Abstract
We describe a strategy to “revive” putatively pathogenic T cells from frozen specimens of human inflammatory target organs. To distinguish pathogenic from irrelevant bystander T cells, we focused on cells that were (i) clonally expanded and (ii) in direct morphological contact with a target cell. Using CDR3 spectratyping, we identified clonally expanded T cell receptor (TCR) β-chains in muscle sections of patients with inflammatory muscle diseases. By immunohistochemistry, we identified those Vβ-positive T cells that fulfilled the morphological criteria of myocytotoxicity and isolated them by laser microdissection. Next, we identified coexpressed pairs of TCR α- and β-chains by a multiplex PCR protocol, which allows the concomitant amplification of both chains from single cells. This concomitant amplification had not been achieved previously in histological sections, mainly because of the paucity of available anti-α-chain antibodies and the great heterogeneity of the α-chain genes. From muscle tissue of a patient with polymyositis, we isolated 64 T cells that expressed an expanded Vβ1 chain. In 23 of these cells, we identified the corresponding α-chain. Twenty of these 23 α-chains were identical, suggesting antigen-driven selection. After functional reconstitution of the αβ-pairs, their antigen-recognition properties could be studied. Our results open avenues for combined analysis of the full TCR α- and β-chain repertoire in human inflammatory tissues.
Keywords: autoimmunity, immunopathology, myositis, repertoire, single-cell PCR
In T cell-mediated immune diseases, specific target organs are attacked by (auto-)aggressive T lymphocytes. These lymphocytes have been isolated and functionally studied in animal models (1–3), but it has been much more difficult to characterize human autoaggressive T cells. Fresh-biopsy human tissue is usually unavailable, and frozen or fixed tissue does not allow the isolation of viable T cells (4). Here we describe a strategy for the “reconstruction” of viable, T cell receptor (TCR)-expressing cells from histological specimens of human inflammatory tissue lesions.
The antigen-reactive TCR consists of two hypervariable α- and β-chains, each of which contributes to antigen recognition (5, 6). In previous studies we successfully amplified TCR β-chains from individual, morphologically characterized, tissue-infiltrating T cells from brains of patients with multiple sclerosis and from muscles of patients with myositis (7–9). Although these studies provided insight into intralesional TCR β-chain repertoires, they did not allow the identification of complete TCR αβ-heterodimers expressed by tissue-infiltrating T cells because information on the coexpressed α-chains was lacking, a result of the very limited number of available anti-α-chain antibodies and higher variability of α-chain genes (10, 11).
In animal models, αβ-pairs were characterized from single cells isolated from cell suspensions by flow cytometry (12–14), but this approach is obviously not feasible with frozen histological specimens of human tissues. To “revive” putatively autoaggressive T cells from frozen histological samples, we had to overcome a major obstacle: identifying the matching α- and β-chains from individual T cells. To this end, we combined molecular and morphological information to identify the β-chains expressed by tissue-infiltrating T cells. We then microdissected single T cells that expressed this particular Vβ element, and we amplified their corresponding α-chains by a multiplex PCR protocol. We successfully applied this method to muscle tissue from patients with inflammatory muscle diseases, which are known to be mediated by myocytotoxic CD8+ T cells (15–17). This protocol, which eventually allows reconstruction of viable, TCR-expressing cells from frozen archival tissue, will be applicable to other inflammatory tissues, including reactions in transplants and tumors.
Results
Design of an RT-PCR Protocol for Identification of Matched TCR α- and β-Chains from Single Cells in Human Tissue Sections.
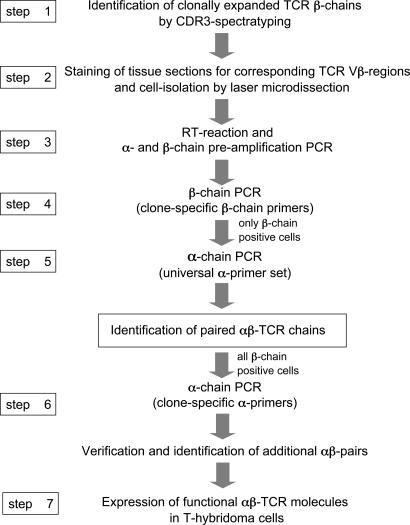
The steps of our protocol are summarized in Fig. 1. First, we identified expanded T cell clones by CDR3 spectratyping (18) of TCR β-chains (step 1). Clonal expansion indicates an antigen-driven response, and therefore it can serve as one criterion for identification of putatively pathogenic T cells. Next, we stained TCR molecules with appropriate anti-Vβ antibodies to detect Vβ+ T cells that make direct morphological contact with a target cell (step 2). This process provides independent, morphological evidence for pathogenic relevance. Steps 1 and 2 identify pathogenic T cells, and they also reduce the complexity of the subsequent multiplex PCR because the full sequence of the β-chain is already established. Therefore, multiplex single-cell PCR is required only for the α-chain. The putatively pathogenic T cells were then isolated by laser microdissection and analyzed by single-cell RT-PCR (a sketch of the relative positions of the primers is shown in Fig. 4, which is published as supporting information on the PNAS web site). After reverse transcription and preamplification of α- and β-chains (step 3), we identified those T cells that express the expanded TCR β-chain (step 4). Positive cells were further examined for matching α-chains by multiplex PCR (step 5) using a set of universal primers that allows the unbiased amplification of any α-chain sequence. After this first round of identification of α-chain sequences, we reinvestigated all cells containing the correct β-chain with clone-specific α-chain primers (step 6). Step 6 confirms the findings of step 5, and it may lead to the identification of additional α-positive cells because the clone-specific primers are more sensitive than the universal primer set. Finally, the α- and β-chains can be reconstituted to functional proteins (step 7).
