. Decker yielded negative results on BgHc, BgRp, and BgHb. cDNA analysis confirmed hemocyanin expression in B. glabrata and, moreover, revealed that in at least one of its oxygen-binding functional units, an active site histidine is substituted for glutamine (see Fig. 2). This mutation possibly will impair the ability of this putative hemocyanin to bind oxygen. Nevertheless, the presence of hemocyanin provides convincing evidence that the Planorbidae did descend from blue-blooded ancestors. A co-occurrence of hemoglobin and hemocyanin is highly unusual and has been reported only in the amphipod Cyamus scammoni, in the phylum Arthropoda (15).
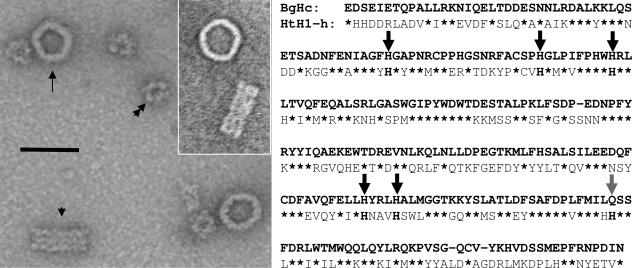
Fig. 2.
Identification of BgHc. (Left) Electron microscopy of BgHc molecules in the top view (arrow; note the lack of internal collar complex) and side view (arrowhead; the larger diameter is pretended by a flattening effect), together with contaminating BgRp molecules (double arrowhead; the views differ from that in Fig. 1b). (Scale bar: 50 nm.) (Left Inset) Two BgHc molecules from another preparation. (Right) The polypeptide encoded by the partial cDNA sequence of BgHc shares many identical residues (asterisks) with the functional unit HtH1-h (1) (and also with the other functional units of Haliotis tuberculata hemocyanin), but only five of the six copper-binding histidines are conserved (black arrows); one histidine is substituted for glutamine (gray arrow). (More BgHc sequences are available upon request.)
By a combination of EST data and RT-PCR with primers based on the known sequence of B. glabrata myoglobin (16), we generated and sequenced cDNAs encoding two complete hemoglobin polypeptides, termed BgHb1 and BgHb2. The overlap between the different clones was at least 150 bp to ensure the correct concatenation of fragments. Instead of 10 domains as proposed, both BgHb polypeptides comprise 13 different globin domains (termed BgHb1-a to BgHb1-m and BgHb2-a to BgHb2-m; each containing 155–157 aa, Mr 17.3–17.8 kDa). The presence of potential N-glycosylation sites in BgHb1 and BgHb2 is concordant with previous analyses of sugar modifications (17, 18). To our surprise, we found ahead of the first globin domain a further domain that is rather small (90 aa in the case of BgHb1; Mr = 10.7 kDa); this domain is unrelated to globins and has no similarity with any other sequence in the data banks. When reducing agents are omitted in SDS/PAGE, a 360-kDa band is observed instead of the 180-kDa hemoglobin chain (5–10), and there is also direct biochemical evidence for disulfide bonds (19). Therefore, it was rather confusing that the 13 globin domains completely lacked any cysteine, but this puzzle was solved when we finally detected three cysteines in the small N-terminal domain (in BgHb1 and BgHb2, a fourth cysteine is in the putative signal peptide and, therefore, is posttranslationally removed). It is likely that this domain functions, in an unanticipated fashion, as a plug connection for subunit dimerization (see below); we therefore termed it “plug domain” (BgHb1-p and BgHb2-p). In 2D-PAGE, only partial separation of hemoglobin polypeptides could be achieved (data not shown), but for BgHb1, it could be confirmed by N-terminal protein sequencing that we indeed sequenced a full-length clone. Also, MALDI-MS was performed to correlate the sequences to the protein spots (data not shown). We also sequenced two globin domains of a third Hb polypeptide (termed BgHb3). The question whether these three polypeptides represent different hemoglobin isoforms (homooligomers) or different subunits of the same hemoglobin (heterooligomer) still is open.
