Abstract
Somatic hypermutation of Ig genes is initiated by transcription-coupled cytidine deamination in Ig loci. Error-prone processing of the resultant DNA lesions is thought to cause extensive mutagenesis, but it is presently an enigma how and why error-prone rather than error-free repair pathways are recruited. During DNA replication, recruitment of error-prone translesion polymerases may be mediated by Rad6/Rad18-mediated ubiquitination of proliferating cell nuclear antigen, a major switchboard controlling the fidelity of DNA lesion bypass in eukaryotes. By inactivation of Rad18 in the DT40 B cell line, we show that the Rad6 pathway is involved in somatic hypermutation in these cells. Our findings imply that targeted recruitment of mutagenic polymerases by the Rad6 pathway contributes to the complex process of somatic hypermutation and provide a framework for more detailed mechanistic studies of the mutagenesis phase of secondary Ig diversification.
Keywords: proliferating cell nuclear antigen, ubiquitination, Rad6 pathway
Given the many challenges that damage cellular DNA daily, the faithful maintenance of genetic information requires the interplay of multiple DNA repair pathways. In most cases, these mechanisms ensure restoration of the original DNA sequence, preventing mutations or aberrations that may lead to impairment of cellular function. In some instances, though, a certain imprecision may be tolerated or even favored by the cell to allow for survival in critical situations. DNA repair mechanisms that are inherently imprecise are the basis of the spontaneous mutability of all genomes and may be recruited and adapted for situations where high genetic variability is mandatory for survival.
The adaptive immune system of vertebrates has been shaped in coevolution with pathogenic organisms of high genetic diversity and variability. The high diversity of the primary immune repertoire established during V(D)J recombination in B and T cells generates sufficient potential for recognition of any pathogen, but the affinity of binding is often not sufficient for effective neutralization. Therefore, a second wave of diversification during acute infections ensures that the binding affinity of the antibodies produced by B lymphocytes is fine-tuned to the task at hand. The underlying mechanism in humans is somatic hypermutation, which modifies the antigen binding region by targeted mutagenesis in the variable region of the Ig genes (1). An alternative diversification mechanism prevalent in chicken and some mammals, Ig gene conversion alters the same region by targeted homologous recombination with upstream Ig pseudogenes (2).
Somatic hypermutation and Ig gene conversion are related processes (3) originating from the same initial DNA lesion. The transcription-coupled deamination of cytosines by activation-induced cytidine deaminase (AID) leads to uracil that may be processed to abasic sites or strand breaks by the excision repair pathway, i.e., uracil-N-glycosylase (4, 5). The resultant DNA lesions may be repaired by mutagenesis or recombination, leading to hypermutation or gene conversion, respectively. The basis for the choice between these pathways and many aspects of the mechanism of somatic hypermutation are not well understood. It is likely, though, that the B cells undergoing diversification of their Ig genes have adapted general mechanisms for mutagenesis and recombination in mammalian cells and the regulatory pathways that govern their function and interplay.
Recent models of somatic hypermutation postulate multiple independent mutagenesis pathways (4, 5): (i) replication over the uracils generated by AID, leading to transition mutations, (ii) uracil-triggered recruitment of an A/T mutator by components of the mismatch repair pathway, and (iii) translesion synthesis over abasic sites formed by uracil excision, generating transversion mutations and transitions. It is unknown how mutagenic polymerases are recruited during these processes. Eukaryotic genomes encode multiple DNA polymerases that differ in their processivity, function, and fidelity. Members of the Y family of translesion polymerases are characterized by a flexible catalytic site that allows them to bypass bulky DNA lesions, but at the same time impairs their accuracy while copying undamaged DNA (6). The low fidelity and hence mutagenic capacity of translesion polymerases such as Polη and Rev 1 is exploited during somatic hypermutation (7, 8). Polη serves as an A-T mutator in this process. Rev1 cooperates with Polζ, a B family polymerase that effectively elongates mismatched primer termini, thus being responsible for most damage-induced mutagenesis in yeast cells (6). In higher eukaryotes, Polζ likely performs other functions during replication and is considered a specialized replicative polymerase, but it has also been implicated in somatic hypermutation (9).