Fig. 1.
Strategy to identify paired TCR α- and β-chains from microdissected tissue-infiltrating T cells. In the first step, we investigated clonal T cell expansions by CDR3 spectratyping of the TCR β-chains. Then, in step 2, we stained sections from frozen biopsy specimens with appropriate anti-Vβ antibodies and isolated positive cells by microdissection. After cDNA preparation and a preamplification PCR for α- and β-chains (step 3), we tested the microdissected cells for expression of the expanded β-chain by PCR using clone-specific β-primers (step 4). Cells that expressed the correct sequence, i.e., the sequence identified by CDR3 spectratyping, were examined for the matching α-chains with a universal primer set that allows amplification of all TCR α-chain sequences (step 5). The preamplification product served as template. Based on the α-chain sequences identified in step 5, we designed clone-specific α-chain primers and reexamined all β-chain-positive cells (step 6), which allowed identification of additional αβ-pairs because clone-specific primers are more efficient than the universal primer set. For functional studies, the corresponding α- and β-chains were expressed on the surface of a T hybridoma cell line (step 7).
Efficiency of the Single-Cell Multiplex PCR Protocol.
We tested the efficiency of our protocol by using 58α−β− mouse T hybridoma cells transfected with a known combination of human Vα2.3-Vβ17 TCR cDNA. We immobilized the transfected cells by Cytospin on the same type of membrane used for microdissection of tissue sections, isolated individual cells by microdissection, and analyzed them by single-cell PCR. As for tissue sections, we used clone-specific PCR primers for β-chain amplification together with our universal primer set for α-chains. To estimate the influence of staining procedures, we compared the yields of TCR α- and β-chains from unlabeled transfectants with the yields from transfectants that had been labeled with anti-Vβ17 antibodies and stained with alkaline phosphatase (AP) (Table 2, which is published as supporting information on the PNAS web site). In 49 of 89 unlabeled cells, we found the correct β-chain, which reflects the loss of cells or RNA during laser capture or fixation. The β-yield decreased to 14 of 90 cells after staining with an anti-Vβ17 antibody and AP. When we analyzed β-chain-positive cells for the corresponding α-chains, we recovered the α-chain from 48 of 49 unlabeled cells and from 6 of 14 stained cells. The combined yield of α- and β-chains, based on all investigated cells, was 48 of 89 unlabeled cells (54%) and 6 of 90 antibody- and AP-labeled cells (6.7%), which shows that our method efficiently amplifies corresponding TCR αβ-pairs from single cells. As expected, the yields decrease after labeling and staining procedures.
Next, we investigated whether our protocol can detect two α-chains in individual T cells because “dual-α T cells” might account for up to 30% of all human T cells (19). We used two different TCR-transfected hybridoma cell lines: one with a Vα2.3-Vβ17 and one with a Vα22-Vβ9 TCR; microdissected individual cells from Cytospin preparations; and catapulted two cells, one from each hybridoma, into one PCR tube to test whether we could amplify the Vβ17-chain together with both α-chains. In 60 of 90 tested cell pairs, we detected the β-chain and both α-chains, indicating that our protocol can detect dual TCR α-chains with high yield.
Analysis of TCR β-Chain Expansions in Muscle Biopsies.
We applied our protocol to muscle-biopsy specimens from patients with inflammatory myopathies, polymyositis (PM), and inclusion body myositis (IBM), which can be considered as a paradigm for T cell-mediated tissue destruction (15, 17, 20). Step 1, screening of the TCR β-chain repertoire by CDR3 spectratyping, revealed prominent clonal expansions of Vβ1-Jβ1.2 T cells in patient PM16488, and of Vβ23-Jβ1.1 in patient IBM15551 (Fig. 5, which is published as supporting information on the PNAS web site). To ensure that the observed peaks represent single T cell clones, we sequenced the corresponding PCR products. The derived β-chain amino acid sequences are listed in Table 1.
Table 1.
Deduced amino acid sequences of paired TCR α- and β-chains of microdissected T cells from muscle biopsy tissue
| Patient | Chain and sequence |
No. of cells with TCR identified by |
|||||
|---|---|---|---|---|---|---|---|
| V | N(D)N | J | Universal primer set | Clone-specific primer | |||
| PM16488 (1,230 cells analyzed) | |||||||
| β-Chain | 1.1 | CASSV | GGL | YGYTF | 1.2 | 64 | |
| α-Chains | 2.2 | CAM | SQGAQKLVF | 54 | 6 | 20 | |
| 11.1 | CAV | GS | NARLMF | 31 | 1 | 1 | |
| 20.1 | CL | LSS | NTGKLIF | 37 | 1 | 1 | |
| 23.1 | CAV | D | SGTYKYIF | 40 | 1 | 1 | |
| IBM15551 (250 cells analyzed) | |||||||
| β-Chain | 23.1 | CASS | SDRE | NTEAFF | 1.1 | 8 | |
| α-Chains | 4.2 | CI | ASV | YGNKLVF | 47 | 1 | 1 |
| 23.1 | CA | VL | NAGNMLTF | 39 | 1 | 1 | |
In a first step, we detected TCR β-chain sequences of expanded clones by CDR3 spectratyping. The deduced amino acid sequences are shown. Then, biopsy sections were stained by appropriate anti-Vβ antibodies, and Vβ-positive cells were isolated by microdissection. We tested these cells for the expression of the correct, expanded TCR β-chains by single-cell PCR using clone-specific primers. We identified 64 cells from patient PM16488 and 8 cells from patient IBM15551 that had identical sequences as determined by CDR3 spectratyping and single-cell PCR. These β-chain-positive cells were then tested for α-chain expression by using a universal primer set that allows amplification of all possible TCR α-chains. The sequences of the α-chains and the numbers of cells identified are listed. Based on the sequences of these α-chains, all β-chain-positive cells were reexamined for α-chains by using clone-specific α-primers. In all cells in which we had identified an α-chain with the universal primer set, we confirmed expression of this α-chain with clone-specific primers. In patient PM16488, we identified the Vα2.2-Jα54 sequence in 14 additional cells.
