Abstract
Temporal restriction of feeding can phase-shift behavioral and physiological circadian rhythms in mammals. These changes in biological rhythms are postulated to be brought about by a food-entrainable oscillator (FEO) that is independent of the suprachiasmatic nucleus. However, the neural substrates of FEO have remained elusive. Here, we carried out an unbiased search for mouse brain region(s) that exhibit a rhythmic expression of the Period genes in a feeding-entrainable manner. We found that the compact part of the dorsomedial hypothalamic nucleus (DMH) demonstrates a robust oscillation of mPer expression only under restricted feeding. The oscillation persisted for at least 2 days even when mice were given no food during the expected feeding period after the establishment of food-entrained behavioral rhythms. Moreover, refeeding after fasting rapidly induced a transient mPer expression in the same area of DMH. Taken in conjunction with recent findings (i) that behavioral expression of food-entrainable circadian rhythms is blocked by cell-specific lesions of DMH in rats and (ii) that DMH neurons directly project to orexin neurons in the lateral hypothalamus, which are essential for proper expression of food-entrained behavioral rhythms, the present study suggests that DMH plays a key role as a central FEO in the feeding-mediated regulation of circadian behaviors.
Keywords: circadian rhythm, feeding, mouse, obesity, period
For animals to survive effectively, the timing of feeding-related behaviors should be appropriately coordinated both with environmental conditions, such as food availability and risk of predation, and internal physiological states, such as gastrointestinal function and energy balance. The circadian oscillator in the suprachiasmatic nucleus (SCN), which is regarded as the master clock in mammals, orchestrates multiple circadian rhythms in the organism and is regulated according to environmental light/dark cues conveyed from the eye (light-entrainable oscillator) (1, 2). However, when food availability is restricted to a single period scheduled at a fixed time of the day [restricted feeding (RF)] animals adapt to this condition within a few days by feeding during the period of food availability and increasing food-seeking activity in the preceding hours [food-anticipatory activity (FAA)] (3, 4). Such anticipatory behaviors are often accompanied by increases in body temperature, adrenal corticosterone secretion, and gastrointestinal motility. These RF-induced biological rhythms persist even when SCN function is physically or genetically ablated, indicating the presence of a food-entrainable oscillator (FEO), that is separate from and independent of SCN.
Expression of certain molecular components of circadian clocks, such as Per1 and Per2, has been found oscillating (usually entrained by the SCN master clock) in many peripheral tissues and brain regions outside of the SCN (1, 2, 5–8). Previous studies demonstrated that RF entrains and shifts the circadian oscillators in the peripheral tissues and certain non-SCN regions of the brain, whereas rhythmicity in the SCN remains phase-locked to the light/dark cycle (9–12). These observations suggest a phase uncoupling of multiple circadian oscillators outside of the SCN from the master clock under RF.
We and others previously reported that neurons producing orexins, neuropeptides critical for promoting wakefulness, convey an efferent signal from the putative FEO in the brain to increase wakefulness and locomotor activity rather than act as the FEO itself (13, 14). Despite the accepted existence of the FEO and efforts to find its anatomical substrate, a long list of lesions to candidate neural and neuroendocrine tissues have failed to identify the locus of the FEO (3, 4). A caveat for lesion-based approach is that these experiments will not tell whether the lesioned area is the site of the FEO, afferent to the FEO, or efferent from the FEO, even if the lesion eliminates food-entrainable rhythms (15, 16). In the present study, we performed an unbiased search for brain region(s) that exhibit a de novo rhythmic expression of mPer1 and mPer2 under RF to identify a candidate locus for the FEO.
