Abstract
Epoxide hydrolases are essential for the processing of epoxide-containing compounds in detoxification or metabolism. The classic epoxide hydrolases have an α/β hydrolase fold and act via a two-step reaction mechanism including an enzyme–substrate intermediate. We report here the structure of the limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis, solved using single-wavelength anomalous dispersion from a selenomethionine-substituted protein and refined at 1.2 Å resolution. This enzyme represents a completely different structure and a novel one-step mechanism. The fold features a highly curved six-stranded mixed β-sheet, with four α-helices packed onto it to create a deep pocket. Although most residues lining this pocket are hydrophobic, a cluster of polar groups, including an Asp–Arg–Asp triad, interact at its deepest point. Site-directed mutagenesis supports the conclusion that this is the active site. Further, a 1.7 Å resolution structure shows the inhibitor valpromide bound at this position, with its polar atoms interacting directly with the residues of the triad. We suggest that several bacterial proteins of currently unknown function will share this structure and, in some cases, catalytic properties.
Keywords: crystal structure/enantioselectivity/epoxide hydrolase/mechanism/monoterpene degradation
Introduction
Epoxide hydrolases (EHs) catalyse the hydrolysis of epoxides to the corresponding diols. In nature, these enzymes have three main functional roles: detoxification, synthesis of signal molecules, and metabolism which allows some bacteria to use epoxides or their alkene and halohydrin precursors as carbon sources. EHs have also gained increasing attention due to their potential for kinetically resolving epoxide enantiomers and thus serving as selective catalysts in the industrial production of enantiomerically pure chemicals.
The most extensively investigated group of EHs can convert highly reactive, and therefore often harmful, epoxides into less toxic and more easily excreted diols; these enzymes usually act with rather broad substrate specificity (Armstrong, 1987). The structures of three related vicinal diol-producing EHs from bacterial, mammalian and fungal sources have been solved to date (Argiriadi et al., 1999; Nardini et al., 1999; Zou et al., 2000). All have a characteristic α/β hydrolase fold (Heikinheimo et al., 1999), combined with a helical lid domain. The catalytic triad of this family is situated on the α/β domain, and consists of an aspartic acid nucleophile and a histidine–acidic-residue pair that acts as a charge relay (Franken et al., 1991). Two tyrosines from the lid help to position the substrate and encourage ring opening through hydrogen bonds to the epoxide oxygen (Argiriadi et al., 2000; Rink et al., 2000; Yamada et al., 2000). The nucleophile attacks primarily at the less-substituted carbon of the oxirane ring to form an enzyme–substrate intermediate. The intermediate is then attacked by a water molecule activated by the charge relay pair.
The structure of human leukotriene A4 hydrolase, a bifunctional enzyme with epoxide hydrolase activity, has recently been solved (Thunnissen et al., 2001). This metalloenzyme catalyses the biosynthesis of leukotriene B4, a potent lipid chemoattractant involved in inflammatory and other responses. The catalytic unit has a fold similar to thermolysin, i.e. two domains with structures very different from those of the α/β hydrolase EHs. The enzyme has an atypical EH activity, in the sense that its product is not a vicinal diol; the hydroxyl group introduced is distant from the oxirane ring. The zinc atom in the active site is proposed to activate the epoxide to form a carbocation intermediate that is delocalized through a conjugated system, thus enabling an activated water molecule to attack at the opposite end of that system.
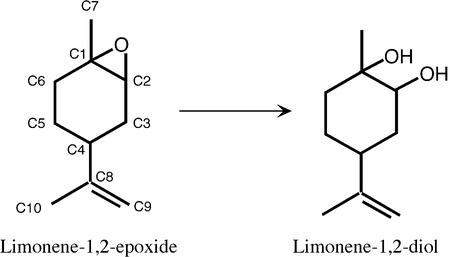
The subject of the present study, limonene-1,2-epoxide hydrolase (LEH) (EC 3.3.2.8), catalyses the reaction shown in Figure 1. This 16 kDa protein is part of a metabolic system that allows the bacterium Rhodococcus erythropolis to grow on limonene as a sole source of carbon and energy (van der Werf et al., 1999c). Since the enzyme is much smaller than, and lacks sequence similarity to, the other types of epoxide hydrolases, it was immediately suspected that it would have a different fold (Barbirato et al., 1998; van der Werf et al., 1998). Furthermore, LEH has been shown to use a different (acid-catalysed) mechanism with different stereochemical consequences (van der Werf et al., 1999a). No detectable enzyme–substrate intermediate was found in studies where enzyme was incubated with 14C-labelled substrate (M.Arand, unpublished results). In addition to its structural and mechanistic interest, LEH has many potential applications in industrial synthesis, particularly since its reaction is enantioconvergent. For example, the (1S, 2R, 4S) and (1R, 2S, 4S) isomers of limonene epoxide are both converted to the (1R, 2R, 4S) diol, while the (1S, 2R, 4R) and (1R, 2S, 4R) isomers are each converted to the (1S, 2S, 4R) diol (van der Werf et al., 1999b). These results indicate that a water molecule must be able to attack at either C1 or C2 of the epoxide ring (van der Werf et al., 1999a).
Fig. 1. The reaction of LEH with its natural substrate. Carbon atoms are numbered.
We report here the structure of LEH from R.erythropolis, the first representative of this third type of epoxide hydrolase, at 1.2 Å resolution. It has a similar topology to several other enzymes of diverse function but very different reaction mechanisms. As for all but one of these enzymes, the active site of LEH lies at the bottom of a deep pocket that is lined primarily with hydrophobic residues. The catalytic importance of several polar residues found at the bottom of the pocket was confirmed by site-directed mutational studies, as well as by a 1.75 Å crystal structure of a complex with the inhibitor valpromide. The combined results suggest a one-step push–pull reaction mechanism. Further, we were able to suggest a function for four putative EHs that show sequence similarity to LEH, including conserved catalytic residues. So far these related proteins have been found only in pathogens, and may represent part of their defence against foreign compounds.
Results
Overall structure
The structure of LEH was solved by single-wavelength anomalous dispersion (Hendrickson and Teeter, 1981) using a selenomethionine-substituted (Se-Met) protein expressed in Escherichia coli. Statistics describing the X-ray diffraction data and the final refined native LEH model are provided in Tables I and II. There is no electron density for the first four residues of either molecule in the asymmetric unit. Of the remainder, only residues 14–17 of one molecule (molecule A in our PDB deposition) have relatively poor electron density.