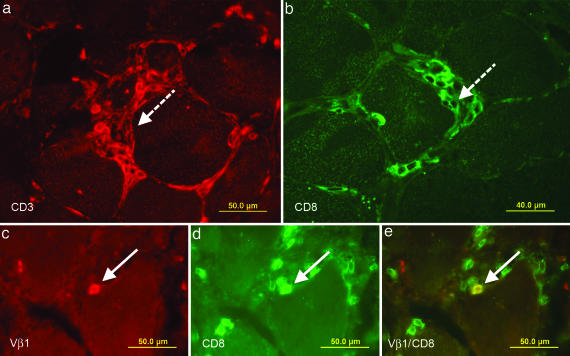
For step 2, we stained biopsy sections with appropriate anti-TCR Vβ mAbs. Fig. 2 shows focal invasion of muscle fibers by CD3+CD8+ T cells (patient PM16488). The dashed arrows in Fig. 2a (anti-CD3 staining) and Fig. 2b (anti-CD8 staining) indicate invasive T cell infiltrates, as they are typically seen in IBM and some cases of PM (15, 17, 20). Dense infiltrates like the ones shown in Fig. 2 a and b are not suitable for isolation of individual cells. For microdissection, we instead focused on distinct cells that make direct contact with a muscle fiber but are not part of an invasive-cell cluster. A typical example is shown in Fig. 2 c–e, where a cryosection was double-stained with an anti-Vβ1 (Fig. 2c) and an anti-CD8 antibody (Fig. 2d). A double-positive cell appears in yellow and is indicated by an arrow (Fig. 2e). Several CD8+ cells appear Vβ1-negative. Some of them are “bystander” cells without contact with the muscle fibers. This finding is consistent with the results from CDR3 spectratyping, which revealed expansion of additional Vβ-sequences, and it agrees with previous observations that the T cell infiltrates in inflammatory myopathies are usually oligoclonal rather than monoclonal (16, 21, 22).
Fig. 2.
Immunolocalization of T cells in muscle tissue of patient PM16488. Cryosections of a frozen biopsy specimen were stained in red with a Cy3-labeled antibody to CD3 (a) and in green with an FITC-labeled antibody to CD8 (b). Dense focal invasions of T cells into muscle fibers are indicated in both figures by dashed arrows. Focal invasions are one of the diagnostic criteria of PM. However, such closely packed T cell aggregates are not suited for the isolation of single T cells by microdissection. We therefore focused on distinct single cells that indent or directly contact a muscle fiber. (c–e) Double stainings of TCR Vβ1 and CD8. Biopsy sections were stained in red with a Cy3-labeled anti-Vβ1 antibody (c) and in green with an FITC-labeled anti-CD8 antibody (d). (e) A double-positive cell in direct contact with a muscle fiber appears in yellow and is indicated by an arrow.
Identification of Paired TCR α- and β-Chains.
We examined individual Vβ-positive T cells for expression of the expanded β-chain cDNA and coexpressed α-chain(s). We stained biopsy sections with appropriate anti-Vβ antibodies (step 2), isolated positive cells by laser microdissection, prepared cDNA, and applied a preamplification step for α- and β-chains (step 3). We used clone-specific primers for the β-chains and a pool of primers for the α-chains (Fig. 4). Then we determined which of the Vβ-positive cells expresses the expanded β-chain (step 4). Specifically, we investigated cells expressing the β-chains Vβ1.1-GGL-Jβ1.2 (patient PM16488) and Vβ23.1-SDRE-Jβ1.1 (patient IBM15551). The total numbers of microdissected cells and of cells with the correct V-N(D)N-J β-chain sequences are shown in Table 1. We found the correct β-chain in 64 of 1,230 microdissected cells of patient PM16488. This yield is in the expected range, as explained in Discussion.
Positive cells were further tested for expression of a corresponding α-chain (step 5). We identified α-chains in 9 of 64 cells that contained the correct β-chain from patient PM16488. The sequences of the paired β- and α-chains are listed in Table 1. Six of these nine cells showed identical Vα2.2-Jα54 chain sequences, whereas three cells each contained a unique α-chain. Next, we reexamined all 64 β-chain-positive cells by using clone-specific primers for the four already identified α-chain sequences (step 6). In all cells in which we had identified α-chains with the universal primer set, we could confirm the presence of these α-chains with the appropriate clone-specific primers. Further, with the clone-specific primers, we identified 14 additional cells with the most frequently occurring (Vα2.2-Jα54) α-chain but no additional cells with the other sequences (Vα11, Vα20, Vα23). In none of the cells did we identify dual α-chains. Thus, the Vα2.2-Jα54 chain is the dominant α-chain partner of the expanded Vβ1-GGL-Jβ1.2 β-chain in muscle of patient PM16488.