Results
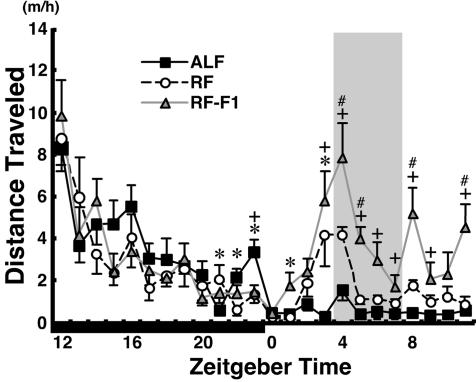
To reveal behavioral rhythms entrained by feeding, we examined the hourly distribution of locomotor activity under ad libitum feeding (ALF) or 4-hr RF in the middle of the light phase [zeitgeber time (ZT) 4–8, ZT0 is defined as the lights-on time and ZT12 as the lights-off time; see Methods]. As reported (3, 4, 13), mice demonstrated under RF a robustly increased locomotor activity for 2 h before food availability (Fig. 1). Temporal pattern of locomotor activity under RF, once established, was maintained with a sharp rise of FAA, even when mice were given no food during the expected feeding period (RF-F1). This finding is consistent with the hypothesis that a circadian oscillator (FEO) regulates food-entrainable rhythms, rather than that FAA is simply triggered each day when energy depletion reaches some threshold before food availability (3, 4).
Fig. 1.
Food-anticipatory locomotor activity under RF. Hourly plots of distance traveled under ALF, RF on day 9, and RF followed by fast (RF-F1, see Results and Methods). The period of food availability is shaded. The dark phase is indicated by a solid horizontal bar. ∗, P < 0.05 ALF vs. RF; +, P < 0.05 ALF vs. RF-F1; #, P < 0.05 RF vs. RF-F1; by one-way repeated-measures ANOVA and Turkey post hoc tests.
To seek for location(s) of the putative FEO(s), we then performed an unbiased search for such brain region(s) that exhibit under RF a strong rhythmic expression of mPer2, a core component of circadian clocks. We collected brains at four time points (ZT1, ZT7, ZT13, and ZT19) on the ninth day under ALF or RF. In situ hybridization using 35S-labeled antisense riboprobe for mPer2 was carried out on a series of stereotaxic coronal sections covering the entire rostrocaudal axis from the olfactory bulb to the medulla oblongata.
In the SCN, we observed weak expression of mPer2 at ZT1, the highest levels at ZT7, moderate levels at ZT13, and essentially no expression at ZT19. As reported (9–12), RF did not appreciably change the mPer2 rhythm in SCN. In contrast, the diffuse mPer2 expression in the striatum and cerebral cortex was higher in the dark phase (ZT13 and ZT19) under ALF, whereas expression was highest at ZT7 under RF (during food availability) (13). These temporal patterns of mPer2 expression are consistent with previous reports and indicate a phase uncoupling of circadian oscillators outside of the SCN from the SCN clock under RF (9–12).
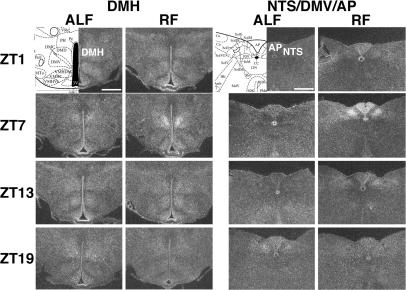
In addition to these previously reported regions, we found a notable, de novo rhythmic expression of mPer2 under RF in the dorsomedial hypothalamic nucleus (DMH), the nucleus of the solitary tract (NTS), and the area postrema (AP) (Fig. 2). Rhythmic expression in the DMH was mostly restricted to the compact part of the DMH (DMHc) located in the medio-central part of the caudal DMH and was most robust at ZT7 under RF. We did not observe strong expression of mPer2 in the DMH at any time points examined under ALF. Expression in the NTS was robust in its medial aspect at ZT7, much weaker at ZT13, and essentially undetectable at ZT1 and ZT19 under RF, whereas only a weak expression was observed at ZT19 under ALF. The temporal expression in the AP was similar to that in the NTS except that a weak expression was also seen at ZT1 under RF.
Fig. 2.
Circadian expression of mPer2 mRNA in the DMH and NTS/AP of mice under RF. Serial coronal brain sections, prepared from mice under ALF or RF at ZT1, ZT7, ZT13, and ZT19 on day 9, were hybridized in situ to a 35S-labeled mPer2 antisense probe. Representative sections from duplicate mice at the level of the DMH and NTS/DVM/AP are shown, together with coronal diagrams from the mouse brain atlas (39). (Scale bar: 0.5 mm.)