Table I. Data collection statistics.
| Native LEH | Se-Met LEH | Valpromide | |
|---|---|---|---|
| Unit cell dimensions (Å) | a = 45.6 | a = 45.4 | a = 45.6 |
| b = 47.6 | b = 47.6 | b = 47.7 | |
| c = 129.7 | c = 129.3 | c = 129.2 | |
| Wavelength (Å) | 0.931 | 0.934 | 0.944 |
| Resolution range (Å) | 64.6–1.2 | 38–1.8 | 30.0–1.75 |
| (1.20–1.21) | (1.8–1.9) | (1.75–1.80) | |
| No. of unique reflections | 88 916 | 26 770 | 29 009 |
| Multiplicity | 4.1 (2.3) | 7.0 (7.1) | 3.9 (3.6) |
| Completeness (%) | 99.7 (94.1) | 100 (100) | 99.3 (97.5) |
| Rsym (%)a | 6.3 (27.2) | 6.6 (32.0) | 5.6 (10.7) |
| Average I/σ(I)b | 28.2 (3.5) | 8.8 (2.3) | 20.7 (12.2) |
| Figure of meritc | – | 0.20 (0.09) | – |
| Cullis R-factor, anomalousc,d | – | 0.83 (0.98) | – |
| Figure of merite | – | 0.44 (0.15) | – |
All forms of LEH crystallized in space group P212121. Values in parentheses refer to data in the outer resolution shell.
aRsym = ∑∑I|II – <I>|/∑<I>, where <I> is the mean intensity of the N reflections with intensities II and common indices h,k,l.
bAs reported by TRUNCATE (French and Wilson, 1978).
cAs reported by MLPHARE (Otwinowski, 1991).
dCullis R-factor = (|FPHi(+) – FPHi(–)| – 2FHi × sin(PHIx)/|FPHi(+) – FPHi(–)|, where PHIx is the protein phase.
eOutput by RESOLVE (Terwilliger, 2000).
Table II. Statistics for the final refined model.
| Model | ‘apo’ | Valpromide |
|---|---|---|
| Resolution range (Å) | 50–1.2 | 29–1.75 |
| No. of reflections | 84 491 | 27 485 |
| No. of reflections used for Rfree calculation | 4306 | 1469 |
| R value, Rfree value (%) | 14.5, 17.2 | 15.8, 19.5 |
| No. of non-hydrogen atoms | 2712 | 2643 |
| No. of riding hydrogen atoms | 3073 | 2932 |
| No. of solvent waters | 389 | 330 |
| Mean B-factor, protein (Å2)a | 15.5 | 14.7 |
| Average B-factor, solvent atoms (Å2)a | 29.1 | 26.0 |
| Average B-factor, ligand atoms (Å2) | 24.3 | 25.7 |
| Ramachandran plot outliers (%)b | 0 | 0 |
| R.m.s.d. bond length from ideal (Å)c | 0.025 | 0.018 |
| R.m.s.d. bond angle from ideal (deg)c | 2.0 | 1.9 |
R.m.s.d., root mean square deviation.
aProtein atoms were modelled using anisotropic temperature factors, and solvents were modelled with isotropic temperature factors.
bUsing a stringent boundary Ramachandran plot (Kleywegt and Jones, 1996).
cIdeal values as defined by Engh and Huber, 1991.
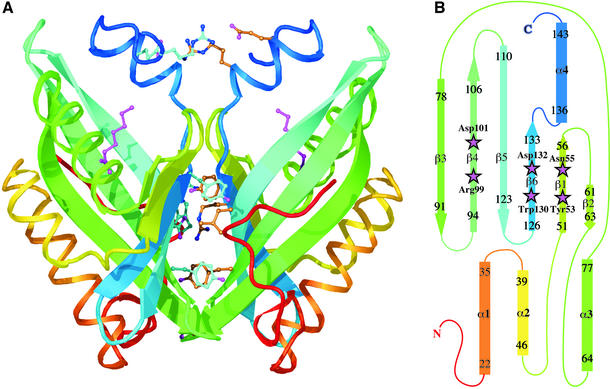
The main fold consists of a six-stranded mixed β-sheet, with three α-helices packed on to one side (Figure 2). Three of the strands (β3–β5) are long and highly curved. In β3, the curvature is achieved by bulges at residues 81 and 86, while in β4 and β5 it is the result of gradual changes in the (φ, ψ) angles of successive residues. The packing of the three N-terminal helices onto the concave face of the sheet creates a pocket that extends ∼15 Å into the protein core. A fourth helix lies beside the long strands of the sheet in such a way that it acts as a structural extension of the three shorter strands and a rim to the pocket. The residues lining this pocket are mainly hydrophobic, with the exception of a cluster of polar and charged residues at its deepest point (Figure 3).
Fig. 2. Structure of LEH. (A) Ribbon drawing showing the dimer, with each subunit coloured going through the rainbow from red at the N-terminus to blue at the C-terminus. Some of the residues contributing to the dimer interface, as described in the text, are shown as ball-and-stick representations. The endogenous ligand is also shown (magenta) in both subunits. (B) Topology diagram of the LEH subunit, using the same rainbow scheme. Residues included in each secondary structural element are numbered; helix α2 is irregular. Active-site residues are indicated by magenta stars.
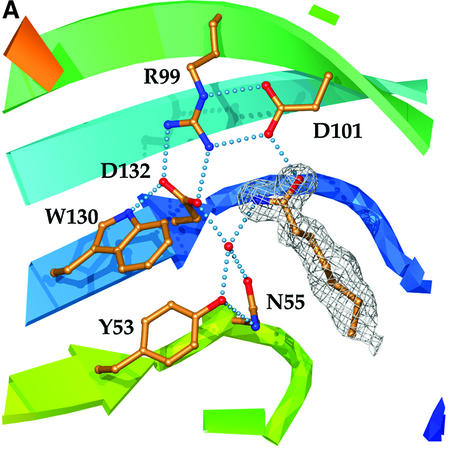
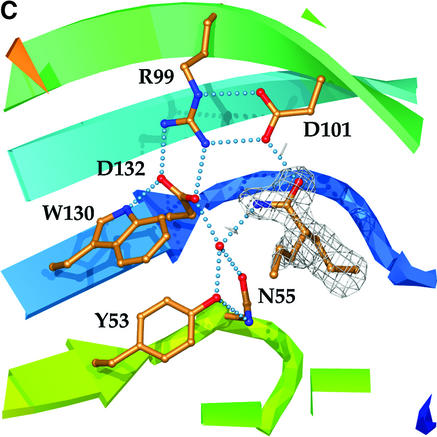
Fig. 3. The active site. (A) Catalytic residues, showing their relationship to each other and supporting side-chains, as well as to the water molecule and endogenous ligand found in the LEH active site. Hydrogen-bonding interactions are shown by dotted lines. Colouring of the ribbon portions follows the rainbow scheme defined in Figure 2. The electron density of the endogenous ligand (modelled as heptanamide) in the final 2Fo – Fco map is contoured at a level of 1σ. (B) Hydrogen-bonding interactions between active-site groups and the endogenous ligand. The figure shows Asp132 acting as the base and Asp101 as the acid. Where the donor–acceptor relationship is not clear from the available data, hydrogen bonds are indicated by double-headed arrows. (C) Complex with valpromide. The electron density of the final 2Fo – Fco map is contoured at a level of 1σ.