Expression of Functional TCR Molecules.
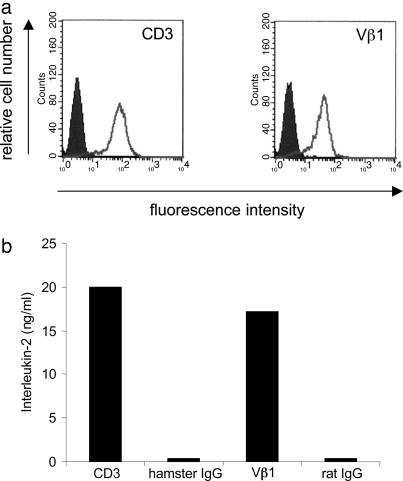
To show that the dominant α- and β-chains indeed yield a functional TCR protein, we reconstructed both full-length chains from the PCR fragments of the four αβ-pairs of patient PM16488 (Table 1), cloned them into expression vectors, and transfected them into the T hybridoma line 58α−β−, which lacks endogenous TCR α- and β-chains. FACS analysis shows surface expression of CD3 and Vβ molecules of the dominant Vα2.2-Vβ1 TCR (Fig. 3a). The TCR-transfected T hybridoma cells specifically secreted IL-2 when activated by immobilized anti-CD3 and anti-Vβ1 antibodies, but not after incubation with control antibodies (Fig. 3b), demonstrating that functional TCR molecules can be reconstituted from frozen biopsy samples. Comparable results were obtained for the three other αβ-pairs (data not shown). To test whether these TCR molecules recognize a broadly expressed self-antigen or a muscle autoantigen, we confronted the TCR transfectants with autologous EBV-transformed B cells or with the human muscle cell line TE671 transfected with all HLA class I molecules of patient PM16488. In none of these experiments could we observe activation of the TCR transfectants (data not shown), indicating that the TCR molecules recognize neither a widely expressed self-antigen (as would be expressed by autologous B cells) nor a muscle-related autoantigen (as would be expressed by TE671).
Fig. 3.
Characterization of the Vα2.2-Vβ1-TCR reconstituted in the T hybridoma cell line 58α−β−. (a) Cell-surface expression as determined by flow cytometry. The TCR transfectants were stained positive with antibodies to mouse CD3 (a Left) and human TCR Vβ1 (a Right) molecules (white areas). The transfectants were not stained with the corresponding isotype control antibodies hamster IgG and rat IgG, respectively (shaded areas). Clone A5 is shown as a representative of three independent clones. These data provide evidence that the reconstituted TCR is expressed at the cell surface of 58α−β− cells. (b) Antibody-induced activation of the reconstituted Vα2.2-Vβ1-TCR clone A5. Antibodies to mouse CD3, Vβ1, and the corresponding isotype control antibodies were coated on microtiter plates, the TCR transfectants were added, and IL-2 was measured in the supernatants by ELISA. The transfectants produced IL-2 in response to anti-CD3 and anti-Vβ1 antibodies but not to the respective control antibodies. These results show that the reconstructed, revived TCR is functional.
Discussion
We identified paired TCR α- and β-chains from individual, putatively autoaggressive T cells directly from human tissue. This approach leads beyond traditional TCR β-chain repertoire analysis because it allows the identification of complete αβ-TCR complexes from morphologically characterized single cells. Previous TCR repertoire studies have focused mainly on β-chains. Analysis of complete αβ-TCR complexes has been impeded by several factors. First, in most cases, tissue from human target organs is available only in frozen or fixed form, precluding the isolation of viable T cells. Second, only a few anti-α-chain antibodies are available, and moreover, α-chains are genetically even more complex than β-chains (10, 11). Third, identification of TCR αβ-pairs requires technically demanding single-cell techniques.
Until now, identification of paired TCR chains was possible only under exceptional circumstances. For example, TCR chains were amplified from intact cells by using a small number of α-chain primers for detection of few stainable or previously known α-chains (13, 23). Baker and colleagues (12) investigated pancreas-infiltrating T cells in prediabetic stages of NOD mice. They isolated living cells by flow cytometry from tissue homogenates and identified the TCR chains coexpressed in individual cells by multiplex RT-PCR. In this favorable situation, the early tissue infiltrates were likely to contain many more autoaggressive than bystander T cells. Therefore, it was unnecessary to identify the T cells morphologically before analysis.
With human tissue-biopsy samples, the situation is different. In most cases, the diagnosis is made long after disease initiation, at a time when the target tissue is infiltrated not only by pathogenic effector cells but also by many different types of bystander cells. The different cell populations can be distinguished only by morphological criteria, i.e., by single-cell analysis. Only in very rare cases of monoclonal expansions (24–27) can paired TCR chains be identified from tissue homogenates. Early pioneering studies described the Ig repertoire of B cells in single-cell experiments (28), but unbiased amplification from single-tissue-infiltrating T cells has so far been achieved only for TCR β-chains (7, 8, 29). Both the α- and β-chain may select from almost 50 different V elements. The β-chain may choose from 13 different J elements, but the α-chain may use 50 (10). Therefore, if rearranged genomic DNA is used as PCR template, “only” 13 Jβ-specific reverse primers are required (7, 8, 29), whereas a comparable α-chain analysis would require 50 reverse J primers. The advantage of DNA as a more stable template than RNA is therefore outweighed by the disadvantage of the huge number of primers that would be required.