In physiological repair situations, translesion synthesis or recombinogenic mechanisms represent alternative ways to repair critical DNA lesions, and the cell has evolved pathways that control the decision between these two. The prototypic example is the Rad6 pathway of postreplication repair in yeast, which regulates the employment of mutagenic translesion synthesis and/or recombinogenic activity during the bypass of unrepaired DNA lesions encountered by a replication fork (10). The enzymes of the Rad6 pathway mediate ubiquitination of proliferating cell nuclear antigen (PCNA), a sliding clamp and processivity factor for DNA polymerases and other DNA repair enzymes (11). Monoubiquitination of PCNA by Rad6, which is recruited to the site of damage by the RING finger E3 ubiquitin ligase Rad18, serves as a signal for recruitment of the error-prone translesion polymerases Polζ and Polη (12, 13) and may thus lead to mutagenic lesion bypass. Subsequent Lys-63-linked polyubiquitination of PCNA by Ubc13/Mms2/Rad5, on the other hand, initiates a pathway of error-free lesion bypass that is likely based on homology-mediated template switch of replication to the undamaged sister chromatide.
Components of the Rad6 pathway are conserved in higher eukaryotes and have been implicated in the regulation of mutagenic and recombinogenic DNA modifications (14, 15). In particular, they have also been implicated in the regulation of translesion polymerases in postreplication repair (16, 17), but the function of these polymerases in other genetic processes in eukaryotes may also be regulated by different means, i.e., independently of the Rad6 pathway (18–20). In the present study, we show that the crucial initiator factor of the Rad6 pathway, Rad18, plays a role during somatic hypermutation in the DT40 B cell line. These findings suggest that to specifically recruit mutagenic repair factors during nontemplated Ig diversification B cells have adapted a generic DNA repair pathway that was designed to bypass fork blocking and thus potentially lethal damage in replicating DNA. Our study leads to a refined model of the mutagenesis phase of somatic hypermutation and also provides a system for investigating why error-prone rather than error-free DNA repair is used to process AID-induced DNA damage in hypermutating Ig genes.
Results
Inactivation of Rad18 in DT40 Cells.
To investigate whether activation of mutagenesis or recombination by the Rad6 pathway is involved in somatic hypermutation or Ig gene conversion, we inactivated the Rad18 gene in the chicken bursal B cell line DT40 that constitutively diversifies its endogenous Ig loci by both processes (3). As parental cells for targeting, we used DT40Cre1 cells that show enhanced Ig diversification activity and contain a tamoxifen-regulatable MerCreMer recombinase for recycling of loxP-flanked resistance markers and transgenes. The targeting strategy shown in Fig. 1A replaces exons 3 and 4 and part of exon 5 of the Rad18 gene by loxP-flanked expression cassettes for antibiotic resistance markers (21). This modification is expected to abolish Rad18 function and hence inactivate the Rad6 pathway, as these exons (corresponding to amino acids 46–209) code for essential and conserved parts of the RING domain that is important in recruiting Rad6 to chromatin to perform PCNA monoubiquitination (13). Targeted integration was verified by Southern blot analysis (Fig. 1B), and deletion of the respective exons was confirmed at the mRNA level (Fig. 1C). The resultant clones are suitable for quantitative comparative analysis of Ig diversification, as unlike human hypermutating B cells, DT40 cells do not show substantial clonal variation in AID protein expression levels (Fig. 1D).
Fig. 1.
Inactivation of Rad18 in DT40 cells. (A) Targeting strategy. The first 7 of 10 exons of the Rad18 gene (boxes), the primers for amplification of the targeting arms (small arrowheads), and the loxP (large arrowheads)-flanked resistance genes (puro, puromycin; bsr, blasticidin) are shown. A presumptive exon 2 (hatched, coding for amino acids 19–45) is not contained in the present genomic sequence files. The exons marked in gray are deleted in the mRNA resulting from the knockout allele. NcoI restriction sites (N) and the Southern probe (black box) are indicated. (B) Southern blot analysis, indicating replacement of the WT alleles by resistance marker cassettes, as well as excision of the resistance marker (−/−Δ). (C) RT-PCR for the complete Rad18 coding region indicating loss of the targeted exons in the mRNA. Rad18−/−R, clone reconstituted with Rad18 expression cassette; Ψ−, pseudogene-deficient cells; HPRT, hypoxanthine phosphoribosyltransferase. (D) Expression levels of the endogenous (lanes 1–5) and transgenic (lanes 7 and 8) AID in DT40Cre1 and Ψ−DT40 cells. (E) PCNA ubiquitination upon methylmethanesulfonate (MMS) treatment in WT and Rad18-deficient DT40 cells. The position of PCNA and its ubiquitinated form in the blot are indicated.