As predicted (Barbirato et al., 1998; van der Werf et al., 1998), the LEH structure does not resemble those of the epoxide hydrolases described previously. However, the folding topology is not new; a search of the Protein Data Bank (PDB) (Bernstein et al., 1977; Berman et al., 2000) identified four similar proteins (Table III). The most closely related structures are 3-oxo-Δ5-steroid isomerase (Kim et al., 1997; Cho et al., 1998) and a nuclear transport factor NTF2 (Bullock et al., 1996). Those of scytalone dehydratase (Lundqvist et al., 1994) and the β subunit of an indoaromatic-ring-hydroxylating naphthalene 1,2-dioxygenase (Kauppi et al., 1998) are more distantly related. Each protein exhibits the distinctive pocket formed when the helices pack onto the β-sheet. All show insignificant sequence conservation to LEH, and could only be identified with the structure in hand.
Table III. Structural alignments to LEH.
| PDB code | Name | No. of atoms matching | Aggregation state | RMS distance | % ID of matched | At Tyr53 | At Asn55 | At Arg99 | At Asp101 | At Asp132 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1OPY | 3-oxo-Δ5-steroid isomerase (Pseudomonas putida) | 107/123 | Dimer | 1.54 | 17.8 | Val38 | Asp40 | Phe86 | Val88 | Ala118 |
| 1ASK | NTF2 | 99/125 | Dimer | 1.35 | 8.2 | Leu39 | Trp41 | Gly87 | Leu89 | Phe119 |
| 4STD | Scytalone dehydratase | 88/164 | Trimer | 1.75 | 9.1 | Ile48 | Tyr50 | Leu106 | Val108 | Pro149 |
| 1NDO | Naphthalene 1,2-dioxygenase | 86/143 | α3β3 hexamer | 1.66 | 5.9 | Tyr564 | Val566 | Val641 | Leu643 | Val678 |
Although LEH was previously reported to be a monomer on the basis of gel filtration experiments (van der Werf et al., 1998), it appears as a dimer in the asymmetric unit of the crystal (Figure 2A). The subunits are related by 179°, corresponding to a nearly perfect 2-fold axis. Only the residues at the N-terminus (i.e. those preceding the main fold) deviate significantly from the dimer symmetry. After least-squares alignment, the Cα atoms of residues 20–149 match with an RMS distance of 0.2 Å; the RMS difference of their independently refined temperature factors is 1.1 Å2.
The β-sheets of the two subunits are packed perpendicular to each other, burying a total solvent accessible area of 3100 Å2. The surfaces have a highly complementary shape and a large number of side-chain–side-chain interactions are formed. The observed temperature factors for atoms in this interface are very similar to those found in the core of the protein, and significantly lower than those of surface residues. The Leu117 side-chains are completely buried at the centre, each making direct van der Waals contact with its equivalent, as well as with Tyr133 of the opposing subunit. A number of interactions result from the juxtaposition of the N-terminal ends of the α4s. The Asp135 side-chains hydrogen bond to the main-chain nitrogens of residues 136. Each Arg137 stacks onto Arg148 of the other subunit, and the arginines simultaneously make bidentate salt-link interactions with Glu140 and Glu141, respectively, across the interface. The two Tyr96 residues stack onto each other, while guanidino-aromatic stacking interactions are formed by Arg9 and Tyr62, as well as Arg131 and Trp10; Arg131 simultaneously forms a salt link with Glu98 across the dyad axis. Several water molecules are also found as part of the interface, some of which obey the 2-fold symmetry. No main-chain–main-chain hydrogen bonds are observed between the two subunits.
Active site
The residues responsible for LEH catalysis had not been identified in any previous study. Our inspection of the proteins with similar structure indicated that, in all but one case, the active site is found in the deep pocket between the β-sheet and the three N-terminal α-helices. The exception is the β subunit of naphthalene 1,2-dioxygenase, where the C-terminus of the polypeptide chain is inserted into the pocket and the active site is situated elsewhere (Carredano et al., 2000). These structurally related proteins have widely diverse functions, and the residues lining the pockets are not conserved; as a result they do not offer further guidance for the analysis of the LEH mechanism.
Although most of the side-chains lining the LEH pocket are hydrophobic, a cluster of polar residues is found at its deepest point where the side-chain of Arg99 interacts with the carboxylate groups of Asp101 and Asp132 (Figure 3). In the experimentally phased map for the Se-Met-substituted enzyme, electron density for an unexpected ligand was observed to stretch from these polar residues to the surface of the protein (Figure 3A). Clear density persisted during the high-resolution refinement of the native protein. The hydrogen-bonding interactions linking the various groups are summarized in Figure 3B. A short distance between the side-chain of Asp101 and the observed ligand (2.5 Å) indicates a strong hydrogen-bonding interaction. A second short hydrogen bond (2.6 Å) links the ligand and a well-defined water molecule, which in turn interacts with the side-chains of Tyr53, Asn55 and Asp132. All these side-chains are contributed by the β-sheet (Figure 2B). In the final stages of the refinement, the ligand was modelled as heptanamide, which is an excellent match for the shape of the electron density and the chemical character of this pocket. However, we have not yet been able to identify the actual compound experimentally. The native and Se-Met proteins were expressed in different hosts and purified using distinct procedures, yet both show clear density for this ligand. The precipitant in the crystallization represents the sole common experimental parameter, and so it seems likely that the ligand is an undocumented impurity of the polyethylene glycol (PEG) 6000 used. The electron density is not consistent with PEG itself. With an added concentration of 30% (w/v) PEG, the impurity need not be present in large quantities if its binding constant is sufficiently favourable. For example, if the Kd were 100 µM, a 0.03% impurity would be adequate for 90% occupancy of the site under our crystallization conditions. Inhibition studies with high concentrations of PEG 6000 were precluded for technical reasons, but PEG 1500 (which might be expected to have similar impurities) does indeed inhibit, with an effective Ki of 7% (w/v). We have not yet been able to obtain diffracting crystals without PEG.
Based on the above information, we tested the inhibition of LEH activity with four commercially available compounds expected to have chemical characteristics similar to the endogenous ligand. Hexanamide, hexylamine and valpromide (dipropylacetamide) were found to act as competitive inhibitors of LEH (Ki values of 2 mM, 35 µM and 100 µM, respectively). Hexanoic acid did not inhibit significantly at a concentration of 1 mM, which is probably not surprising given the expected repulsion between its negatively charged carboxylate group and those of the active site.