We integrated morphological and molecular evidences for the pathogenic relevance of the microdissected T cells according to two criteria: (i) clonal T cell expansion in situ was demonstrated by CDR3 spectratyping; (ii) cytotoxic behavior was indicated by morphology. Together, these criteria allowed us to identify the complete β-chain sequence before single-cell PCR. Therefore, multiplex primers are needed only for the α-chains. The yields of our protocol, <1%, are in the expected range. First, minor losses were caused by fixation and inefficient cell capture. Second, antibody-labeling and AP staining significantly decreased the αβ-yields to 6 of 90 cells (Table 2). Third, the yield is further reduced by a factor of 2 by geometrical constraints: Considering 10-μm sections, half of the microdissected cells will represent only a “calotte” containing just a small fraction of the cytoplasm. Fourth, only a proportion of Vβ-stained cells contain the correct (clonally expanded) N(D)N-J regions. Immunohistochemical staining may just detect particular Vβ elements, but only a small fraction of these Vβ-positive T cells have the relevant N(D)N-Jβ regions.
We applied our protocol to muscle specimens of patients with PM or IBM. In both diseases, muscle fiber injury is partly mediated by CD8+ myocytotoxic T cells that contact and invade muscle fibers (16, 30, 31). These contacts are characterized by intimate membrane apposition and vectorial orientation of cytotoxic, perforin-containing T cell granules toward the target muscle fiber (32), which is reminiscent of an immunological synapse (33, 34). Therefore, pathogenic effector T cells and innocent-bystander cells can be easily distinguished by morphology. We have previously demonstrated that the individual myocytotoxic T cells are clonally restricted, whereas the bystander T cells express a more diverse TCR repertoire (21, 35). In subsequent studies, we isolated individual autoaggressive T cells by microdissection from muscle-biopsy sections, and we identified their individual TCR β-chain sequences by single-cell PCR from rearranged genomic DNA (8). Although these previous TCR β-chain studies provided insight into the T cell repertoire within lesions, they did not allow the identification of a complete TCR from biopsy specimens.
In our present study, patient PM16488 was especially informative. We found that in this patient’s muscle, the majority of clonally expanded Vβ1-positive autoaggressive T cells coexpressed a dominant Vα2.2-chain. In none of the cells could we detect a second α-chain, although we have shown that our method was sensitive enough to identify dual α-chain expression (Table 2). Furthermore, we reexamined all Vβ1-positive cells by using clone-specific α-chain primers, and we still never found dual α-chain T cells.
Apart from the dominant Vα2.2-Vβ1 TCR, we identified three T cells that expressed the same TCR β-chain, in each case together with a different functional α-chain. Interestingly, the CDR3 amino acid sequences of these α-chains were similar, suggesting antigen-driven selection. A comparable situation was previously observed in immunized mice in which several different, although structurally related, α-chains were associated with the same expanded β-chain (13). This study demonstrated that one antigen may recruit similar “half-sibling” αβ-TCR complexes, indicating that a common ancestral β-chain may combine with different α-chains during T cell diversification. Our data indicate that similar mechanisms might operate in the myositis patient. The TCR transfectants did not recognize either autologous EBV-transformed B cells or muscle cells transfected with all six HLA class I molecules of patient PM16488, consistent with the possibility that the muscle-invasive T cells recognize a yet-unknown foreign (e.g., viral) antigen, rather than an autoantigen. A viral cause of PM and IBM has long been suspected but never formally proved (17). Regardless of the particular disease paradigm studied here, our method for the analysis and reconstruction of tissue-infiltrating αβ-T cells can be applied to inflammatory lesions in other human diseases.
Materials and Methods
Clinical Samples and Cell Lines.
Muscle-biopsy specimens were obtained for diagnostic reasons independent of the current study. Patients with inflammatory myopathies were diagnosed according to published criteria (20, 31): PM16488 (female, 64 years) and IBM15551 (male, 62 years). The muscle blocks were stored under liquid nitrogen or at −80°C. The study was approved by institutional review boards of Ludwig Maximilians University and the Mayo Clinic.
The T hybridoma cell line 58α−β− (36) was transfected with α- and β-chains of human T cell lines BBC9 (37) and COP-1/17 (Vβ17.1 … tgt gcc agt agt GGG AGA CAG GGC CTt gaa … Jβ1.1; Vα2.3 … tgt gtg gtg aac CCC CCC CGG ggc … Jα37; N(D)N nucleotides are capitalized) as described in ref. 26. Nomenclature of the TCR V regions is taken from Arden et al. (38) throughout. An EBV-transformed B cell line was generated by standard procedures. Human TE671 rhabdomyosarcoma cells (American Type Culture Collection, Manassas, VA) were transfected individually by electroporation with all class I HLA molecules of patient PM16488 (HLA-A*0201, -A*2601, -B*0801, -B*3801, Cw0701, Cw1203). Human CD8 α- and β-cDNAs were linked by an internal ribosome entry site (IRES) sequence, inserted into the plasmid pLPCX (Clontech, Mountain View, CA), and transfected into the packaging line GP+E (American Type Culture Collection), which produces the αβ-CD8 retrovirus.
