Abstract
The bacterium Vibrio cholerae has two chromosomes. The origin of replication of chromosome I is similar to that of Escherichia coli. The origin-containing region of chromosome II (oriCII) resembles replicons of plasmids such as P1, except for the presence of an additional gene, rctA [Egan, E. S. & Waldor, M. K. (2003) Cell 114, 521–530]. The oriCII region that includes the initiator gene, rctB, can function as a plasmid in E. coli. Here we show that RctB suffices for the oriCII-based plasmid replication, and rctA in cis or trans reduces the plasmid copy number, thereby serving as a negative regulator. The inhibitory activity could be overcome by increasing the concentration of RctB, suggesting that rctA titrates the initiator. Purified RctB bound to a DNA fragment carrying rctA, confirming that the two can interact. Although rctA apparently works as a titrating site, it is nonetheless transcribed. We find that the transcription attenuates the inhibitory activity of the gene, presumably by interfering with RctB binding. RctB, in turn, repressed the rctA promoter and, thereby, could control its own titration by modulating the transcription of rctA. This control circuit appears to be a putative novel mechanism for homeostasis of initiator availability.
Keywords: checkpoint control, DNA–protein interactions, homeostasis, replication control
Our knowledge of DNA replication control in bacteria comes largely from studying the chromosome and plasmids of Escherichia coli (1, 2). In general, controlling proteins that initiate replication, initiator proteins, seems to be the central mechanism to regulate replication. A variety of mechanisms have been found to control the initiator.
Transcriptional autoregulation of the initiator gene is common in plasmids with iterated initiator-binding sites (iterons), where the initiator serves as the repressor of its own synthesis (3). The E. coli initiator, DnaA, also is autoregulated (4). In addition, the dnaA promoter is inactivated by sequestration to the cell membrane for approximately one-third of the cell generation (5). More traditional modes of transcriptional control using repressors and activators also have been found in phage λ and plasmid RK2 (6, 7).
Initiator translation is regulated by antisense RNAs in a variety of plasmids, the classic examples being plasmids R1 and pT181 (8, 9). The antisense RNA either blocks the ribosome-binding site of the initiator gene directly or of a regulatory peptide for the initiator gene (10).
Most other mechanisms operate posttranslationally either by titrating or inactivating the initiator protein. In E. coli, there are ≈300 DnaA-binding sites scattered all over the chromosome (11, 12). Titration of the initiator to these sites is believed to reduce availability of the protein after replication initiation, because new sites are created during replication elongation.
Initiator inactivation is the most widely used mechanism to prevent premature replication reinitiation. The pT181 initiator is inactivated by covalent modification at the end of one round of replication (13). E. coli DnaA is converted from an active ATP bound form to an inactive ADP form after replication (14–16). A similar mechanism also operates in Bacillus subtilis (17). The initiators of iteron-carrying plasmids are inactivated by dimerization, the active form being monomers (18). Dimerization not only reduces monomer concentration, the dimers also participate in inactivating origins (19–21). Eukaryotic initiation factors such as Cdt1 are inactivated after replication initiation by one or more of several mechanisms, such as inhibitory phosphorylation, proteolysis, titration by an inhibitor (geminin), and exportation out of the nucleus (22).
We are interested in studying DNA replication control in Vibrio cholerae. Like eukaryotes, bacteria also can have multiple chromosomes (23). V. cholerae is one such case with two chromosomes (chrI and chrII) (24). The origin of chrI (oriCI) is largely similar to that of the E. coli chromosome (25). The origin of chrII (oriCII) appears similar to those of plasmids with iterons (25). Replication from oriCII, however, initiates at the same time as oriCI, unlike the situation in plasmids (26, 27). Thus, a plasmid like origin seems to have evolved into a chromosomal one in V. cholerae. The most distinguishing feature of the oriCII region is the presence of a gene, rctA, which has no counterpart in other chromosomal or plasmid replicons. This feature prompted us to study this previously undescribed gene.
We find that rctA is not required for replication but can be a strong inhibitor of replication. The presence of rctA in cis or trans decreased copy number of oriCII-based plasmids in E. coli. Clones of rctA-inhibited growth of V. cholerae and generated DNA-less and/or elongated cells, indicating defects in cell cycle events. Interestingly, the gene served best as an inhibitor when not transcribed and functioned as DNA by providing binding (titrating) sites for the oriCII-specific initiator, RctB. The transcription of rctA apparently attenuates its inhibitory activity by interfering with RctB binding. The transcription, in turn, is controlled by RctB, which acts as a repressor of the rctA promoter (28). RctB thus controls its own titration by controlling transcription of rctA, a putative novel mechanism of initiator control. Transcriptional interference of DNA–protein interactions recently has been implicated in controlling recombination in the ribosomal operon of Saccharomyces cerevisiae (29).