We performed multiple protein sequence alignments of the various BgHb domains together with several bivalve hemoglobins and gastropod myoglobins (Fig. 3). As expected from the insular occurrence of planorbid hemoglobin, and from very different quaternary structures (see literature cited in ref. 20), the bivalve hemoglobin sequences are too remote to serve as suitable outgroups (sequence identity with BgHb ≈17%). Even sea hare (Aplysia) myoglobin shares only 17 ± 2% identity with the BgHb domains. However, myoglobin from B. glabrata, and also that from Lymnaea stagnalis (pond snail; possessing hemocyanin), show 54 ± 4% identity with the BgHb domains; similar values are obtained if the paralogous BgHb domains are compared. Orthologous domains from the different BgHb polypeptides, however, show 71 ± 7% identity. Phylogenetic trees constructed from such alignments (Fig. 4) strongly suggest that planorbid hemoglobin evolved by multiple gene duplication and gene fusion events from an ancient pulmonate myoglobin, as predicted in ref. 21. Moreover, it is clear that BgHb1 and BgHb2 originated from a single gene duplication event, which occurred after evolution of the complex molecular organization of the initial hemoglobin subunit. Relative rate tests were performed that showed that some heterogeneity in the evolutionary rates can be observed when the different domains are compared, but their mean value supports a rather homogenous evolutionary rate. The oldest fossil record of a land snail (Dawsonella meeki) stems from the Carboniferous, 325 million years ago (22), and has been used here to define the origin of the pulmonates; the oldest fossil of a planorbid dates back 175 million years (23) and defines here the origin of BgHb. On this basis, we calibrated a molecular clock; it suggests that the BgHb1/BgHb2 split occurred 110 ± 49 million years ago in the late Mesozoic. (A somewhat earlier duplication event apparently created BgHb3.)
Fig. 3.
Multiple sequence alignment of the different globin domains. The aligned sequences are from BgHb1, BgHb2, and BgHb3 (Upper) and other molluscan heme-containing globins (Lower). The first two lines show the N-terminal “plug” domains BgHb1-p and BgHb2-p, each containing three conserved cystein residues (pink) and an N-terminal signal peptide (italic). Three potential N-glycosylation sites are marked in yellow. Note that all proximal histidines are replaced by glutamines (left double arrow), whereas the distal histidine (right double arrow) and two phenyalanines are strictly conserved (red). Other conserved residues are marked in blue (80%) or green (60%). A presumptive flexible linker region joining the 13 domains is marked by a bracket. The well known secondary structure of hemoglobin/myoblobin is added for better orientation. LsMb, L. stagnalis contig of CN810207/CN810223/CN810610/CN810699/CN810837/CN811024; BgMb, Biomphalaria U89283; NmMb, Nassarius (Nassa) mutabilis P31331 (Neogastropoda); BuMb, Buccinum undatum A44588 (Neogastropoda); SiHb, Scapharca inaequivalvis hemoglobin A chain S83524 (Bivalvia); BlHb, Barbatia lima α chain of the tetrameric (intracellular) hemoglobin D63933 (Bivalvia); AjMb, Aplysia juliana AB003277 (sea hare).
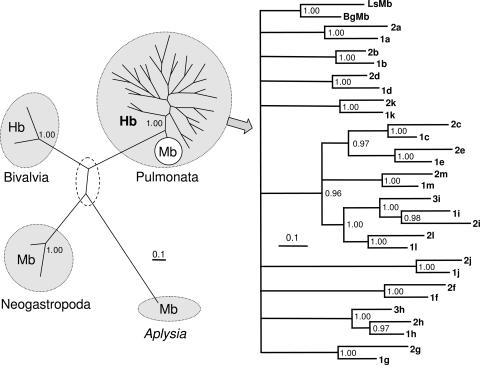
Fig. 4.