CDR3 Spectratype Analysis.
CDR3 spectratype analysis (18) of TCR β-chains was performed as described in refs. 8 and 9. Briefly, RNA was extracted from muscle tissue by TRIzol–LS reagent (GIBCO/BRL, Karlsruhe, Germany). cDNA was synthesized by using oligo(dT) primer and SuperScript II reverse transcriptase (GIBCO/BRL) and subjected to TCR Vβ gene family-specific PCR with 26 Vβ-specific and a Cβ-specific reverse primer (39). From each PCR product, a runoff reaction was performed with fluorescence-labeled Jβ-specific primers (40) and analyzed on an ABI377 DNA sequencer (Applied Biosystems, Darmstadt, Germany). Clonal expansions appear as distinct peaks of a defined CDR3 length above a Gaussian-like background of polyclonal cells. Candidate Vβ-Jβ populations were reamplified and sequenced directly (9). These candidates were regarded as clonally expanded only if readable sequences were obtained.
Immunohistochemistry and Laser Microdissection.
To detect muscle-infiltrating T cells in biopsy specimens, we used cryostat sections fixed with 4% paraformaldehyde. For single-color immunofluorescence analysis, we used an affinity-purified rabbit polyclonal anti-CD3 antibody (DAKO, Hamburg, Germany; diluted 1:50 in 2% BSA/5% donkey serum) detected with multilabel Cy3- or rhodamine-labeled donkey anti-rabbit IgG (1:1,000 and 1:400; Jackson ImmunoResearch, West Grove, PA) and the FITC-labeled anti-CD8 antibody B9.11 (Beckman Coulter, Krefeld, Germany), diluted 1:20 in 2% goat serum/2% BSA in PBS. For double-color analysis of PM16488, we used the FITC-labeled mouse anti-CD8 antibody B9.11 at a 1:20 dilution and the rat anti-Vβ1 antibody BL37.2 at a 1:200 dilution in the above buffer. The anti-Vβ1 antibody was visualized with the Cy3-conjugated AffiniPure goat anti-rat IgG (H+L) antibody (Jackson ImmunoResearch) at a 1:100 dilution.
To isolate T cells from muscle-biopsy specimens, we used acetone-fixed 10-μm cryostat sections. The TCR Vβ regions were labeled with antibodies and stained by standard AP procedures (LSAB2 system; DAKO). We used the anti-Vβ1 antibody BL37.2 (Beckman Coulter) for patient PM16488 and the anti-Vβ23 antibody AF23 (Beckman Coulter) for patient IBM15551. Both antibodies were diluted 1:100 in 2% goat serum in Tris-buffered saline containing 3% Prime RNase inhibitor (Eppendorf, Hamburg, Germany). Because the BL37.2 is of rat origin, we replaced the biotinylated anti-mouse antibody from the LSAB2 kit by the biotinylated sheep, anti-rat IgG (H+L) antibody AAR10B (Serotec, Dusseldorf, Germany) in 2% BSA. Stained cells were isolated by laser microdissection (Molecular Machines & Industries, Zurich, Switzerland) as described in ref. 8. For control experiments, unstained TCR-transfected T hybridoma cells were centrifuged onto PEN films (1.35 μm; PALM Microlaser, Bernried, Germany) in a Cytospin centrifuge device (Heraeus, Hanau, Germany) and isolated by laser microdissection. For some experiments, control TCR (clone COP-1/17)-transfected hybridoma cells were stained with anti-Vβ17 and AP before isolation.
Amplification of TCR α- and β-Chains from Microdissected Cells by RT-PCR.
mRNA of the TCR α- and β-chains from microdissected cells was transcribed for 30 min at 50°C into cDNA by using a one-step RT-PCR kit (Qiagen, Hilden, Germany) and constant region-specific RT primers at 0.625 μM each (see Fig. 4; for all primer sequences see Data Set 1, which is published as supporting information on the PNAS web site). All primers were HPLC-purified (Metabion, Martinsried, Germany). Then primers for the simultaneous preamplification of α- and β-chains were added (step 3 in Fig. 1). We used clone-specific primers for the β-chains that hybridize to the Vβ and Jβ regions at 0.6 μM each: a pool of 24 primers (0.062 μM for each primer) that covers all Vα genes, and a reverse Cα-specific primer (0.6 μM). After a 2-min incubation at 94°C, touchdown PCR was run. The annealing steps were for 1 min each for 4 cycles at 61°C, 4 cycles at 58°C, 4 cycles at 56°C, and 40 cycles at 53°C. Denaturation and extension steps were for 1 min at 94°C and 1 min at 72°C. Without further analysis, 1-μl aliquots were used for β-chain-specific, double-nested PCR (step 4) by using clone-specific primers hybridizing to the Vβ and N(D)N-J regions (0.5 μM for each primer). Only cells that yielded the correct β-chain as verified by sequencing of the PCR products were further analyzed for the corresponding α-chain (step 5). As template, we used five aliquots of 1 μl each of the preamplification PCR product. In total, 36 forward primers were grouped into five pools of 7 or 8 primers each. The final concentration of each primer was 0.5 μM. All forward primers and the Cα-specific reverse primer (0.5 μM) were nested relative to the outer primers used in the preamplification reaction 3. Candidate PCR products were sequenced directly. In independent experiments, we confirmed that our universal primer set amplifies the entire panel of TCR Vα-chains with whole-blood cDNA as template. Finally, we reexamined all β-chain-positive cells by using clone-specific α-chain primers (0.5 μM) in a standard PCR (step 6).