A complex of LEH with valpromide was obtained through crystal soaking experiments and refined at 1.75 Å resolution. This inhibitor bound in the same manner in the two subunits (Figure 3C), with clear electron density for its amide group appearing in an identical location to the putative amide group of the endogenous ligand (compare Figure 3A). A water molecule is again firmly positioned by hydrogen bonds to the side-chains of Tyr53, Asn55 and Asp132. In both active sites, the strongest electron density was for the propyl group that follows the path of the fatty acid tail of the endogenous ligand. The side-chain conformation of Leu74 was altered, apparently to accommodate the second propyl group of valpromide. The conformation of Leu103 was also different, and a similar conformational change in Leu147 is accompanied by a more general movement of the C-terminal residues 145–149 in both subunits. The side-chain conformation of Phe139 is changed slightly in both active sites. When combined, these changes illustrate how the hydrophobic lining of the active-site pocket of LEH can be adapted to fit differently shaped substrates.
Site-directed mutations
The structural and functional importance of the Asp– Arg–Asp triad and nearby residues was further evaluated using site-directed mutations. In total, 12 mutants were constructed, targeting the five polar residues that could reasonably have direct roles in catalysis, namely Tyr53, Asn55, Arg99, Asp101 and Asp132 (Table IV). Some of the mutant proteins formed inclusion bodies when expressed at 30°C, but growth of the bacteria at 26°C allowed the production of apparently soluble mutant protein in each case. However, folding was aberrant for some mutant proteins, as judged by their behaviour during ion exchange chromatography where they eluted at much higher salt concentrations than the wild-type LEH (Table IV). For some mutants, improper folding was further suggested by a slow-growth phenotype and somewhat reduced expression (Table IV). Mutations at Arg99 were particularly prone to these types of behaviour.
Table IV. Comparison of wild-type and mutant LEH enzymes.
| Residue | Replaced with | Growth | Expression level (% of total protein) | [NaCl] (mM) needed to elute from ion-exchange column | kcat (s–1) | Km (mM) | kcat/Km (s–1M–1) |
|---|---|---|---|---|---|---|---|
| WT | – | Normal | 20–40 | 210 | 0.470 | 1.4 | 340 |
| Tyr53 | Phe | Normal | 20–40 | 210 | 0.170 | 3.7 | 46 |
| Asn55 | Ala | Normal | 20–40 | 215 | 0.002 | 1.4 | 1.64 |
| Asn55 | Asp | Normal | 20–40 | 580 | ND | ND | ND |
| Arg99 | Ala | Slow | 3–10 | 560 | ND | ND | ND |
| Arg99 | Lys | Slow | 3–10 | 530 | ND | ND | ND |
| Arg99 | Gln | Slow | 3–10 | 500 | ND | ND | ND |
| Arg99 | His | Slow | 3–10 | 545 | ND | ND | ND |
| Asp101 | Ala | Normal | 20–40 | 170 | ND | ND | ND |
| Asp101 | Asn | Normal | 20–40 | 215 | ND | ND | ND |
| Asp132 | Ala | Normal | 20–40 | 455 | ND | ND | ND |
| Asp132 | Asn | Normal | 20–40 | 195 | ND | ND | ND |
| Asn55/Asp132 | Asp/Asn | Normal | 20–40 | 240/480a | ND | ND | ND |
Mutants that showed any measurable activity in an initial assay with 60 µM of racemic styrene-7,8-oxide were characterized further.
ND, not detectable. The threshold of detectable activity was ∼0.1% of that of the wild-type enzyme.
aTwo peaks were observed for this mutant.
The purified recombinant wild-type enzyme converts racemic styrene-7,8-oxide with a turnover number of 0.47 s–1. Although this substrate is somewhat poorer than the natural limonene-1,2-epoxide, which has a turnover number of 23.4 s–1 (van der Werf et al., 1998), use of styrene oxide allowed us to perform a partition assay that is faster, easier and much more sensitive than the traditional gas chromatographic measurement of limonene epoxide hydrolysis. The mutant activity results, summarized in Table IV, are in complete agreement with the proposal that the central pocket of LEH contains its active site. The members of the Asp101–Arg99–Asp132 triad are clearly most important. Both the aspartic acid residues are essential for enzyme activity. Although all mutations of Arg99 were also catalytically inactive, the structural roles of this residue mean that its contribution to catalysis cannot be viewed in isolation. However, its intimate associations with Asp101 and Asp132 (four hydrogen bonds) strongly imply that it will act together with them. The residues that position the observed water molecule are also important, although less central to activity. Replace ment of Tyr53 with phenylalanine resulted in a partially active mutant. Mutation of Asn55 to the isosteric aspartic acid generated an inactive enzyme, probably due in part to a local negative charge overload; the biochemical data suggest that some of this mutant protein may be misfolded (Table IV). A double mutation Asn55Asp/Asp132Asn that restores the local charge rescues folding, but not activity, and again illustrates the importance of Asp132 in catalysis. Mutation of Asn55 to the smaller side-chain of alanine left a small residual activity, possibly because of non-optimal positioning of the catalytic water. A similar explanation may lie behind the partial activity of the Tyr53Phe mutant.
In summary, the mutagenesis data, like the structural observations, are consistent with the proposal that the water molecule associated with Asp132, Asn55 and Tyr53 is the hydrolytic water and Asp101 is the acid catalyst that protonates the epoxide oxygen.
Comparison with other proteins of unknown structure
Searches of the sequence databases identified several hypothetical proteins with sequence similarity to LEH (Figure 4). The sequences fall into two groups.
Fig. 4. Sequence alignments of LEH with proteins of unknown function. Sequence alignment was performed using hidden Markov models (Karplus et al., 1998), with some small manual adjustments using the LEH structure as a guide. Every tenth residue in the LEH sequence is marked. Residues preceding α1 are poorly conserved or missing in the other proteins, and so the alignments shown omit those segments. The roles that conserved residues play in LEH are indicated using the following code: a, active site; c, core or other structure; d, dimer; s, surface. Members of the first group of proteins, which appear to have both structural and functional relationships to LEH, are gi|2145793| (hypothetical protein B2235_F3_140 from Mycobacterium leprae), gi|2624262| (hypothetical protein Rv2740 from Mycobacterium tuberculosis), gi:13424591| (hypothetical protein from Caulobacter crescentus) and gi:17547308 (conserved hypothetical protein from the plant pathogen Ralstonia solanacearum). Members of the second group, which are presumed to have a similar structure, but distinct function, are gi|11345638| (hypothetical protein VC1118 from Vibrio cholerae) and gi|11350018| (hypothetical protein PA3856 from Pseudomonas aeruginosa). A few regions may contain small shift errors, which will require new sequence/structural data to locate and correct.