As shown before, Rad18 deficiency did not adversely affect cell viability or proliferation (15, 22). Treatment with methylmethanesulfonate resulted in efficient PCNA monoubiqitination in WT cells (Fig. 1E), whereas in Rad18-deficient cells a clear defect was observed. To our surprise, we noted a residual ubiquitinating activity in the mutant cells. It will be interesting to see whether this activity represents a DT40-specific phenomenon or a more ubiquitous alternative E3 ubiquitin ligase for PCNA in vertebrates.
Ig Gene Conversion in Rad18-Deficient Cells.
The DT40Cre1 cells used in this study do not express surface Ig, because of a frameshift mutation in the rearranged light chain gene (23). Repair of the frameshift may occur during homologous recombination with upstream pseudogenes, and the generation of Ig-positive cells is therefore a direct measure of Ig gene conversion activity. Clones with the desired genotypes were subject to limiting dilution subcloning, and 23–24 derivative subclones were analyzed for surface Ig expression after 10 days of culture. As shown in Fig. 2, the reversion frequency of the frameshift mutation was not significantly affected by Rad18 deficiency (P = 0.958), averaging ≈0.5% in each genotype analyzed. In agreement with a recent study (22), we conclude that Rad18 deficiency has no adverse impact on Ig gene conversion activity in DT40.
Fig. 2.
Ig gene conversion in Rad18-deficient DT40 cells. The frequency of Ig+ cells among 10,000 total cells in 23–24 clones of the indicated genotypes is given. Two independently generated Rad18−/− clones were analyzed. The bar indicates the average frequency of Ig+ cells.
Somatic Hypermutation of Transgenes in DT40.
DT40 cells mainly diversify their Ig genes by Ig gene conversion, which is accompanied by a lower degree of somatic hypermutation. The relative activity of hypermutation may be enhanced by impairment of homologous recombination in cis [i.e., by deletion of the pseudogene locus (3)] or in trans [e.g., by inactivation of XRCC2/3 (24)]. The latter approach is not suitable in the present context, as combined inactivation of homologous recombination and the Rad6 pathway is synthetic lethal in DT40 (15).
We reasoned that a transgene containing Ig enhancer elements (25, 26) but lacking upstream pseudogenes should also be subject to hypermutation in DT40. To test this idea, we transfected DT40 cells with the hypermutation reporter shown in Fig. 3A, which contains a CMV-promoter-driven yellow fluorescent protein (YFP) gene rendered nonfunctional by a premature TAG stop codon, and also includes an enhancer cassette derived from the human κ locus (26). Hypermutation of this reporter causes reversion of the stop codon by point mutations and expression of functional YFP that is detectable by FACS. Most of the transfected subclones indeed contained revertant cells (Fig. 3 B and C), with an average revertant frequency of 3.5 × 10−5. Hypermutation of this construct is clearly AID-dependent, as in AID−/− DT40 cells (23) only one single revertant cell was observed among all clones analyzed (Fig. 3C). We conclude that transfected genes containing appropriate enhancer elements are subject to somatic hypermutation in DT40, providing a sensitive tool for simultaneously studying hypermutation and gene conversion in these cells.
Fig. 3.
Hypermutation of transgenes in DT40 cells. (A) The hypermutation reporter. The YFP is driven by a CMV promotor and contains a premature TAG stop codon. The enhancer cassette contains intronic and 3′ enhancers and matrix attachment region of the human Igκ locus. (B) FACS analyses of DT40 cells transfected with the hypermutation reporter (Left) and a respective negative control (Right). (C) AID-dependent hypermutation of a transgene in DT40. The pie charts indicate the fraction of clones with the respective number of revertants among 500,000 cells analyzed. The total number of analyzed clones is indicated in the middle of each chart.