Molecular phylogeny of hemoglobin domains (Hb) and myoglobins (Mb) from mollusks. Note in the left unrooted tree the very large phylogenetic distance between pulmonate hemo/myoglobins on the one hand and other molluscan hemo/myoglobins on the other hand. (The branching order within the dashed field is not well supported.) Note that in the right tree, which has been rooted with the two Mb sequences, the topologically corresponding globin domains from the two polypeptides BgHb1 and BgHb2 are more similar to each other than any two domains within either BgHb1 or BgHb2, thereby indicating that the two polypeptides diversified by a single gene duplication event. The two as-yet-sequenced additional domains indicate a third polypeptide (BgHb3). The trees suggest that planorbid hemoglobin evolved by a series of gene duplications and fusions from an ancestral planorbid myoglobin. Posterior probabilities >0.95 are indicated at each node of the rooted tree. Nodes with lower supporting values are collapsed. The different BgHb domains are labeled according to the alignment of Fig. 3. Bg, B. glabrata; Ls, L. stagnalis; 1a–1m, domains BgHb1a to BgHb1m; 2a –2m, domains BgHb2a to BgHb2m; 3h and 3i, domains BgHb3h and BgHb3i
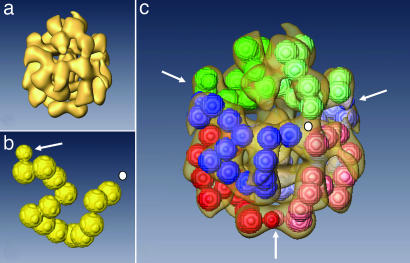
The calculated molecular mass of the BgHb1 polypeptide is 238 kDa instead of the ≈180 kDa as deduced from SDS/PAGE (see Fig. 1e), which means that the subunit dimer mass is 476 kDa. It has been suggested from small angle x-ray scattering that the native 1.4–1.8 MDa molecule is a tetramer of dimers (4 × 2-mer) with point group D2 symmetry (10). To prove this model, we performed transmission electron microscopy-based single particle analysis (3) to provide a preliminary 3D reconstruction at 3 nm resolution (1/2-bit criterion). For the latter, 1,818 negatively stained molecules representing very different views were arranged into 333 final classes. Initially, a C1 and then a C2 symmetry was assumed (and confirmed by evaluating the quality of reprojections), and a 3D reconstruction at 3 nm resolution was retrieved (Fig. 5). This reconstruction suggests the presence of six repeating units arranged in pairs (3 × 2-mer/D3 symmetry), and challenges the earlier 4 × 2-mer/D2 symmetry model (10). It allowed a preliminary fit of six similar copies of a BgHb chain model, together representing 78 globin domains plus six plug domains (Fig. 5; see also Fig. 6, which is published as supporting information on the PNAS web site). This configuration comes very close to the 80 globin domains assumed earlier on the basis of eight subunits and 10 domains for each subunit. From the reconstruction, a maximum diameter of the BgHb molecule of 17.5 nm was calculated. With the negatively stained images used here, we failed to further improve this 3D reconstruction, and cryoelectron microscopy (3) can be applied to prove the new model.
Fig. 5.
Three-dimensional reconstruction of the quaternary structure of BgHb. (a) 3D reconstruction of native BgHb (resolution, 3 nm). (b) Model of the BgHb polypeptide subunit with 13 globular masses for the heme domains and N-terminally a smaller mass for the plug domain (arrow). (c) The same view as in a, incorporating six polypeptide subunits as modeled in b, distinguished by different colors. By bringing together the N-terminal plug domains (arrows) of pairs of polypeptide chain subunits, this model is consistent with the occurrence of disulfide-bridged dimers of polypeptide chains, as was demonstrated biochemically. Additionally, subunits form triplets by joining C-terminal domains (white dot). Also see Fig. 6.