In the first are four proteins with 25–30% sequence identity to LEH. These are probably related in function, as both the LEH structure and catalytic apparatus appear to be intact. Insertions and deletions fall neatly between elements of secondary structure, with the exception of deletions that are likely to fall in or just before α3. As this helix provides one wall of the active-site pocket, structural changes here would provide a simple means of altering substrate specificity (as was illustrated above by the changes in LEH structure on binding different ligands). The most highly conserved segment is near the end of β6, where the side-chains alternate between roles in the active site/protein core and in the dimer interface (Figure 2A). Similar conservation patterns appear in strands β1 and β4 of LEH, and also include active-site groups. The residues implicated in catalysis (Tyr53, Asn55, Arg99, Asp101, Trp130 and Asp132) are very highly conserved. The only exceptions are the Asp132Asn change in the Mycobacterium leprae sequence and the Tyr53Trp change in the Ralstonia solanacearum case. More subtle details involving the active site are also conserved. For example, the Arg99–Asp132 salt link lies over strand β5 between Leu116 and Gly118. The distance between Asp132 OD1 and the Cα atom of Gly118 is only 3.3 Å, and so all other residue types would be unsuitable at this position. Most of the remaining conserved residues are found in the core of the enzyme or at the dimer interface. There are changes in several hydrophobic residues that line the walls of the active-site cavity. Introduction of other non-polar or even polar side-chains at these positions presumably reflects differing characteristics of the natural substrates. Another interesting replacement is that of Leu117 in the dimer contact surface of LEH (Figure 2A) with cysteine in the two mycobacterial sequences, since this could indicate an inter-subunit disulfide bond in these instances.
In the second group, with 20–25% amino acid sequence identity, the fold is apparently the same, but some important aspects of the active site have been changed. Only the residue equivalent to Asp132 is conserved, but substitutions of other residues suggest potential new constellations of polar side-chains. However, the low level of sequence identity in some regions (and possibly locally incorrect alignments) makes it impossible to predict exactly how these active sites will look. As with the first group of LEH homologues, the dimer interactions appear to be preserved.
Discussion
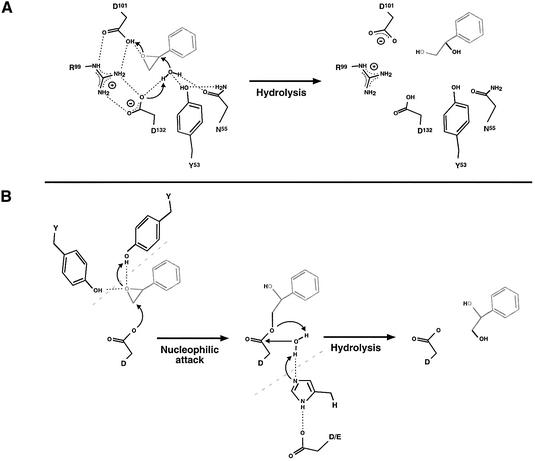
H218O-labelling studies suggested that LEH would behave as an acid catalyst, i.e. that it would act directly without an enzyme–substrate intermediate (van der Werf et al., 1999a). Furthermore, we have looked for formation of an intermediate by precipitation of the enzyme after incubation with 14C-labelled substrate but could not detect any incorporation of the label into the protein. Therefore we propose a single-step mechanism. Such a mechanism generally includes two components: (i) activation of water (by removing a proton) for its nucleophilic attack on the epoxide, and (ii) a means of encouraging ring opening (generally by donation of a proton to the ring oxygen). Water must always attack the limonene ring at the face opposite that with the oxirane ring in order to explain the reported enantiomeric results with limonene-1,2-epoxide (van der Werf et al., 1999a,b); thus water attack and ring opening must be coupled and simultaneous. Our mutagenesis results clearly indicate central roles for Arg99, Asp101 and Asp132, and important contributions from Tyr53 and Asn55. Combined with our structural results, they suggest the reaction mechanism outlined in Figure 5A. In this scheme, the crystallographically observed water molecule is activated for its attack on the epoxide ring by Asp132. At the same time, the epoxide is encouraged to open by the donation of a proton from Asp101. The arginine side-chain plays a central role, interacting with and positioning the carboxylate groups of both catalytic aspartates. Each carboxylate group will maintain a pair of hydrogen bonds with the guanidino group during the reaction, but the salt linkage (i.e. the charge) will flip from one side of the guanidino group to the other. Tyr53 and Asn55 play a subsidiary role, but are needed for correct positioning of the catalytic water. Modelling studies show that the exact docking of a particular substrate with respect to the water molecule would be sufficient to determine whether it is attacked at C1 or C2. An explanation for the observed enantioconvergence is thus encompassed by this mechanism.
Fig. 5. Mechanisms of epoxide hydrolases. (A) The mechanism of LEH indicated by the experimental results, using styrene oxide as an example. The catalytic water molecule is held in place and activated by hydrogen bonding to residues Asp132, Asn55 and Tyr53. The activated water molecule forces epoxide ring opening by nucleophilic attack at one of the ring carbons. At the same time, Asp101 activates the epoxide ring by donation of a proton to the epoxide oxygen (acid catalysis). Thus the formation of the diol from the epoxide proceeds in a single step by a push–pull mechanism. After this step, Asp132 should be in the protonated state and Asp101 should be charged, which can be rapidly reversed with the aid of Arg99 as a proton shuttle. The hydrogen bond donor/acceptor atoms for Tyr53 and Asp55 cannot be proved using current information, and only one of the two possibilities is drawn. (B) Reaction mechanism of α/β fold EHs. In these enzymes, two tyrosines position the epoxide oxygen by hydrogen bonding and activate the epoxide for nucleophilic attack by an aspartic acid residue. This first step leads to the formation of an enzyme–substrate ester intermediate. Subsequent hydrolysis of the intermediate is achieved by a water molecule activated by a His-Asp/Glu charge relay system. The hydrolysis leads to product formation and reconstitution of the active enzyme. The tyrosines and charge relay system are only shown in the present scheme where they contribute to the mechanism.
There are some general similarities of the LEH mechanism to that of ketosteroid isomerase, an enzyme that is related in structure but carries out a very different chemical reaction. Asp40 (the equivalent of LEH’s Asn55) is thought to act as a general base, abstracting a proton from the steroid substrate. Asp103 of the isomerase is structurally equivalent to Gly118 of LEH, but its side-chain carboxylate group is placed in a similar position to Asp132; this residue (pKa ≈ 9.5) is thought to stabilize the dienolate intermediate of the isomerase. No other catalytic residues of the structurally related proteins have any apparent analogy with those of LEH.