Results
RctB Suffices for oriCII Plasmid Replication in E. coli.
A plasmid carrying the two neighboring V. cholerae genes, rctA and rctB, and the intergenic region, ig2, replicates in E. coli (Fig. 1; ref. 25). The rctB gene codes for an initiator protein. The rctA gene is transcribed but not translated. The role of transcription in replication requirement is not known. In V. cholerae, the right half of ig2 suffices for oriCII plasmid replication, the rctA and rctB genes being present in the chromosome. The left half of ig2, when present in multicopy plasmids, strongly inhibits cell growth, possibly by inhibiting oriCII function.
Fig. 1.
Functional elements of the region governing oriCII plasmid replication. The region contains two genes, rctB and rctA, transcribed divergently (arrows) and an intergenic region (ig2) carrying several functional elements as described in ref. 25. The six tandem 12-mers on the right part of ig2 have been compacted into one for convenience. Relative copy numbers were determined in E. coli (BR8706; ref. 30) by cloning origin fragments (black lines) into vector pGP704 and supplying RctB from an arabinose-inducible source of RctB (pTVC11) (the system is shown schematically on the bottom left corner). The arabinose concentration was 0.002%, which produced RctB at a level approaching that of V. cholerae (31). For pTVC20 and pTVC25, the inducer concentration also was 0.2%. This increased plasmid yield as found in ref. 31, making quantification of plasmid bands more reliable. The copy numbers were normalized relative to that of pTVC22, also determined at two different arabinose concentrations. The numbers below the lines are coordinates of fragment ends and they were obtained from GenBank: AE003853. NA, not applicable, because no transformants were obtained in these cases.
To further define the elements that control chrII replication, we embarked on a systematic deletion analysis of the region (Fig. 1). The entire ig2 region and the adjoining rctA gene were cloned into a suicide vector, pGP704 (32). The cloned region was flanked by rpoC transcription terminators to protect the region from extraneous transcription. The vector is based on R6Koriγ, which requires the cognate initiator π for function. In the absence of π, replication of chimeric plasmids requires functioning of a cloned origin. Such was the case with the ig2- and rctA-carrying plasmid pTVC20, because its replication depended on RctB in the absence of π (Fig. 1; data not shown). However, deletion of rctA still allowed replication and, in fact, significantly improved it (Fig. 1, pTVC20 vs. pTVC21 and pTVC22). Further deletions of the left half of ig2 allowed replication as long as the DnaA box was intact (pTVC31 and pTVC32, Fig. 1). Deletions from the opposite end showed that the first putative A+T rich 13-mer is not essential for oriCII function but the second one is (Fig. 1, pTVC33 and pTVC34). We conclude that RctB is the only V. cholerae-transacting factor required for oriCII function in E. coli. There are multiple negative regulatory elements in the left half of ig2, as was seen in ref. 25. Their effects were not always additive. For example, although a single 11-mer was inhibitory (because pTVC29 copy number was lower than pTVC30, Fig. 1), the other three 11-mers at the left end of ig2 appeared to nullify some of the inhibitory activity present elsewhere (because pTVC22 copy number was higher than that of pTVC25, Fig. 1). The finding that rctA gene served only to inhibit replication interested us the most because the results were unexpected.
To rule out the possibility that the vector (pGP704) sequences could somehow obviate the need for rctA, a plasmid was constructed by adding only the V. cholerae sequences present in plasmid pTVC31 to a drug resistance gene, bla, flanked by transcription terminators. The resulting plasmid, pTVC35, replicated in E. coli by requiring only RctB (data not shown). This finding confirmed the initiator to be the only V. cholerae-specific requirement for oriCII function in E. coli. pTVC35 was chosen for further studies on regulation of oriCII function.
rctA Transcription Is Unnecessary for Its Inhibitory Activity.