Impaired Hypermutation in Rad18-Deficient DT40 Cells.
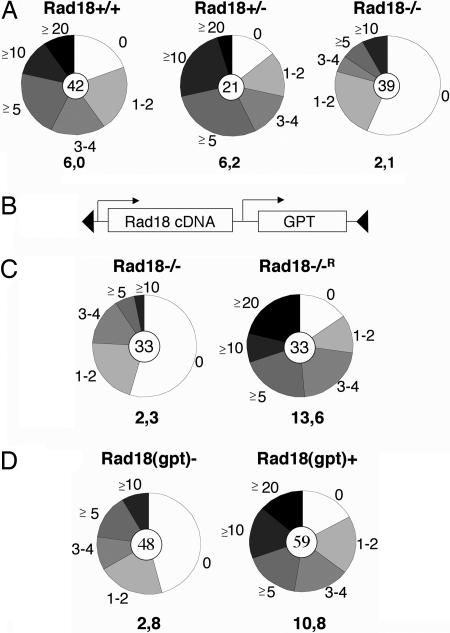
The effect of Rad18 deletion on somatic hypermutation was assessed by transfection of the hypermutation reporter into cells of the respective genotypes and analysis of antibiotics-resistant subclones for revertant cells after 2 weeks of culture. In WT DT40, most of the subclones contained revertant cells, as observed before (Fig. 4A). A similar situation was seen in Rad18+/− cells. In contrast, in Rad18−/− cells fewer revertants could be identified, and more than half of the clones contained no revertants at all (Fig. 4A). A corresponding decrease in the average frequency of revertant cells per clone clearly indicates that the deletion of Rad18 impairs somatic hypermutation in DT40 (P = 0.002).
Fig. 4.
Rad18 deficiency impairs somatic hypermutation of reporter constructs. (A) Cells of the indicated genotypes were transfected with the hypermutation reporter and analyzed as before. The average frequency of revertant cells per clone is given below each pie chart. (B) Schematic view of the loxP-flanked Rad18 cDNA expression cassette containing a gpt (mucophenolic acid) resistance marker. (C) Rescue of the Rad18−/− phenotype by Rad18 overexpression. Cells were assessed for their hypermutation potential as before. (D) Loss of Rad18 expression impairs somatic hypermutation. A clone containing both the hypermutation reporter and the loxP-flanked Rad18 expression cassette was subject to deletion of the loxP-flanked Rad18 expression cassette in a fraction of the cells by limited activation of the MerCreMer recombinase. Subclones were independently assessed for Rad18(gpt) positivity and revertant cells.
To confirm that the observed phenotype was indeed caused by Rad18 inactivation, Rad18−/− cells were transfected with a vector containing a loxP-flanked Rad18 cDNA expression cassette linked to a resistance marker (Fig. 4B) and analyzed for their hypermutation capacity as above. The results shown in Fig. 4C indicate that the re-expression of Rad18 in these cells indeed rescues the phenotype of Rad18 inactivation (P = 0.006). To exclude the possibility that the observed effects are caused by integration or copy number effects of the transfected reporter system, we activated the MerCreMer recombinase in one of the reconstituted subclones containing the integrated hypermutation reporter to achieve deletion of the loxP-flanked Rad18 in a fraction of the cells. Clones obtained after limiting dilution were independently assessed for revertant cells and Rad18(gpt) positivity. The comparison of revertant frequencies in the Rad18(gpt)+ versus Rad18(gpt)− clones again confirmed that its presence increases hypermutation activity (Fig. 4D; P = 0.002).
Defective Hypermutation of Endogenous Ig Genes in Rad18-Deficient Cells.
Finally, we wanted to ascertain whether inactivation of the Rad6 pathway also affects somatic hypermutation on the endogenous Ig locus. To this end, we inactivated the Rad18 gene in DT40 cells lacking the pseudogenes required for gene conversion (Ψ−DT40), as in these cells AID-induced lesions in the Ig loci are processed by mutagenesis (3). Deletion of the respective exons was confirmed by RT-PCR (Fig. 1C), and equal expression levels of the transgenic AID were verified by Western blot analysis (Fig. 1D).