But why did these snails change their respiratory protein, from hemocyanin to hemoglobin, when hemocyanin has successfully performed its task in countless mollusks for ≈740 million years (1)? The only other known gastropod that has hemoglobin is the deep-sea vent caenogastropod Alviniconcha hessleri, which harbors the red pigment in its gill to support endosymbiotic bacteria (24). Functional analysis has revealed that BgHb exhibits a high oxygen affinity (p50 ≈6 mmHg; 1 mmHg = 133 Pa); this feature is interpreted as an adaptation of the snails to their pond habitat, where they experience extreme variation in oxygen gas tension and temperature (20); functional studies of Helisoma (Planorbella) trivolvis Hb domains and intact Hb point into the same direction (25). Other freshwater snail species, such as the Lymnaeidae that employ hemocyanin for oxygen transport, also prosper in similar habitats. However, because of the comparatively higher affinity of their hemoglobin for oxygen, planorbids have a more efficient physiological exploitation of the pulmonary O2 store, yielding a substantially increased diving potential, compared with lymneids (20).
Also, various clam species possess high-affinity hemoglobin that is closely related to their respective myoglobin, but most hemoglobins in bivalves are small and intracellular; the only known exceptions are the huge 14- to 24-domain rodlike hemoglobins in Astarte and Cardita (26, 27). In the deep-sea clam Calyptogena kaikoi, the major physiological role of hemoglobin is apparently storage of oxygen under low-oxygen conditions rather than circulating of oxygen (28). As suggested by Mangum (29), hemocyanin probably is not able to evolve into high-affinity forms, in contrast to hemoglobin. An alternative scenario is suggested by the existence of the truncated BgHc (see Fig. 2). Of course, the underlying mutations might have happened after hemocyanin was replaced by hemoglobin, but what if these mutations occurred first? This incomplete hemocyanin still may have served for oxygen transport in a limited way, which was once sufficient for a terrestrial life but became a handicap, needing to be replaced when the terrestrial ancestors of the planorbids colonized freshwater habitats (20).
Regardless of which reason hemocyanin was abandoned as an oxygen transporter in planorbids, the evolution of a high molecular mass extracellular hemoglobin (in adaptation to colloid-osmotic, rheological, and functional constraints; refs. 20 and 29) from a small intracellular myoglobin, usually present in the gastropod radula muscle (26), is strikingly elegant. This hemoglobin is ingeniously designed in that multiple gene duplications and fusions, plus the introduction of a simple plug connection, enabled this intriguing multimolecular association to occur without major changes of the globin domains. Interestingly, the prime attribute of molluscan hemocyanin, the large multidomain polypeptide, is also a structural feature in planorbid hemoglobin. Moreover, the higher-order quaternary structure of both proteins is achieved by using the subunit dimer as the repeating unit. Remarkably, gastropods synthesize both proteins in the same cell type, the so-called pore cells or rhogocytes, from which they are released by merocrine secretion (30, 31). It therefore appears that the planorbid snails use the same cellular machinery as their gastropod relatives to express red blood as an alternative to the phylogenetically older blue blood.
Materials and Methods
Animals and Hemolymph Collection.
B. glabrata were obtained as a gift from R. Geyer and M. Dornhoff (University of Giessen, Giessen, Germany). Before bleeding, animals were cooled on ice. Hemolymph was collected by inserting a thin needle into the foot of the animals, and immediately after bleeding, protease inhibitor (Pefabloc; Roth, Karlsruhe, Germany) was added to a final concentration of 1 mM. Blood cells and cellular debris were removed by centrifugation at 10,000 × g for 10 min at room temperature.
Purification of Hemolymph Proteins.