Because the rctA gene is transcribed, we wanted to determine how transcription might affect the regulatory activity of the gene. The gene lacking its natural promoter was cloned under PBAD promoter in a low-copy number (pSC101-derived) vector and was fused to a downstream luciferase gene (operon fusion), so that the latter can serve as a reporter of transcription. The promoter activity in the resultant plasmid (pTVC38) was controlled either by adding arabinose, which induced the operon, or by glucose, which repressed the operon. The effect of rctA transcription on the copy number of an oriCII plasmid, pTVC35, was measured by supplying a constant level of RctB from a separate plasmid, pTVC15. Irrespective of the level of transcription, the oriCII plasmid copy number decreased in all cases, confirming that the gene is a negative regulator of oriCII (Table 1). Unexpectedly, the inhibitory effect of rctA was observed even when the gene was apparently not transcribed (in the presence of glucose) or transcribed in the wrong direction, as in pTVC128. In fact, transcription decreased the inhibitory activity. Similar results were obtained with a different oriCII plasmid, pTVC36, which lacks the rctB leader region proposed to be a target for rctA acting as an antisense RNA (ref. 31; data not shown). The V. cholerae sequence present in pTVC36 is identical to those present in pTVC33 (Fig. 1). The rctB leader region also was absent in the plasmid that supplied RctB (pTVC15). These results suggest that the inhibitory activity of rctA is not mediated through rctB expression. Because transcription of rctA was not required to inhibit oriCII, the gene could be acting as DNA.
Table 1.
Effect of rctA transcription on oriCII plasmid copy number in E. coli
| Plasmid* in trans | Glucose/arabinose (0.2%) | Luciferase activity, arbitrary units | Relative copy number of oriCII plasmid pTVC35† |
|---|---|---|---|
| pTVC12 (PBADluc) | Glucose | <0.1 | 1.0 ± 0.38 |
| None | 47 ± 34 | 1 | |
| Arabinose | 4,830 ± 383 | 1.0 ± 0.19 | |
| pTVC38 (PBADrctA-luc) | Glucose | <0.1 | 0.29 ± 0.15 |
| None | 129 ± 33 | 0.44 ± 0.18 | |
| Arabinose | 4,787 ± 268 | 0.68 ± 0.11 | |
| pTVC128 (PBADAtcr-luc) | Glucose | <0.1 | 0.38 ± 0.14 |
| None | 78 ± 47 | 0.57 ± 0.07 | |
| Arabinose | 6,524 ± 474 | 0.89 ± 0.22 |
*Cells also had pTVC15 to supply RctB constitutively. Its level was similar in all cases tested (Fig. 5, which is published as supporting information on the PNAS web site).
†In pTVC35, oriCII fragment is identical to that in pTVC31 except that it has no vector origin (Fig. 1).
rctA Functions as DNA to Inhibit Replication.
To test whether rctA could act as DNA, a 159-bp fragment spanning the predicted 44-codon ORF was cloned in a transcriptionally silent region (TSR) of a pBR322-derived vector pTVC61, generating plasmid pTVC43 (Table 5, which is published as supporting information on the PNAS web site). The vector is identical to pRLM167 (courtesy of R. McMacken, Johns Hopkins University, Baltimore, MD), except for the changed drug-resistance gene. The TSR region is composed of phage λ sequences (21226–22346), believed to be devoid of promoters, and is flanked on both sides by four tandem ρ-independent T1 terminators from the E. coli rrnB operon. A strong inhibitory activity again was seen from the cloned rctA ORF (pTVC43-pTVC45, Table 2). The orientation of the cloned fragment did not make a difference, whereas having two tandem copies of the ORF made the inhibitory activity stronger. We conclude that rctA acts as DNA to inhibit oriCII function.
Table 2.
rctA acts as DNA to inhibit oriCII function
| rctA plasmid in trans (insert description) | Relative copy number of oriCII plasmid pTVC35 |
|
|---|---|---|
| Low [RctB]* | High [RctB]* | |
| pTVC61 (no fragment) | 1 | 1.30 ± 0.20 |
| pTVC43 (rctA ORF →)† | <0.1 | 0.15 ± 0.03 |
| pTVC44 (rctA ORF ←) | <0.1 | 0.13 ± 0.04 |
| pTVC45 (rctA ORF →)‡ 2× | NA | <0.1 |
*RctB is expressed from PBADrctB in pTVC11 in the presence of either 0.002% (Low) or 0.2% (High) arabinose.
†The arrow shows the direction of rctA fragment.
‡pTVC45 has two copies of rctA. It could not transform cells with pTVC35 and low RctB. NA, not applicable.
rctA Binds RctB in Vitro.