To analyze hypermutation activity, subclones of Rad18+/+ and Rad18−/− Ψ−DT40 cells were cultured for 6 weeks, the rearranged light chain loci were amplified and cloned, and for each genotype >100 plasmids (derived from two WT and three mutant subclones) were sequenced (Fig. 5). In Rad18-proficient cells, an average mutation frequency of 2.5 × 10−3 mutations per bp was observed, and these base changes focused mainly on dG/dC residues, as noticed before in DT40 and other in vitro systems. Rad18-deficient cells showed a significantly decreased mutation frequency of 0.9 × 10−3 mutations per bp (P < 0,001), because of a decrease in all types of mutations at dG/dC residues (Fig. 5). Also, an increase in deletions was notable in the sequence database established from the mutant cells (Fig. 5A). We conclude that Rad18 plays a critical role during somatic hypermutation in DT40 cells, while it is dispensable for Ig gene conversion.
Fig. 5.
Impaired hypermutation of endogenous Ig genes in Rad18-deficient cells. (A) Frequency of sequences with the indicated number of mutations in Rad18-proficient and -deficient Ψ−DT40 cells. The total number of sequences analyzed is given in the middle of the pie chart. Average mutation frequencies for the individual subclones (mutations per 103 bp) were 2.0 and 3.0 for WT and 0.8, 0.9, and 1.2 for mutant clones. (B) Pattern analysis for the total numbers (Upper) and percentages (Lower) of mutations in WT and mutant Ψ−DT40 cells.
Discussion
The present study identifies Rad18, the initiator protein of the Rad6 pathway, as a key factor in somatic hypermutation in DT40 B cells. It thereby suggests a model for how error-prone DNA repair factors are recruited to AID-induced lesions in Ig genes. We propose that bypass of the lesions generated by AID by mutagenic translesion polymerases may be triggered by activation of the Rad6 pathway, which ubiquitinates PCNA as a major cellular switchboard for regulation of DNA repair processes. This modification would serve as a signal for the switch from error-free DNA synthesis to error-prone lesion bypass, and hence mutagenesis. In sum, this model infers that the targeted activation of translesion synthesis by the Rad6 pathway is a major mechanistic contributor to somatic hypermutation in DT40.
Rad18 deficiency led to a decrease in mutagenesis in two independent hypermutation assays and targeting approaches in our study. As recent models of somatic hypermutation postulate several parallel pathways of mutagenesis (27), such incomplete inhibition is certainly not surprising. Partial phenotypes are in fact a hallmark of factors involved in the DNA repair and mutagenesis phase of somatic hypermutation (5, 7, 9, 27, 28, 30, 31), because inhibition of one erroneous processing pathway may channel AID-induced DNA lesions into others. This functional redundancy may occlude mechanistic analyses of some pathways in certain experimental settings, in particular if semiquantitative detection systems are used. Indeed, the role of Rad18 in hypermutation has been missed in a previous approach using a different targeting strategy and a combined hypermutation/gene conversion assay that selects for deleterious changes in Ig genes (22). Given the high complexity and redundancy of the hypermutation mechanism, the successful detection of its subpathways may demand systems that are not subject to antigenic or experimental selection or the use of specialized reporters focusing on certain mutagenesis modes.