Reconstruction, Expression, and Characterization of Vα2.2-Vβ1 TCR Proteins in T Hybridoma Cells.
Full-length cDNA clones of the Vα2.2-Jα54 and Vβ1-Jβ1.2 TCR chains were reconstructed from the PCR products of the single-cell PCR and from unrelated lymphocyte cDNA. Typically, we ligated three fragments: The first contained a 5′ SalI restriction site before the leader sequences and reached as far as a restriction site within the downstream region of the V regions. The second fragment reached from this site to a unique site in the conserved region. This fragment covered the N(D)N-J regions of the respective chain. The third fragment is part of a cloning cassette already contained in our expression plasmids pRSVneo and pRSVhygro covering the remainder of the conserved α and β regions. After the stop codon, a BamHI restriction site was introduced. The α- and β-chains were inserted into the SalI and BamHI sites of pRSVneo and pRSVhygro, respectively, cotransfected into the mouse T hybridoma cell line 58α−β−, and selected by G418 and Hygromycin B (Sigma, Deisenhofen, Germany) as described for a γδ-TCR molecule (26, 27). Individual clones were isolated and characterized. Surface expression of CD3 and Vβ1 chains was monitored by flow cytometry as described in ref. 26. We used the FITC-labeled anti-CD3ε mAb 145-2C11 (BD PharMingen, Heidelberg, Germany), the anti-human Vβ1 mAb BL37.2 (Beckman Coulter), and corresponding isotype control mAbs FITC-labeled hamster IgG 554234 (BD Pharmingen) and rat IgG IM300039 (Beckman Coulter). The Vα2.2 chain of our TCR could not be stained because the available Vα2.3 antibody does not recognize Vα2.2. To test whether the transfectants were capable of producing IL-2 when activated by TCR, the above antibodies were diluted 1:500 in PBS and coated for 2 h at 37°C on 96-well flat-bottom plates (Costar, Corning, NY). Alternatively, 2 × 105 EBV-transformed B cells from patient PM16488 or confluent HLA-transfected TE671 cells were used as antigen-presenting cells. For these experiments, αβ-TCR transfectants were transformed with αβ-CD8 retrovirus and selected by 0.5 μg/ml puromycin. The TCR transfectants were added at 40,000 cells per well in 150 μl of RPMI medium 1640/10% FCS and incubated for 14 h at 37°C. Secreted IL-2 was measured by using an eBioscience mouse IL2 ELISA kit (NatuTec, Frankfurt, Germany).
Supplementary Material
Acknowledgments
We thank Tjalf Ziemssen for providing T cell lines, Ingrid Eiglmeier for expert technical assistance, and Dieter Jenne and Alexander Flügel for helpful comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 571-A1, Chinese State Commission for Overseas Studies Grant 2003850073, and National Institutes of Health Grant NS6277 (to A.G.E.).
Abbreviations
- AP
alkaline phosphatase
- IBM
inclusion body myositis
- PM
polymyositis
- TCR
T cell receptor.
Footnotes
References
- 1.Steinman L. J. Exp. Med. 2003;197:1065–1071. doi: 10.1084/jem.20030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen I. R. Immunol. Today. 1992;13:490–494. doi: 10.1016/0167-5699(92)90024-2. [DOI] [PubMed] [Google Scholar]
- 3.Bach J.-F., Chatenoud L. Annu. Rev. Immunol. 2001;19:131–161. doi: 10.1146/annurev.immunol.19.1.131. [DOI] [PubMed] [Google Scholar]
- 4.von Herrath M. Nature. 2005;435:151–152. doi: 10.1038/435151a. [DOI] [PubMed] [Google Scholar]
- 5.Krogsgaard M., Davis M. M. Nat. Immunol. 2005;6:239–245. doi: 10.1038/ni1173. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson M. J., Hahn M., Wucherpfennig K. W. Immunity. 2005;23:351–360. doi: 10.1016/j.immuni.2005.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babbe H., Roers A., Waisman A., Lassmann H., Goebels N., Hohlfeld R., Friese M., Schröder R., Deckert M., Schmidt S., et al. J. Exp. Med. 2000;192:393–404. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofbauer M., Wiesener S., Babbe H., Roers A., Wekerle H., Dornmair K., Hohlfeld R., Goebels N. Proc. Natl. Acad. Sci. USA. 2003;100:4090–4095. doi: 10.1073/pnas.0236183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skulina C., Schmidt S., Dornmair K., Babbe H., Roers A., Rajewsky K., Wekerle H., Hohlfeld R., Goebels N. Proc. Natl. Acad. Sci. USA. 2004;101:2428–2433. doi: 10.1073/pnas.0308689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefranc M.-P., Lefranc G. The T Cell Receptor Facts Book. London: Academic; 2001. [Google Scholar]