Hemolymph proteins were purified by HPLC (Gilson, Middleton, WI) by using a Q-Sepharose anion-exchange column with a bed volume of 20 cm3 (Amersham Pharmacia, Germany). Protein (20–30 mg) was applied to the column and eluted with a step gradient of NaCl (0–0.5 M) in “stabilization buffer” (50 mM Tris·HCl/5 mM CaCl2/5 mM MgCl2, pH 7.4). The flow rate was 0.5 ml/min. Two-milliliter fractions were collected, containing either purified BgHb (>95% of total protein) or BgRp/BgHc (<5% of total protein). The latter fractions were combined, concentrated to a volume of 0.5 ml, and applied to a Sephacryl S-400 gel permeation column (Amersham Pharmacia, Freiburg, Germany) with a bed volume of 30 ml. Proteins were eluted with stabilization buffer, and 0.5-ml fractions were collected.
Protein Analysis.
Negatively stained specimens for transmission electron microscopy were prepared from hemolymph proteins by the single-droplet procedure as described in ref. 4; before specimen preparation, the protein sample was treated with 0.05% glutaraldehyde, because otherwise structural alterations of the particles later identified as hemoglobin were observed. Three-dimensional reconstruction by single-particle analysis of 1,818 individual hemoglobin molecules by using the IMAGIC software package was done as described in ref. 3. This analysis was possible from negatively stained particles because they were present on the grids in many different views. The reconstruction was visualized by the AMIRA software package, and the model hemoglobin chain was fitted by hand. SDS/PAGE in 10% gels and crossed immunoelectrophoresis also were performed as described in ref. 4. Rabbit anti-Biomphalaria total hemolymph protein antibodies were used. N-terminal sequencing and peptide pass mapping by MALDI-TOF mass spectrometry were done by commercial services (Hans Heid, Deutsches Krebsforschungszentrum, Heidelberg, Germany, and Christian Hunzinger, ProteoSys, Mainz, Germany, respectively).
RNA Preparation.
RNA from whole tissue was isolated by using the PolyATtract mRNA Isolation System (Promega) and performed according to the manufacturer’s instruction manual. The bound RNA finally was eluted by using diethyl pyrocarbonate-treated water. The RNA was stored at −20°C.
RT-PCR, Electrophoresis, and Cloning.
Reverse transcription was performed with the Transcriptor First-Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) by using 100 ng of mRNA, according to the proposed protocol of the manufacturer. First, primers were depicted from short sequence data, which were obtained by screening a cDNA library. Subsequent primers were designed from the obtained data and combined with first primers, which strongly cross-reacted within other domains. PCR was done by using a three-step PCR protocol with an initial denaturing step of 94°C for 2 min and a final extension step of 10 min at 72°C. Cycling was performed for 35 times with two different primer annealing temperatures (53°C and 60°C for 30 sec). Denaturation was 10 sec, and the polymerization step was 2 min. The 5′ end of the cDNA of BgHb1 and BgHb2 was obtained by chance, whereas the 3′ end of BgHb1 and BgHb2 was obtained by using a standard oligo(dT)-primer in combination with a specific primer depicted from BgHb1-m and BgH2-m, respectively. PCR products were separated in a standard 0.8% agarose gel by using 1× TBE (89 mM Tris/90 mM boric acid/ 2 mM EDTA, pH 8) as the electrophoresis buffer. Bands were extracted by using the Spin Gel Extraction Kit from Qiagen (Hilden, Germany) and cloned into Topo TA (Invitrogen, Karlsruhe, Germany) or sequenced directly. Recombinant plasmid-containing clones were processed by using the plasmid Miniprep Spin Kit from Peqlab (Erlangen, Germany) or PCR verified. cDNAs were sequenced by commercial services from both ends by using M13 forward/reverse primers. Direct sequencing and primer walking was done with gene-specific primers purchased from Sigma-Ark (Darmstadt, Germany). Cycle sequencing reactions were performed by using the Taq DyeTerminator system.
Phylogenetics.