For factors involved in a defined subpathway of hypermutation, the analysis of mutational patterns may also give decisive clues, as the three major subpathways are characterized by transitions, transversions, and A/T mutations, respectively (27). The DT40 system, as most other in vitro hypermutation models, does not allow substantial analyses of A/T mutagenesis (3). Concerning G/C-biased mutagenesis, Rad18 might function specifically during translesion synthesis over abasic sites, which is required for G/C transversions but may also insert transitions (15, 27), and/or might be involved in the recruitment of mismatch extenders such as Polζ and Polθ that appear to affect the overall mutation load rather than specific subpathways (9, 32–34). Clearly, all types of mutations are negatively affected by Rad18 deficiency (Fig. 5B). We do also note a stronger decrease in transversion than transition mutations, mostly because of substantial drops in C-G and G-T changes (coupled to a relative increase in G-A changes), features that have previously been reported for Rev1 and Polθ, respectively (34, 35). In the case of DT40, however, in-depth pattern analyses must be interpreted with caution, as the consensus sequences of clones differ slightly because of ongoing mutagenesis in the parental cultures, and not all of the individually scored base changes may be unique events, especially at hotspots. The most prominent pattern change was indeed an increase in deletion events (from ≈1% to ≈10% of total events, even though the analysis was focused on close to full-length PCR products), which may be caused by strand breaks arising when the processing of abasic sites by translesion synthesis is impaired. It is evident, though, that transversions indicative of abasic site bypass may still occur in the absence of Rad18, and also a reporter specialized in transversions (preferentially measuring TAG to TAC/TAT mutations in G/C-biased systems) shows a clear, but partial, defect in Rad18-deficient cells (Fig. 4). We surmise that this phenomenon is caused by Rad18-independent translesion polymerases that either play a role in the normal hypermutation process or compensate in the absence of Rad18 function. It will hence be interesting to investigate which of the mutagenic polymerases involved in hypermutation requires Rad18 function. In particular, an analysis of Rad18 effects on the A/T mutator Polη is warranted (7), as Polη is known to be regulated by the Rad6 pathway during replication in yeast and mammalian cells (14).
Rad18 is the only factor of the Rad6 pathway for which no role in other cellular functions has been described to date. Our findings therefore suggest that monoubiquitination of PCNA serves as a signal for specific recruitment of error-prone DNA polymerases to AID-induced lesions in Ig loci. Formal proof for PCNA as the target of Rad18 activity during hypermutation and for dispensability of PCNA polyubiquitination will, however, be required. In fact, the clear-cut decision between error-prone and error-free lesion bypass in the Rad6 pathway offers a tractable model system for studying the major mechanistic mystery of somatic hypermutation: why error-prone rather than error-free DNA repair pathways are recruited to process AID-induced DNA lesions in Ig loci.
Given the well established function of the Rad6 pathway during replication (36), it will be interesting to determine whether mutagenic bypass of AID-induced lesions occurs during DNA replication, or whether Rad18 may also recruit error-prone polymerases during other repair processes. These two possibilities are not mutually exclusive as evidence for AID-dependent mutagenesis in G1 (37) and AID-dependent DNA lesions in G2 has been presented (29). Independently of the cell-cycle phase of Rad18 function during hypermutation, its dispensability for Ig gene conversion also marks it as a critical player in the apparent competition of mutagenesis and recombination during processing of AID-induced lesion by hypermutation and Ig gene conversion (3, 24).
In conclusion, our study identifies an important factor involved in somatic hypermutation and points at a signal for recruitment of error-prone DNA repair during this process. These findings provide an experimental system and conceptual framework to precipitate and facilitate more detailed mechanistic studies of the DNA repair and mutagenesis phase of secondary antibody diversification.
Materials and Methods
Cell Culture.
The DT40Cre1 line (21) was cultured as described (23). AID−/− cells were obtained from Hiroshi Arakawa (GSF-Research Center for Environment and Health) (23). Transfections were performed as described (23) with a BioRad (Hercules, CA) gene pulser set at 50 μF and 800 V. Deletion of loxP-flanked cassettes was achieved by overnight culture in the presence of 2 μM 4′-OHT (H7904; Sigma, St. Louis, MO), followed by limiting dilution subcloning and genotypic and/or resistance analysis of resultant subclones.
Targeted Inactivation of Rad18, Expression Analysis, and Reconstitutions.