Some features are common to all three epoxide hydrolysis mechanisms proposed to date. In each case, the epoxide ring is activated by interactions of an acidic group with its ring oxygen (through either protonation or metal binding), while the attack of a base at one of its carbon atoms forces ring opening. In the two-step mechanism of the α/β hydrolase family of enzymes (Figure 5B), a protein group acts as the attacking base, thus creating an enzyme–substrate intermediate. This mechanism combines both broad specificity and a low Km, which are useful characteristics for systems intended to process a large number of structurally diverse compounds, each present at low concentration. LEH’s restrictive pocket provides an excellent basis for tight substrate specificity and enantioselectivity. Its one-step mechanism is simpler than that of the α/β hydrolase type of EH and the reaction is faster than observed for most enzymes of that class. In the biological setting, this combination is useful for the rapid processing of a limited number of substrates for use as sources of carbon.
Epoxide hydrolases have already attracted wide interest in industrial applications (Archelas and Furstoss, 2001), and a new class of enzyme will provide additional tools for the bioorganic chemist. LEH is particularly interesting in this context because of its enantioconvergent reaction, its small stable scaffold and the fact that expression in E.coli is already vastly more efficient than that of the previously applied epoxide hydrolases. Under medium-density fermentation conditions (OD600 15–20), LEH expression of 3 g/l cell culture is routinely obtained. LEH’s actual specificity is well suited to biotechnological applications as it is one of the few enzymes that can hydrolyse trisubstituted epoxides. Although the applications for limonene-1,2-epoxide itself are also useful, it would be advantageous to be able to design LEH-like enzymes that can act on other substrates as well. The alignment of the LEH sequence with those of several other related proteins offers further ideas for substitutions that should give rise to stable active proteins with differing specificities.
LEH provides some clues to the function of several enzymes present in some pathogen genomes, but apparently not in mammalian ones. These enzymes may prove to be useful targets for drug design, since a genomic and metabolic analysis suggests that they will play a role in the defence of the microorganisms against toxic epoxides. Another tempting possibility is suggested by the structural similarity of LEH to the 3-oxo-Δ5-steroid isomerase. Previous data had hinted at a relationship between LEH and the mammalian cholesterol 5,6-oxide hydrolase, an enzyme of as yet unknown sequence and structure. Like LEH, the cholesterol oxide hydrolase is small (Watabe et al., 1986) and uses an acid-catalysed mechanism that lacks a covalent intermediate (Müller et al., 1997). Further, both enzymes possess the unusual capability of hydrolyzing trisubstituted epoxides and both show enantioconvergence with their diastereomeric epoxide substrates. That the active site of LEH could easily be redesigned to accommodate a substrate of this nature is shown by its relationship to the steroid isomerase. We have tested LEH itself for cholesterol epoxide hydrolase activity with a method sensitive down to the femtomole/milligram/minute range, and have not found any evidence that such activity exists (M.Arand, unpublished results). Nevertheless, the LEH results have provided us with new ideas for the cloning and exploration of a key mammalian enzyme that has so far resisted numerous attempts at purification and characterization.
Materials and methods
Cloning, expression and purification
Rhodococcus erythropolis strain DCL14 was originally isolated from a fresh-water sediment sample (van der Werf et al., 1999c). LEH was purified from its native host as described previously (van der Werf et al., 1998).
A recombinant expression system for producing Se-Met substituted (and, later, mutant) protein was set up in E.coli. The limA gene was amplified from the plasmid pLEH2 (Barbirato et al., 1998) using Pfu DNA polymerase (Stratagene). The introduction of NcoI restriction sites changed the second codon of the LEH gene from ACA to GCA, leading to the replacement of Thr2 with alanine in the recombinant enzyme. The amplified fragment was digested with NcoI and cloned into the expression vector pGEF II (Arand et al., 1999). The resulting plasmid, pGEF-LEH, was transformed into the methionine-auxotroph E.coli strain B834(DE3). To produce native protein, cells grown to an initial OD600 of 0.6–1.0 were induced for 2 h with 0.4 mM ispropyl-β-d-thiogalactopyranoside (IPTG) [Luria broth (LB) medium, 100 mg/l ampicillin, 30°C]. LEH was expressed as 30–40% of the total cell protein. For production of Se-Met LEH, a modified M9 minimal medium was used instead, supplemented with amino acids (5 mg/l each of tryptophan and tyrosine, 50 mg/l of Se-Met and 50 mg/l of each of the other 17 amino acids), 4 g/l glucose, 1% (v/v) LB and 20 mg/l ampicillin, in a volume of 250 ml. Cells were harvested by centrifugation, resuspended in 30 ml of 100 mM sodium phosphate buffer pH 7.4 and 1 mM dithiothreitol (DTT), and then broken by a single pass through a French press at 30 000 psi. After centrifugation at 10 000 g for 20 min, the supernatant was diluted with four volumes of 1 mM DTT, passed through a 0.2 µm filter and then applied to a UNO Q12 column (Bio-Rad). The column was developed with a 100–240 mM sodium chloride gradient in 20 mM sodium phosphate pH 7.4 and 1 mM DTT. LEH eluted at a salt concentration of 180 mM, essentially free from protein contaminants. Fractions containing LEH activity were pooled, concentrated to 500 µl with a Centricon YM 10 filter (Amicon) and applied to a BioPrep S1000/17 size exclusion column (Bio-Rad) previously equilibrated with 10 mM HEPES pH 7.0 and 1 mM DTT. LEH eluted as a single homogenous peak. Fractions were pooled, concentrated as described above and stored at 4°C after addition of sodium azide to a final concentration of 0.02%.
Site-directed mutagenesis and mutant protein production
Mutations were introduced into pGEF–LEH using the Quikchange™ Site Directed Mutagenesis Kit (Stratagene). Mutants were prescreened using diagnostic restriction sites introduced or deleted together with the desired mutation and verified by DNA sequencing of the expression construct. Mutant LEH was expressed and purified as described for the native protein above, with two differences: Escherichia coli strain BL21(DE3) was used as the host, and growth temperature was reduced to 26°C to minimize the production of inclusion bodies. The protein concentration in the final purified enzyme solutions was measured according to the Bio-Rad Bradford assay.
Enzyme assays
The activity of wild-type and mutant LEH was assayed in the presence of 14C-labelled styrene-7,8-oxide in 100 mM sodium phosphate buffer pH 7.4 in a total volume of 100 µl. After incubation for 5–10 min, the mixture was extracted with an equal volume of chloroform to remove the remaining substrate. Product formation was assessed by measuring the radioactivity in the aqueous phase using liquid scintillation counting. Under these conditions, virtually all styrene oxide was extracted into the organic phase while ∼65% of the reaction product, styrene glycol, remained in the aqueous phase. For the determination of the Ki values, inhibitors were added at varying concentrations (generally from ethanol stock solutions, with a constant 1% ethanol concentration in the final assay mixture) and their influence on Km and Vmax was assessed.