Multiple amino acid sequence aligments were performed by using ClustalX 1.83 (ref. 32; ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) and manually optimized by using Genedoc (ref. 33; www.psc.edu/biomed/genedoc). Tree reconstructions were done by the Bayesian Monte Carlo Markov Chain method implemented within MrBayes (http://mrbayes.csit.fsu.edu). The best model was identified by using the ProtTest (ref. 34, http://darwin.uvigo.es/software/prottest_server.html). Four chains were run simultaneously for 600,000 generations, burn-in was 60,000 generations, and trees were sampled every 100 generations. The consensus tree was edited and visualized by using Treeview 1.6.6 (ref. 35; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Supplementary Material
Acknowledgments
We thank Rudolf Geyer and Michael Dornhoff for the snails, Thomas Schubert for technical assistance, Annette Amann for preliminary protein analyses, Dorothea Nillius in the laboratory of Heinz Decker (University of Mainz, Mainz, Germany) for performing the phenol oxidase test, and J. Robin Harris (University of Mainz) for critical reading of the manuscript and valuable suggestions. This work has benefited from the Biomphalaria genome sequencing that was initiated recently (http://biology.unm.edu/biomphalaria-genome). The German group has been supported by the research fund of Rheinland-Pfalz (J.M.), Deutsche Forschungsgemeinschaft Graduate College Grant GK1043 (to A.M.), and the Feldbausch Foundation (B.L.). The U.S. group has been supported by National Institutes of Health (NIH) Grant R01 AI052363 (to C.M.A.) and the NIH Minority Access to Research Careers program at the University of New Mexico (S.A.S.).
Abbreviations
- BgHb
Biomphalaria glabrata hemoglobin
- BgRp
B. glabrata rosette protein
- BgHc
B. glabrata hemocyanin.
Footnotes
References
- 1.Lieb B., Altenhein B., Markl J. J. Biol. Chem. 2000;275:5675–5681. doi: 10.1074/jbc.275.8.5675. [DOI] [PubMed] [Google Scholar]
- 2.Lieb B., Altenhein B., Markl J., Vincent A., van Olden E., van Holde K. E., Miller K. I. Proc. Natl. Acad. Sci. USA. 2001;98:4546–4551. doi: 10.1073/pnas.071049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meissner U., Dube P., Harris J. R., Stark H., Markl J. J. Mol. Biol. 2000;298:21–34. doi: 10.1006/jmbi.2000.3631. [DOI] [PubMed] [Google Scholar]
- 4.Lieb B., Boisguérin V., Gebauer W., Markl J. J. Mol. Evol. 2004;59:536–545. doi: 10.1007/s00239-004-2646-3. [DOI] [PubMed] [Google Scholar]
- 5.Figueiredo E. A., Gomes M. V., Heneine I. F., Santos I. O., Hargreaves F. B. Comp. Biochem. Physiol. B. 1973;44:481–491. [Google Scholar]
- 6.Wood E. J., Mosby L. J. Biochem. J. 1975;149:437–445. doi: 10.1042/bj1490437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terwilliger N. B., Terwilliger R. C., Schabtach E. Biochim. Biophys. Acta. 1976;453:101–110. doi: 10.1016/0005-2795(76)90254-3. [DOI] [PubMed] [Google Scholar]
- 8.Herskovits T. T., Hamilton M. G. Comp. Biochem. Physiol. B. 1994;107:433–441. doi: 10.1016/0305-0491(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 9.Arndt M. H. L., Santoro M. M. Comp. Biochem. Physiol. B. 1998;119:667–675. [Google Scholar]
- 10.Arndt M. H. L., de Oliveira D. L. P., Regis W. C. B., Torriani I. L., Santoro M. M. Biopolymers. 2003;69:470–479. doi: 10.1002/bip.10367. [DOI] [PubMed] [Google Scholar]