The arms of homology of the targeting vector were amplified with primers 18Rad1 (5′-gggctcgagcagcgggcagtgaggacacctttcc-3′) and 18Rad2 (5′-gggggatcctatacatatgtgtgtgcgcgtgtgtct-3′) for the 5′ arm and 18Rad3 (5′-gggggatccgcaggatcaagagagtgttctgaaggc-3′) and 18Rad4a (5′-gggactagttcctctctgctcagacagctgtccag-3′) for the 3′ arm. PCR products were cut with SpeI, XhoI, and BamHI and cloned into the pBS-KS vector. LoxP-flanked resistance marker cassettes were excised from ploxpuro, ploxbsr, and ploxgpt (Hiroshi Arakawa) with BamHI and cloned into the BamHI site between the two targeting arms. Rad18 inactivation in DT40Cre1 cells was performed with the respective Rad18puro and Rad18bsr targeting vectors, whereas inactivation in pseudogene-deficient DT40Cre1 cells used Rad18bsr and Rad18gpt targeting vectors. After transfection of the vector into DT40Cre1 cells and selection in the presence of the respective antibiotics, lysates from ≈500 cells were screened for targeted integration by a PCR approach (21), using primer 18Rad6 (5′-gaggccggctgtcactcgggccggtc-3′) in combination with the primers in the respective resistance cassette. For Southern blot analysis, 5–10 μg of cellular DNA was cleaved with NcoI, and the resultant membranes were probed with an external probe amplified with the primers 18Rad201 (5′-catgtcagtctggatctgctg-3′) and 18Rad202 (5′-aaagaacagaccttcacctgag-3′). The Rad18 coding region was amplified with the primers 18Rad9 (5′-ggagctagcgccaccaatggccctggcgctgc-3′) and 18Rad10 (5′-gggagatctgaagcacttagccttctgtaccac-3′) from DT40Cre1 cDNA using Pfu polymerase and cut with NheI and BglII for cloning into pExpress (21). The cDNA expression cassette was subsequently excised with SpeI and cloned into the NheI site of ploxgpt (21). For expression analysis of Rad18, amplification of the coding region with primers 19Rad9 and 18Rad10 and of hypoxanthine phosphoribosyltransferase (HPRT) with primers HPRTfor1 (5′-tattgttggaactggaaggacaatg-3′) and HPRTrev1 (5′-actcactgctgtatatattcatcag-3′) used Expand high-fidelity polymerase (Roche, Indianapolis, IN). For analysis of AID expression, 20 μg of cellular protein was used for Western blotting with the EK2–5G9 monoclonal anti-AID antibody as described (38), and loading was controlled with the C2 anti-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). For analysis of PCNA ubiquitination, cells were cultured in the presence of the indicated doses of methylmethanesulfonate (Sigma) for 5 h, and Western blot analysis was performed with the PC10 antibody (Abcam, Cambridge, MA).
Analysis of Ig Diversification Activity.
For analysis of Ig gene conversion, cells were subcloned by limiting dilution, cultured for 10 days, and stained with anti-chicken-IgM-FITC (Bethyl Laboratories, Montgomery, TX) before FACS analysis with a FACStar (Becton Dickinson, Franklin Lakes, NJ). For analysis of somatic hypermutation, cells were transfected with the plasmid pMS-YFP(X) cut with SgrA1 and Xmn1 to allow for random genomic integration of the reporter. After 13–17 days of culture (variations between different experiments, not within one experiment), cells were harvested, and 500,000 viable cells per clone were analyzed for YFP expression by FACS analysis. Control transfections with a functional GFP in the same vector context confirmed that Rad18 deficiency does not significantly affect GFP expression levels (data not shown). Data analysis was performed with CellQuest software, and P values were derived by Student’s t tests. For analysis of hypermutation on the endogenous gene locus, cells of the respective genotypes were subject to limiting dilution subcloning and cultured for 6 weeks. Genomic DNA from the subclones was used for amplification of the rearranged light chain locus with primers Vlam1 (5′-tgggaaatactggtgataggtggat-3′) and C2 (5′-cctccattttttgacagcacttacctggacagctg-3′), using Pfu polymerase. Products were subcloned into the Topo-TA vector, and sequencing was performed with the primer Vlam2 (5′-gagcgcagggagttatttgcatag-3′). Sequence analysis was performed with the Applied Biosystems (Foster City, CA) chromas lite program and ClustalW.
Acknowledgments
We thank André Kutzera, Sandra Klemmer, and Markus Schmidt for technical assistance; Hiroshi Arakawa and Jean-Marie Buerstedde for material and advice; Ralf Küppers and Jerzy Adamski for sequencing options; all members of the J.B., B.J., and Friederike Eckardt-Schupp laboratories at GSF for help and discussion; and Dirk Eick and Georg Bornkamm for generous support. This study was supported by Deutsche Krebshilfe Grant 10-1984-JuI and Deutsche Forschungsgemeinschaft Grant JU 2690/1-1.