An inspection of rctA sequence revealed three stretches resembling 11- and 12-mers of ig2, putative-binding sites for the initiator RctB (Fig. 2A). This finding suggested the possibility that rctA could be acting by titrating RctB. To test for RctB binding to rctA DNA, we used a gel assay (EMSA) with purified components. The mobility shift was evident with a DNA fragment covering the rctA ORF (Fig. 2B). The binding improved in the presence of chaperones as has been found for many plasmid initiators (3). A single retarded band of identical mobility was seen with and without chaperones, although three putative-binding sites were present in the rctA fragment.
Fig. 2.
rctA DNA binds RctB. (A) Three putative sites (I, II, and III) for RctB binding are shown within the rctA ORF (coordinates 112–246), as also their similarities to the 11- and 12-mer consensus sequences of ig2. In the alignment, the identical bases are given in bold letters. (B) EMSA with purified RctB and a PCR fragment covering either rctA ORF (135 bp) or nonspecific sequences (193 bp) from pRLM167. (C) Effect of rctA sites on the oriCII plasmid copy number in vivo. The rectangle represents mutated site II with several base substitutions. Fragment 9 covers coordinates 240–281 of V. parahaemolyticus chrII (GenBank:BA000032). RctB was supplied from pTVC11-containing PBADrctB in the presence of 0.002% (Low) or 0.2% (High) arabinose. (D) EMSA of rctA fragments used in C.
To determine which, if any, of the putative binding sites are functional, they were deleted singly and in pairwise combinations (Fig. 2C). In general, at low RctB concentration, the inhibitory activity toward oriCII (pTVC35) plasmid reduced when the sites were deleted or mutated. Site II, in which five bases of the middle 11-mer had been substituted, retained some activity, because it reduced pTVC35 copy number more than the fragment with site II deletion (fragments 5 vs. 6, Fig. 2C). Fragment 4 was the only case where deletion of a site (site III) did not have an effect on the oriCII plasmid copy number. In all cases, increasing RctB concentration alleviated the inhibitory effect coming from intact or mutated rctA fragments.
If rctA inhibited oriCII function by titrating RctB, we expected the two activities to be correlated. To test this prediction, binding of the mutated rctA fragments to RctB was determined (Fig. 2D). A reasonable correlation between the in vivo and in vitro activities was evident: The fragments that reduced the oriCII plasmid copy number more also bound RctB more (Fig. 2 C and D). The correlation, however, was not perfect in the sense that fragments with single boxes that showed no binding in vitro still were capable of reducing the copy number in vivo (fragments 6–8, Fig. 2 C and D). Overall, the results are consistent with the three sites being relevant and cooperating for binding RctB. To inhibit replication, rctA appears to interact with RctB directly.
rctA Inhibits Growth of V. cholerae.
We next determined whether rctA could inhibit growth of V. cholerae as was found with other negative regulators present in ig2 (25). A recA derivative of the sequenced strain N16961, CVC250 (31), was used for this purpose. A multicopy plasmid carrying the rctA ORF only, pTVC40, could not be introduced into CVC250 (Table 3). The introduction was successful when rctA was cloned under PBAD, as in pTVC58. However, the transformants formed small colonies. The colony size improved significantly when PBAD was activated with arabinose. pTVC40 also could be introduced when additional RctB was provided through a plasmid (pTVC17) before transformation. However, the transformants formed small colonies, and further tests showed that the rctA plasmid was partly deleted (eight colonies tested). As before, colony sizes improved when rctA was cloned under PBAD. In these transformants, the rctA plasmid was intact (10 colonies tested). Thus, extra rctA in trans retards V. cholerae growth, and this inhibitory activity is attenuated either by transcribing the gene and/or providing extra RctB, as in E. coli. These results also showed that transcription of rctA more effectively neutralizes the inhibitory activity of the gene than the oversupply of RctB. In the latter situation, preferential deletion in the rctA plasmid suggests that oversupply of RctB most likely allowed some cell growth for deletion mutations to appear, but there was still enough inhibitory activity left for the preferential growth of cells where the cloned rctA fragment was deleted.
Table 3.
Rescue of V. cholerae growth inhibition due to excess rctA by transcription of the gene or providing excess RctB
| Plasmids (relevant description) | Colony size of transformants |
|---|---|
| pTVC12 (vector for rctB)/pTVC40 (rctAORF) | No transformants |
| pTVC12/pTVC58 (PBADrctA) | Small |
| pTVC12/pTVC58 | Large* |
| pTVC17 (PlacrctB)/pTVC40 | Small†‡ |
| pTVC17/pTVC58 (PBADrctA) | Medium‡ |
| pTVC17/pTVC58 | Large*‡ |
*In the presence of 0.2%.