- 11.Arstila T. P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 12.Baker F. J., Lee M., Chien Y.-H., Davis M. M. Proc. Natl. Acad. Sci. USA. 2002;99:9374–9379. doi: 10.1073/pnas.142284899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamrouni A., Aublin A., Guillaume P., Maryanski J. L. J. Exp. Med. 2003;197:601–614. doi: 10.1084/jem.20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahara H., Fujio K., Araki Y., Setoguchi K., Misaki Y., Kitamura T., Yamamoto K. J. Immunol. 2003;171:2154–2160. doi: 10.4049/jimmunol.171.4.2154. [DOI] [PubMed] [Google Scholar]
- 15.Arahata K., Engel A. G. Ann. Neurol. 1984;16:193–208. doi: 10.1002/ana.410160206. [DOI] [PubMed] [Google Scholar]
- 16.Mantegazza R., Andreetta F., Bernasconi P., Baggi F., Oksenberg J. R., Simoncini O., Mora M., Cornelio F., Steinman L. J. Clin. Invest. 1993;91:2880–2886. doi: 10.1172/JCI116533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalakas M. C. Nat. Clin. Pract. Rheumatol. 2006;2:219–227. doi: 10.1038/ncprheum0140. [DOI] [PubMed] [Google Scholar]
- 18.Pannetier C., Even J., Kourilsky P. Immunol. Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 19.Padovan E., Casorati G., Dellabona P., Meyer S., Brockhaus M., Lanzavecchia A. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 20.Dalakas M. C. Rev. Neurol. 2002;158:948–958. [PubMed] [Google Scholar]
- 21.Bender A., Ernst N., Iglesias A., Dornmair K., Wekerle H., Hohlfeld R. J. Exp. Med. 1995;181:1863–1868. doi: 10.1084/jem.181.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishio J., Suzuki M., Miyasaka N., Kohsaka H. J. Immunol. 2001;167:4051–4058. doi: 10.4049/jimmunol.167.7.4051. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa M., Tong J., Matsui T., Masuko-Hongo K., Yabe T., Nishioka K., Yamamoto K., Kato T. Clin. Exp. Immunol. 2001;123:340–345. doi: 10.1046/j.1365-2249.2001.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohlfeld R., Engel A. G., Ii K., Harper M. C. N. Engl. J. Med. 1991;324:877–881. doi: 10.1056/NEJM199103283241303. [DOI] [PubMed] [Google Scholar]
- 25.Pluschke G., Rüegg D., Hohlfeld R., Engel A. G. J. Exp. Med. 1992;176:1785–1789. doi: 10.1084/jem.176.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiendl H., Malotka J., Holzwarth B., Weltzien H. U., Wekerle H., Hohlfeld R., Dornmair K. J. Immunol. 2002;169:515–521. doi: 10.4049/jimmunol.169.1.515. [DOI] [PubMed] [Google Scholar]
- 27.Dornmair K., Schneider C. K., Malotka J., Dechant G., Wiendl H., Hohlfeld R. J. Neuroimmunol. 2004;152:168–175. doi: 10.1016/j.jneuroim.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Küppers R., Zhao M., Hansmann M.-L., Rajewsky K. EMBO J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roers A., Montesinos-Rongen M., Hansmann M.-L., Rajewsky K., Küppers R. Eur. J. Immunol. 1998;28:2424–2431. doi: 10.1002/(SICI)1521-4141(199808)28:08<2424::AID-IMMU2424>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 30.Engel A. G., Arahata K. Ann. Neurol. 1984;16:209–216. doi: 10.1002/ana.410160207. [DOI] [PubMed] [Google Scholar]
- 31.Engel A. G., Hohlfeld R. In: Myology: Basic and Clinical. Engel A. G., Franzini-Armstrong C., editors. New York: McGraw–Hill; 2005. pp. 1321–1366. [Google Scholar]
- 32.Goebels N., Michaelis D., Engelhardt M., Huber S., Bender A., Pongratz D., Johnson M. A., Wekerle H., Tschopp J., Jenne D., et al. J. Clin. Invest. 1996;97:2905–2910. doi: 10.1172/JCI118749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stinchcombe J. C., Bossi G., Booth S., Griffiths G. M. Immunity. 2001;15:751–761. doi: 10.1016/s1074-7613(01)00234-5. [DOI] [PubMed] [Google Scholar]
- 34.Purbhoo M. A., Irvine D. J., Huppa J. B., Davis M. M. Nat. Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 35.Bender A., Behrens L., Engel A. G., Hohlfeld R. J. Neuroimmunol. 1998;84:86–91. doi: 10.1016/s0165-5728(97)00246-4. [DOI] [PubMed] [Google Scholar]
- 36.Blank U., Boitel B., Mège D., Ermonval M., Acuto O. Eur. J. Immunol. 1993;23:3057–3065. doi: 10.1002/eji.1830231203. [DOI] [PubMed] [Google Scholar]
- 37.Giegerich G., Pette M., Meinl E., Epplen J. T., Wekerle H., Hinkkanen A. Eur. J. Immunol. 1992;22:753–758. doi: 10.1002/eji.1830220319. [DOI] [PubMed] [Google Scholar]
- 38.Arden B., Clark S. P., Kabelitz D., Mak T. W. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 39.Monteiro J., Hingorani R., Peroglizzi R., Apatoff B., Gregersen P. K. Autoimmunity. 1996;23:127–138. doi: 10.3109/08916939608995336. [DOI] [PubMed] [Google Scholar]
- 40.Puisieux I., Even J., Pannetier C., Jotereau F., Favrot M., Kourilsky P. J. Immunol. 1994;153:2807–2818. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.