Abbreviations
- PCNA
proliferating cell nuclear antigen
- AID
activation-induced cytidine deaminase
- YFP
yellow fluorescent protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa H., Buerstedde J. M. Dev. Dyn. 2004;229:458–464. doi: 10.1002/dvdy.10495. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa H., Saribasak H., Buerstedde J. M. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rada C., Williams G. T., Nilsen H., Barnes D. E., Lindahl T., Neuberger M. S. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 5.Di Noia J., Neuberger M. S. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 6.Goodman M. F., Tippin B. Nat. Rev. Mol. Cell Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- 7.Zeng X., Winter D. B., Kasmer C., Kraemer K. H., Lehmann A. R., Gearhart P. J. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 8.Simpson L. J., Sale J. E. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zan H., Komori A., Li Z., Cerutti A., Schaffer A., Flajnik M. F., Diaz M., Casali P. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich H. D. Chembiochemistry. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- 11.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 12.Stelter P., Ulrich H. D. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 13.Ulrich H. D., Jentsch S. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita Y. M., Okada T., Matsusaka T., Sonoda E., Zhao G. Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannouche P. L., Wing J., Lehmann A. R. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 17.Tissier A., Kannouche P., Reck M. P., Lehmann A. R., Fuchs R. P., Cordonnier A. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Kawamoto T., Araki K., Sonoda E., Yamashita Y. M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Takeda S. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Sonoda E., Okada T., Zhao G. Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N. S., van Gent D. C., Takata M., Takeda S. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada T., Sonoda E., Yamashita Y. M., Koyoshi S., Tateishi S., Yamaizumi M., Takata M., Ogawa O., Takeda S. J. Biol. Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 21.Arakawa H., Lodygin D., Buerstedde J. M. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson L. J., Sale J. E. DNA Repair (Amst) 2005;4:503–510. doi: 10.1016/j.dnarep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Arakawa H., Hauschild J., Buerstedde J. M. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 24.Sale J. E., Calandrini D. M., Takata M., Takeda S., Neuberger M. S. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 25.Bachl J., Olsson C. Eur. J. Immunol. 1999;29:1383–1389. doi: 10.1002/(SICI)1521-4141(199904)29:04<1383::AID-IMMU1383>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Polack A., Feederle R., Klobeck G., Hortnagel K. EMBO J. 1993;12:3913–3920. doi: 10.1002/j.1460-2075.1993.tb06069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada C., Di Noia J. M., Neuberger M. S. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Faili A., Aoufouchi S., Flatter E., Gueranger Q., Reynaud C. A., Weill J. C. Nature. 2002;419:944–947. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- 29.Papavasiliou F. N., Schatz D. G. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 30.Rada C., Ehrenstein M. R., Neuberger M. S., Milstein C. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 31.Bardwell P. D., Woo C. J., Wei K., Li Z., Martin A., Sack S. Z., Parris T., Edelmann W., Scharff M. D. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 32.Diaz M., Verkoczy L. K., Flajnik M. F., Klinman N. R. J. Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 33.Zan H., Shima N., Xu Z., Al-Qahtani A., Evinger A. J., Iii, Zhong Y., Schimenti J. C., Casali P. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda K., Ouchida R., Takeuchi A., Saito T., Koseki H., Kawamura K., Tagawa M., Tokuhisa T., Azuma T., O-Wang J. Proc. Natl. Acad. Sci. USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen J. G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulrich H. D. Eukaryotic Cell. 2002;1:1–10. doi: 10.1128/EC.1.1.1-10.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faili A., Aoufouchi S., Gueranger Q., Zober C., Leon A., Bertocci B., Weill J. C., Reynaud C. A. Nat. Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 38.Greiner A., Tobollik S., Buettner M., Jungnickel B., Herrmann K., Kremmer E., Niedobitek G. J. Pathol. 2005;205:541–547. doi: 10.1002/path.1746. [DOI] [PubMed] [Google Scholar]