†Plasmids in transformants were partly deleted.
‡Results were same when rctB was induced with 50 μM IPTG.
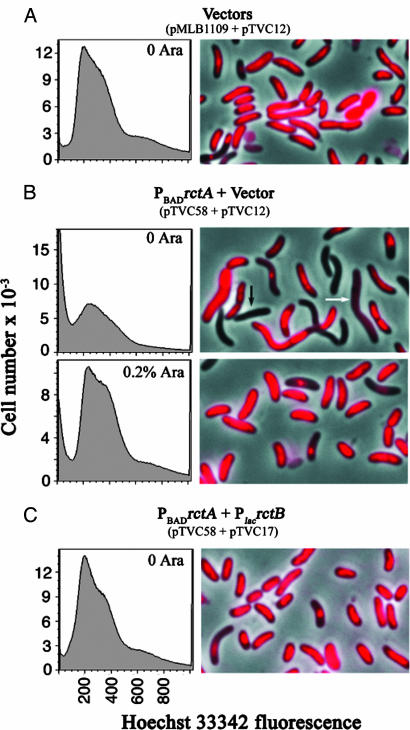
To understand the basis of V. cholerae growth inhibition by rctA, the DNA content per cell was analyzed by flow cytometry. The main peak of DNA-containing cells (with fluorescence values ≈200) reduced when rctA was additionally supplied through pTVC58 (Fig. 3A and B Upper). Simultaneously, cells with less or no DNA (fluorescence values <100) increased significantly. Upon DNA staining, abnormal cells (with less or no DNA, and elongated) were found frequently (Fig. 3B, arrows), indicating severe DNA breakdown and cell division defects. The abnormal cells were less frequent when rctA was transcribed or extra RctB was provided, as was also evident from flow cytometric analysis (Fig. 3 B Lower and C and Table 4). These results are consistent with our expectation from studies in E. coli and additionally provide evidence that interfering with chrII replication can perturb other cell cycle events.
Fig. 3.
(A–C) Flow cytometry and fluorescence microscopy of Hoechst-stained V. cholerae cells (CVC250) showing their DNA contents when additional (plasmid-borne) rctA and rctB genes were present. (A and B) In control cells without the rctA and/or rctB plasmids, the vectors for the missing genes were used. (B) Arrows mark cells that appear empty (black) or largely devoid of DNA (white).
Table 4.
Effect of rctA on DNA content and cell size of V. cholerae
| Plasmids in V. cholerae | % arabinose | % (no.) of cells |
||
|---|---|---|---|---|
| Normal | <50% DNA | ≥3× size* | ||
| Vectors (pMLB1109 + pTVC12) | 0 | 90 (795) | 7.8 (69) | 2.3 (20) |
| PBADrctA + vector | 0 | 45 (273) | 46 (276) | 8.7 (52) |
| (pTVC58 + pTVC12) | 0.2 | 90 (744) | 8.7 (72) | 1.2 (10) |
| PBADrctA + PlacrctB (pTVC58 + pTVC17) | 0 | 94 (464) | 5.2 (26) | 1.2 (6) |
*Cells were largely empty in the majority of cases.
rctA in Other Members of Vibrionacea Family.
Unlike the initiator gene rctB, which is well conserved in the sequenced members of the Vibrionacea family, rctA is not a conserved gene (25). Because our studies suggested that the gene is no more than a few extra binding sites for the initiator RctB, we searched and found weakly homologous sites in the corresponding region in other Vibrio species (Fig. 6, which is published as supporting information on the PNAS web site). The site present in V. parahaemolyticus was tested for function. An oligonucleotide carrying the site (a putative 12-mer) was cloned, and the resultant plasmid (pTVC65) did reduce the V. cholerae oriCII plasmid copy number (Fig. 2C, fragment 9). The oligonucleotide also showed binding to V. cholerae RctB in vitro (Fig. 2D, fragment 9). We note that although the binding was poor, none of the rctA sites of V. cholerae showed binding when provided singly (Fig. 2D). Thus, although rctA is not conserved, the sites for binding RctB seem to be present in other members of the Vibrionaceae family.