Crystallization
Purified protein was crystallized by the vapour diffusion method (McPherson, 1982). The temperature of crystallization was chosen based on the dynamic light-scattering profiles obtained with a DynaPro-801TC (Protein Solutions Inc., Charlottesville, VA). The native wild-type protein behaved as a monodisperse dimer at 6°C and became increasingly polydisperse as the temperature was raised to 37°C. Therefore sitting drops containing 3 µl protein (12.8 mg/ml in 10 mM HEPES buffer pH 7.0) and 3 µl of the reservoir solution (30% PEG 6000, 1 M LiCl and 0.1 M MES buffer pH 6.0) were equilibrated against the reservoir (1 ml) at 4°C. Small octahedral prisms (0.1 mm in the longest dimension) grew in 5–7 days and were stable for more than a year in the drops. The crystallization conditions for the recombinant Se-Met substituted protein were identical to those for the wild-type protein except that the protein concentration was increased to 25 mg/ml and 1 mM DTT was added to the buffer (10 mM HEPES pH 7.0).
Data collection, structure solution and refinement
Prior to flash-freezing in liquid nitrogen, crystals of native LEH were transferred to a cryosolution of 37% PEG 6000, 1 M LiCl and 0.1 M MES buffer pH 6.0 for 10 s. A complete dataset was collected on one frozen crystal at the ESRF ID14:4 beamline. The HKL suite of programs (Otwinowski and Minor, 1997) was used to integrate and scale the images. Statistics describing the X-ray data are presented in Table I. Crystals of Se-Met-substituted LEH were flash frozen as for the native crystals and data were collected at a wavelength of 0.931 Å at beamline ID14:1 at ESRF. Although this wavelength is far from the selenium absorption edge, the anomalous signal was strong enough for successful structure determination. Data were integrated with MOSFLM (Steller et al., 1998) and scaled with SCALA, both from the CCP4 suite of programs (Collaborative Computing Project, 1994). Statistics relating to the quality of the X-ray data are given in Table I. As these data were not isomorphous to the native data set, the single-wavelength anomalous dispersion method (Hendrickson and Teeter, 1981) was used to determine the structure. The positions of eight selenium atoms out of 10 possible Se-Met residues (not including that at the N-terminus) were determined using direct methods as implemented in Shake-and-Bake 2.1 (Weeks and Miller, 1999). Phase estimates were calculated using MLPHARE (Otwinowski, 1991) and then subjected to maximum-likelihood density modification as implemented in RESOLVE (Terwilliger, 2000). The ARP/wARP suite (Perrakis et al., 1997) was used for automatic main-chain tracing. The amino acid sequence was then placed interactively using the program O (Jones et al., 1991). The quality of the experimental map was sufficient to locate residues 23–145 in this initial map with complete confidence. Residual electron density for a small molecule of ∼9 atoms, probably introduced with the crystallization reagents, was also present in the active site pocket. The remaining amino acid residues were added in the course of refinement using the high-resolution native dataset in CNS (Brünger et al., 1998) and REFMAC5 (Murshudov et al., 1997). The chemical nature of the endogenous ligand was deduced based on the electron density and hydrogen-bonding interactions. In the final models, riding hydrogen atoms were added and an anisotropic temperature factor model was used for non-hydrogen protein atoms. Non-crystallographic restraints were not used during the refinement. Final model statistics are presented in Table II. There are no outliers in a stringent-boundary Ramachandran plot (Kleywegt and Jones, 1996).
A complex with valpromide was obtained by soaking a crystal of the Se-Met enzyme with 10 mM inhibitor in MES-containing mother liquor for 1 h prior to freezing. Data were collected at I711, Max-Lab, Lund; data collection and refinement statistics are reported in Tables I and II. One molecule of MES was included in the final model.
The coordinates and structure factor data have been deposited at the Protein Data Bank with identity codes 1NWW and 1NU3 for the apo protein and valpromide complex, respectively.
Sequence and structural analysis
The buried surface area was calculated (Lee and Richards, 1971) using a 1.4 Å probe. Sequences similar to LEH were located using PSI-BLAST (Altschul et al., 1997) and hidden Markov models (Karplus et al., 1998); the alignments obtained from the latter program were adjusted manually to be consistent with the structural data. Structures similar to LEH were found with the programs TOP (Lu, 2000) and DALI (Holm and Sander, 1993). Representative structures were then obtained from the PDB and analysed in detail with LSQMAN (Kleywegt and Jones, 1997) and O (Jones et al., 1991), using a 3.5 Å cut-off in the structural alignment. Homology models were built using SOD (Kleywegt et al., 2001) and O. Figures were prepared using O, Molray (Harris and Jones, 2001), POV-Ray version 3.1 (http://www.povray.org) and Canvas 6 (Deneba Systems Inc.).
Acknowledgments
Acknowledgements
The authors would like to thank Heike Dürk for expert assistance with mutant expression and enzyme assays, and Mats Sandgren, Evalena Andersson, Anna Larsson and Sean McSweeney for help with data collection. This project has been supported by EC contracts BIO4-CT95–0005 (T.A.J., M.A. and J.A.M.dB.) and BIO4-CT95–0049 (M.J.vdW.), the Deutsche Forschungsgemeinschaft (M.A.) and the Swedish Natural Science Research Council (T.A.J.).