- 11.Morgan J. A., Dejong R. J., Adeoye G. O., Ansa E. D., Barbosa C. S., Bremond P., Cesari I. M., Charbonnel N., Correa L. R., Coulibaly G., et al. Mol. Ecol. 2005;14:3889–3902. doi: 10.1111/j.1365-294X.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S. M., Adema C. M., Kepler T. B., Loker E. S. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 13.Harris J. R., Markl J. Eur. J. Biochem. 1994;225:521–528. doi: 10.1111/j.1432-1033.1994.00521.x. [DOI] [PubMed] [Google Scholar]
- 14.Decker H., Tuczek F. Trends Biochem. Sci. 2000;25:392–397. doi: 10.1016/s0968-0004(00)01602-9. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger N. B., Ryan M. C. Biol. Bull. 2006;210:38–50. doi: 10.2307/4134535. [DOI] [PubMed] [Google Scholar]
- 16.Dewilde S., Winnepenninckx B., Arndt M. H. L., Nascimento D. G., Santoro M. M., Knight M., Miller A. N., Kerlavage A. R., Geoghagen N., van Marcke E., et al. J. Biol. Chem. 1998;273:13583–13592. doi: 10.1074/jbc.273.22.13583. [DOI] [PubMed] [Google Scholar]
- 17.Afonso A. M. M., Arrieta M. R., Neves A. G. A. Biochim. Biophys. Acta. 1976;439:77–81. doi: 10.1016/0005-2795(76)90163-x. [DOI] [PubMed] [Google Scholar]
- 18.de Freitas T. V., Afonso A. M., Neves A. G. A. Comp. Biochem. Physiol. B. 1985;81:743–747. [Google Scholar]
- 19.Nascimento M. C. S., Daniel J. P., Heneine I. F. Comp. Biochem. Physiol. B. 1982;73:251–256. [Google Scholar]
- 20.Bugge J., Weber R. E. Am. J. Physiol. 1999;276:R347–R356. doi: 10.1152/ajpregu.1999.276.2.R347. [DOI] [PubMed] [Google Scholar]
- 21.Arndt M. H. L., Nascimento D. G., Xavier L. P., Santoro M. M. Mem. Inst. Oswaldo Cruz. 1998;93(Suppl. I):171–172. doi: 10.1590/s0074-02761998000700026. [DOI] [PubMed] [Google Scholar]
- 22.Kano Y., Chiba S., Kase T. Proc. Biol. Sci.; 2002. pp. 2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benton M. J. The Fossil Record 2. London: Chapman & Hall; 1993. pp. 125–270. [Google Scholar]
- 24.Wittgenstein J. B., Stein J. L. Biol. Bull. 1995;188:5–7. doi: 10.2307/1542061. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger R. C., Terwilliger N. B., Bonaventura C., Bonaventura J. Biophys. Biochim. Acta. 1977;494:416–425. doi: 10.1016/0005-2795(77)90171-4. [DOI] [PubMed] [Google Scholar]
- 26.Weber R. E., Vinogradow S. N. Physiol. Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger R. C., Terwilliger N. B., Schabtach E. Comp. Biochem. Physiol. B. 1978;59:9–14. doi: 10.1016/0300-9629(76)90122-5. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T., Kawamichi H., Ohtsuki R., Iwai M., Fujikura K. Biochim. Biophys. Acta. 2000;1478:152–158. doi: 10.1016/s0167-4838(99)00210-1. [DOI] [PubMed] [Google Scholar]
- 29.Mangum C. P. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1985;248:R505–R517. [Google Scholar]
- 30.Sminia T., Boer H. H., Niemantsverdriet A. Z. Zellforsch. Mikrosk. Anat. 1972;135:563–568. doi: 10.1007/BF00583437. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht U., Keller H., Gebauer W., Markl J. Cell Tissue Res. 2001;304:455–462. doi: 10.1007/s004410100368. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., II Embnew. News. 1997;4:1–4. [Google Scholar]
- 34.Abascal F., Zardoya R., Posada D. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 35.Page R. D. M. Computer Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.