Discussion
Here we show that the rctA gene, which distinguishes the origin region of V. cholerae chrII from other bacterial and plasmid replicons, is a specific inhibitor of replication. The gene functions as DNA by providing binding (titrating) sites for the initiator RctB. We propose that the binding activity contributes to a previously undescribed regulatory system to regulate RctB availability homeostatically (Fig. 4). The proposal is based on the findings that rctA transcription reduces its inhibitory activity probably by interfering with RctB binding and that the transcription, in turn, is repressed by RctB (28). Thus, RctB controls its own titration through rctA transcription. RctB also represses its own transcription (autorepression) (28, 31). These control steps seem suited to adjust for both upward and downward fluctuations in RctB availability and contribute to initiator homeostasis (Fig. 4). When RctB is high, its level is adjusted by reducing new synthesis by autorepression and increasing titration by repressing the rctA promoter. Similarly, at low RctB, its availability is increased by new synthesis and reduced titration because of increased activities of rctA and rctB promoters.
Fig. 4.
A model for homeostasis of RctB availability. The model is based on the facts that RctB represses its own and rctA promoters and that transcription of rctA reduces its ability to titrate RctB. The two mechanisms, transcription and titration, together, in principle, can adjust for RctB fluctuation more sensitively than either mechanism can individually.
The principle of transcriptional interference of DNA–protein interactions as inferred here in the regulation of rctA inhibitory activity, although novel in the field of replication control, was applied first to conditionally inactivate a centromere in S. cerevisiae (33). More recently, a similar mechanism has been proposed to prevent recombination between repeating units of ribosomal operons in S. cerevisiae (29).
To keep their copy number low, iteron-based plasmids use additional iterons outside of the origin (3). V. cholerae also uses repeat sequences outside of the origin related to the origin iterons for down-regulating replication (Fig. 1; ref. 25). As we show here, the rctA gene is also a collection of repeats. Thus, both iteron-based plasmids and chrII employ multiple repeats to control replication. The magnitude of the copy number reduction also appears comparable but not identical in the two cases. Deletion of the control locus in stringently controlled plasmids such as P1 and F increases copy number ≈10-fold. Deletion of rctA alone increases copy number ≈4-fold (Fig. 1, pTVC20 vs. pTVC22), and deletion of ig2 region dedicated for negative control increases copy number another 8-fold (pTVC22 vs. pTVC31). The chrII appears to maintain a higher initiation potential, necessitating a tighter negative control to maintain its copy number, compared with P1 and F.
The RctB-binding sites differ from plasmid iterons in a major way. Iterons are ≈20 bp long and are highly homologous. RctB-binding sites are smaller (12 bp or less) and can be quite degenerate. For example, rctA sites II/III match with the consensus 11-mer sequence at six to seven positions only (Fig. 2A). The 11-mer itself matches the consensus 12-mer sequence in eight positions. RctB-binding sites for autoregulation also show only six/seven matches to the 12-mer (31). A search of RctB repeats outside of ig2 by using the program RSAT (http://embnet.ccg.unam.mx/rsa-tools) identified 36 sites in chrI and 23 sites in chrII, when only up to three mismatches to the 12-mer was allowed (data not shown). There thus can be a large number of RctB sites in the V. cholerae genome, analogous to the situation in E. coli, which has ≈300 sites for binding DnaA. In this sense, chrII is not a typical plasmid and apparently utilizes the entire large genome for initiator titration, like the E. coli chromosome. That the titration plays a role is evident from the fact that increasing [RctB] increases oriCII-based plasmid copy number (Table 2; ref. 31) and from the results of Table 3, where despite having an autoregulatory rctB gene in the chromosome, a gratuitous source of RctB helped cell growth. In view of the presence of a large number of RctB sites, the question arises how only three sites of rctA could have a significant effect in initiation control. Simple titration appears unlikely to be the only role of rctA. The sites most likely participate in higher-order interactions. The same could be true also for the repeats in ig2, because their successive deletion did not increase the oriCII-plasmid copy number correspondingly (Fig. 1).
The presence of higher-order interactions also can be deduced from theoretical considerations. We have argued that controlling replication initiation frequency by initiator autoregulation and titration cannot be sufficient because it can only dampen initiator increase after duplication of the initiator gene upon passage of the replication fork (34). To prevent reinitiation, the initiator level must decrease as a consequence of replication. We suspect that the titrated initiators, including those in rctA, most likely participate in additional interactions, such as handcuffing (3).
Initially, rctA was found to be required for oriCII minichromosome replication in E. coli (25). We, however, find the gene to be a typical negative regulator: It is dispensable, and increasing its number in trans reduces replication more effectively (Fig. 1 and Table 2). The experimental conditions used in the two cases are different, and how they influenced the results remains to be understood. It is possible that controlling RctB availability by rctA may become obligatory under some conditions. It is well known that a fewfold change in iteron and initiator concentrations can reverse their activities from stimulatory to inhibitory in iteron-carrying plasmids (35).