References
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arand M., Hemmer,H., Dürk,H., Baratti,J., Archelas,A., Furstoss,R. and Oesch,F. (1999) Cloning and molecular characterization of a soluble epoxide hydrolase from Aspergillus niger that is related to mammalian microsomal epoxide hydrolase. Biochem. J., 344, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archelas A. and Furstoss,R. (2001) Synthetic applications of epoxide hydrolases. Curr. Opin. Chem. Biol., 5, 112–119. [DOI] [PubMed] [Google Scholar]
- Argiriadi M.A., Morisseau,C., Hammock,B.D. and Christianson,D.W. (1999) Detoxification of environmental mutagens and carcinogens: structure, mechanism and evolution of liver epoxide hydrolase. Proc. Natl Acad. Sci. USA, 96, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiriadi M.A., Morisseau,C., Goodrow,M.H., Dowdy,D.L., Hammock,B.D. and Christianson,D.W. (2000) Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation. J. Biol. Chem., 275, 15265–15270. [DOI] [PubMed] [Google Scholar]
- Armstrong R.N. (1987) Enzyme-catalyzed detoxication reactions: mechanisms and stereochemistry. CRC Crit. Rev. Biochem., 22, 39–88. [DOI] [PubMed] [Google Scholar]
- Barbirato F., Verdoes,J.C., de Bont,J.A. and van der Werf,M.J. (1998) The Rhodococcus erythropolis DCL14 limonene-1,2-epoxide hydrolase gene encodes an enzyme belonging to a novel class of epoxide hydrolases. FEBS Lett., 438, 293–296. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyer,E.T.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bullock T.L., Clarkson,W.D., Kent,H.M. and Stewart,M. (1996) The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2). J. Mol. Biol., 260, 422–431. [DOI] [PubMed] [Google Scholar]
- Carredano E. et al. (2000) Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol., 296, 701–712. [DOI] [PubMed] [Google Scholar]
- Cho H.S., Choi,G., Choi,K.Y. and Oh,B.H. (1998) Crystal structure and enzyme mechanism of delta 5-3-ketosteroid isomerase from Pseudomonas testosteroni. Biochemistry, 37, 8325–8330. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project, Number 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Franken S.M., Rozeboom,H.J., Kalk,K.H. and Dijkstra,B.W. (1991) Crystal structure of haloalkane dehalogenase: an enzyme to detoxify halogenated alkanes. EMBO J., 10, 1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French G.S. and Wilson,K.S. (1978) On the treatment of negative intensity observations. Acta Crystallogr. A, 34, 517–525. [Google Scholar]
- Harris M. and Jones,T.A. (2001) Molray—a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D, 57, 1201–1203. [DOI] [PubMed] [Google Scholar]
- Heikinheimo P., Goldman,A., Jeffries,C. and Ollis,D.L. (1999) Of barn owls and bankers: a lush variety of alpha/beta hydrolases. Structure, 7, 141–146. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A. and Teeter,M.M. (1981) Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulphur. Nature, 290, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Karplus K., Barrett,C. and Hughey,R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- Kauppi B., Lee,K., Carredano,E., Parales,R.E., Gibson,D.T., Eklund,H. and Ramaswamy,S. (1998) Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure, 6, 571–586. [DOI] [PubMed] [Google Scholar]
- Kim S.W. et al. (1997) High-resolution crystal structures of delta 5-3-ketosteroid isomerase with and without a reaction intermediate analogue. Biochemistry, 36, 14030–14036. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1996) Phi/Psi-cology: Ramachandran revisited. Structure, 4, 1395–1400. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1997) Detecting folding motifs and similarities in protein structures. Methods Enzymol., 277, 525–545. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J., Zou,J.Y., Kjeldgaard,M. and Jones,T.A. (2001) Around O. In Rossmann,M.G. and Arnold,E. (eds), International Tables for Crystallography, Vol. F. Kluwer Academic, Dordrecht, The Netherlands, pp. 353–356.
- Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. [DOI] [PubMed] [Google Scholar]
- Lu G. (2000) TOP: a new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr., 33, 176–183. [Google Scholar]
- Lundqvist T., Rice,J., Hodge,C.N., Basarab,G.S., Pierce,J. and Lindqvist,Y. (1994) Crystal structure of scytalone dehydratase—a disease determinant of the rice pathogen, Magnaporthe grisea. Structure, 2, 937–944. [DOI] [PubMed] [Google Scholar]
- McPherson A.J. (1982) Preparation and Analysis of Protein Crystals. John Wiley, New York, NY.
- Müller F. et al. (1997) Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase and their substrates. Eur. J. Biochem., 245, 490–496. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nardini M., Ridder,I.S., Rozeboom,H.J., Kalk,K.H., Rink,R., Janssen,D.B. and Dijkstra,B.W. (1999) The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J. Biol. Chem., 274, 14579–14586. [PubMed] [Google Scholar]
- Otwinowski Z. (1991) Maximum likelihood refinement of heavy atom parameters. In Evans,P. and Leslie,A. (eds), Isomorphous Replace ment and Anomalous Scattering. SERC Daresbury Laboratory, Daresbury, UK, pp. 80–85.
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Sixma,T.K., Wilson,K.S. and Lamzin,V.S. (1997) wARP: improvement and extension of crystallogaphic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D, 53, 448–455. [DOI] [PubMed] [Google Scholar]
- Rink R., Kingma,J., Lutje Spelberg,J.H. and Janssen,D.B. (2000) Tyrosine residues serve as proton donor in the catalytic mechanism of epoxide hydrolase from Agrobacterium radiobacter. Biochemistry, 39, 5600–5613. [DOI] [PubMed] [Google Scholar]
- Steller I., Bolotovsky,R. and Rossmann,M.G. (1998) An algorithm for automatic indexing of oscillation images using Fourier analysis. J. Appl. Crystallogr., 30, 1036–1040. [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunnissen M.M., Nordlund,P. and Haeggstrom,J.Z. (2001) Crystal structure of human leukotriene A4 hydrolase, a bifunctional enzyme in inflammation. Nat. Struct. Biol., 8, 131–135. [DOI] [PubMed] [Google Scholar]
- van der Werf M.J., Overkamp,K.M. and de Bont,J.A.M. (1998) Limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis DCL14 belongs to a novel class of epoxide hydrolases. J. Bacteriol., 180, 5052–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf M.J., de Bont,J.A. and Swarts,H.J. (1999a) Acid-catalyzed enzymatic hydrolysis of 1-methylcyclohexene oxide. Tetrahedron: Asymmetry, 10, 4225–4230. [Google Scholar]
- van der Werf M.J., Orru,R.V.A., Overkamp,K.M., Swarts,H.J., Osprian,I., Steinreiber,A., de Bont,J.A.M. and Faber,K. (1999b) Substrate specificity and stereospecificity of limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis DCL14; an enzyme showing sequential and enantioconvergent substrate conversion. Appl. Microbiol. Biotechnol., 52, 380–385. [Google Scholar]
- van der Werf M.J., Swarts,H.J. and de Bont,J.A. (1999c) Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol., 65, 2092–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T., Ozawa,N., Ishii,H., Chiba,K. and Hiratsuka,A. (1986) Hepatic microsomal cholesterol epoxide hydrolase: selective inhibition by detergents and separation from xenobiotic epoxide hydrolase. Biochem. Biophys. Res. Commun., 140, 632–637. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB, v2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Yamada T., Morisseau,C., Maxwell,J.E., Argiriadi,M.A., Christianson,D.W. and Hammock,B.D. (2000) Biochemical evidence for the involvement of tyrosine in epoxide activation during the catalytic cycle of epoxide hydrolase. J. Biol. Chem., 275, 23082–23088. [DOI] [PubMed] [Google Scholar]
- Zou J., Hallberg,B.M., Bergfors,T., Oesch,F., Arand,M., Mowbray,S.L. and Jones,T.A. (2000) Structure of Aspergillus niger epoxide hydrolase at 1.8 Å resolution: implications for the structure and function of the mammalian microsomal class of epoxide hydrolases. Struct. Fold Des., 8, 111–122. [DOI] [PubMed] [Google Scholar]