A great interest in studying replication control in a two-chromosome bacterium is to determine if and how the chromosomes communicate so that replication of both are completed before cell division. Initial evidence for communication between the chromosomes has been obtained because they initiate replication synchronously (26). Here we show that excess of rctA can arrest cell growth and generate DNA-less cells of normal and elongated sizes. The rctA effect is most likely exerted through blocking chrII replication because the gene alone or together with RctB did not affect oriCI plasmid replication in E. coli (data not shown). It remains to be seen whether blocking chrII replication turns on a checkpoint mechanism that affects chromosome segregation and cell division, thereby generating chromosomeless cells. An additional possibility is that a toxin gets activated in cells born without chrII, where multiple toxin-antitoxin systems are present (36). The toxin then could be causing chromosomal breakdown, a common occurrence in apoptotic eukaryotic cells (37).
Materials and Methods
Strains and Plasmids.
E. coli and V. cholerae strains and plasmids used are listed in Table 5, and details of plasmids constructed in this study are in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Luciferase Assay.
The assay was performed as described in ref. 31.
Plasmid Copy Number.
The copy number was measured from log-phase cells (OD600 ≤ 0.5) as described in ref. 20.
EMSA.
One microgram of gel-purified PCR fragments or oligonucleotides (Sigma-Genosys, The Woodlands, TX) in 50 μl was end-labeled with 50 μCi [γ-32P]ATP (1 Ci = 37 GBq) (Amersham Biosciences, Piscataway, NJ) by using 30 units of T4 polynucleotide kinase (NEB, Beverly, MA). After purification through ProbeQuant G-50 micro columns (Amersham Biosciences), 2 nM DNA was used in a 20-μl binding reaction containing 20 mM Tris-acetate (pH 7.5), 100 mM potassium glutamate (Fluka, Milwaukee, WI), 10 mM magnesium acetate, 5% (vol/vol) glycerol (Invitrogen, Carlsbad, CA), 0.1 mM EDTA, 1 mM DTT (NEB), 100 μM ATP (Amersham Biosciences), 0.1% (vol/vol) igepal CA-630 (Sigma, St. Louis, MO), and 300 ng of poly dI-dC (Amersham Biosciences).
Before binding, 200 ng of DnaJ and 400 ng of DnaK (both from Stressgen, Victoria, Canada) were mixed with various concentrations of purified RctB and left at room temperature for 30 min. After adding DNA, the incubation was continued for another 10 min. Fifteen microliters of the binding reaction was run on 5% polyacrylamide gel in 1× TBE buffer (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3) at room temperature at 25 mM constant current. The gel was dried, and band intensities were determined by using a phosphoimager (FLA-5100; Fujifilm, Edison, NJ).
Flow Cytometry and Fluorescence Microscopy.
For experiments of Fig. 3, V. cholerae CVC250 transformed with desired plasmids was grown in LB with drugs to select for plasmids and 0.2% arabinose to silence the plasmid-borne rctA gene. After ON growth at 37°C, the cultures were diluted to OD600 = 0.001 in fresh medium with and without arabinose. At early log-phase, all cultures grew with a generation time of ≈1 h. However, when rctA was maximally active (no arabinose and no extra RctB; Fig. 3), the growth rate started slowing from an OD600 of ≈0.1. All cultures were processed in parallel after they reached an OD600 of ≈0.2. Flow cytometry and light microscopy were done from different aliquots of the same culture as described in ref. 38, except that for staining cells for cytometry, they were in 0.01 M Tris (pH 7.4)/0.01 M MgSO4. For staining of nucleoids for microscopy, cells were concentrated by using a microfuge at 600 × g for 5 min and washed once with 1× PBS containing 1 mM EDTA. The DNA stain Hoechst 33342 (Molecular Probes) was added to a final concentration of 50 μg/ml, and the cells were left at room temperature for 10 min. Approximately 2.5 μl of cells was placed on a slide and overlaid with a coverslip treated with poly-l-lysine (Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Jessica Koziski for constructing plasmids, Yikang Rong and Michael Yarmolinsky for comments on the manuscript, and Michael Lichten for suggesting that chrI could be getting degraded because of activation of a toxin in cells born without chrII.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Messer W. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 2.Helinski D. R., Toukdarian A. E., Novick R. P. Replication Control and Other Stable Maintenance Mechanisms of Plasmids. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 3.Chattoraj D. K. Mol. Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 4.Messer W., Weigel C. Methods Enzymol. 2003;370:338–349. doi: 10.1016/S0076-6879(03)70030-5. [DOI] [PubMed] [Google Scholar]
- 5.Campbell J. L., Kleckner N. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 6.Szalewska-Palasz A., Wegrzyn A., Blaszczak A., Taylor K., Wegrzyn G. Proc. Natl. Acad. Sci. USA. 1998;95:4241–4246. doi: 10.1073/pnas.95.8.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young C., Prince A. S., Figurski D. H. Proc. Natl. Acad. Sci. USA. 1985;82:7374–7378. doi: 10.1073/pnas.82.21.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M., Diaz-Orejas R. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordstrom K. Plasmid. 2006;55:1–26. doi: 10.1016/j.plasmid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Praszkier J., Pittard A. J. Plasmid. 2005;53:97–112. doi: 10.1016/j.plasmid.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Roth A., Messer W. Mol. Microbiol. 1998;28:395–401. doi: 10.1046/j.1365-2958.1998.00813.x. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa R., Ozaki T., Moriya S., Ogawa T. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasooly A., Novick R. P. Science. 1993;262:1048–1050. doi: 10.1126/science.8235621. [DOI] [PubMed] [Google Scholar]
- 14.Kato J., Katayama T. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boye E., Lobner-Olesen A., Skarstad K. EMBO Rep. 2000;1:479–483. doi: 10.1093/embo-reports/kvd116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camara J. E., Breier A. M., Brendler T., Austin S., Cozzarelli N. R., Crooke E. EMBO Rep. 2005;6:736–741. doi: 10.1038/sj.embor.7400467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noirot-Gros M.-F., Velten M., Yoshimura M., McGovern S., Morimoto T., Ehrlich S. D., Ogasawara N., Polard P., Noirot P. Proc. Natl. Acad. Sci. USA. 2006;103:2368–2373. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickner S., Hoskins J., McKenney K. Proc. Natl. Acad. Sci. USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zzaman S., Bastia D. Mol. Cell. 2005;20:833–843. doi: 10.1016/j.molcel.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Das N., Chattoraj D. K. Mol. Microbiol. 2004;54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- 21.Toukdarian A. E., Helinski D. R. Gene. 1998;223:205–211. doi: 10.1016/s0378-1119(98)00370-9. [DOI] [PubMed] [Google Scholar]
- 22.Blow J. J., Tanaka T. U. EMBO Rep. 2005;6:1028–1034. doi: 10.1038/sj.embor.7400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egan E. S., Fogel M. A., Waldor M. K. Mol. Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- 24.Trucksis M., Michalski J., Deng Y. K., Kaper J. B. Proc. Natl. Acad. Sci. USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egan E. S., Waldor M. K. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 26.Egan E. S., Lobner-Olesen A., Waldor M. K. Curr. Biol. 2004;14:R501–R502. doi: 10.1016/j.cub.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 27.Leonard A. C., Helmstetter C. E. J. Bacteriol. 1988;170:1380–1383. doi: 10.1128/jb.170.3.1380-1383.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan E. S., Duigou S., Waldor M. K. J. Bacteriol. 2006;188:789–793. doi: 10.1128/JB.188.2.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T., Ganley A. R. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 30.Fekete R. A., Chattoraj D. K. Mol. Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- 31.Pal D., Venkova-Canova T., Srivastava P., Chattoraj D. K. J. Bacteriol. 2005;187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller V. L., Mekalanos J. J. J. Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill A., Bloom K. Mol. Cell. Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das N., Valjavec-Gratian M., Basuray A. N., Fekete R. A., Papp P. P., Paulsson J., Chattoraj D. K. Proc. Natl. Acad. Sci. USA. 2005;102:2856–2861. doi: 10.1073/pnas.0409790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park K., Mukhopadhyay S., Chattoraj D. K. J. Biol. Chem. 1998;273:24906–24911. doi: 10.1074/jbc.273.38.24906. [DOI] [PubMed] [Google Scholar]
- 36.Pandey D. P., Gerdes K. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pessina A., Croera C., Savalli N., Bonomi A., Cavicchini L., Turlizzi E., Guizzardi F., Guido L., Daprai L., Neri M. G. Cell Res. 2006;16:306–312. doi: 10.1038/sj.cr.7310038. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava P., Fekete R. A., Chattoraj D. K. J. Bacteriol. 2006;188:1060–1070. doi: 10.1128/JB.188.3.1060-1070.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.