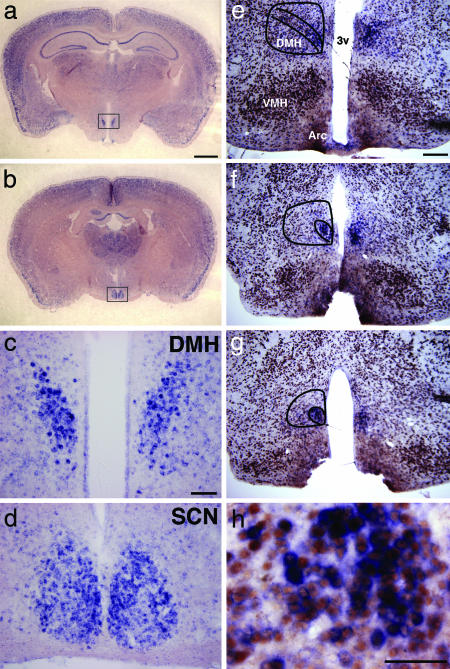
To further delineate the spatial and temporal expression patterns, we used digoxigenin-labeled antisense riboprobes for in situ hybridization. In pilot studies, the mPer1 and mPer2 probes both demonstrated temporal patterns of expression in the DMH essentially identical to the results shown in Fig. 2. Because the mPer1 probe appeared to have better sensitivity under our experimental conditions, we used mPer1 as a marker of circadian clocks for further study. During the period of food availability (ZT5 and ZT7) under RF, mPer1-expressing cells were scattered around the entire DMH and in many other areas in the brain. The most striking expression was seen in clusters of intensely labeled cells in the caudal DMH (Fig. 3a). When observed in coronal sections at low magnification, we could easily delineate the bilateral expression domains in the DMH. The intensity of mPer1 expression in this cluster appeared comparable to that in the SCN, although the total number of mPer1-expressing cells was fewer in the DMH than in the SCN (Fig. 3 c and d). In situ hybridization coupled with immunohistochemistry for a neuron-specific antigen NeuN demonstrated that this cluster consisted of neurons covering three to four sections 90 μm apart within the ventromedial part of the DMHc (Fig. 3 e–h).
Fig. 3.
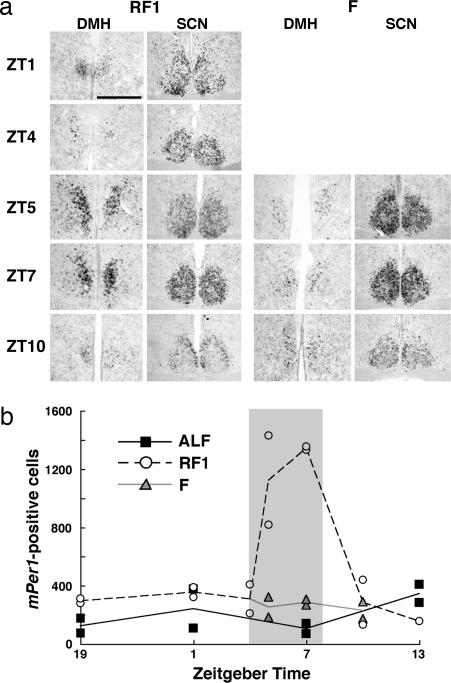
Localization of mPer1-expressing neurons in the DMH of mice under RF. Coronal brain sections were hybridized in situ to digoxigenin-labeled mPer1 antisense probe. (a and b) Low-magnification view of coronal sections containing the DMH (a) and the SCN (b) from a mouse under RF followed by fast (RF-F1) at ZT7 on day 7. (c and d) Representative higher-magnification images of mPer1-expressing cells in the DMH (c) and the SCN (d) in the fields shown by rectangles in a and b, respectively. (e–h) Coronal sections, prepared from a mouse under RF at ZT5, were dual-labeled by in situ hybridization for mPer1 (blue staining in perinuclear area) coupled with immunohistochemistry for a neuron-specific antigen NeuN (brown staining in nuclei). Cells with intense mPer1 expression are clustered in the ventromedial region of the DMHc (e–g in successive sections). Designated in lines are the whole DMH and its compact part. Arc, arcuate nucleus; VMH, ventromedial hypothalamic nucleus; 3v, third ventricle. (h) A representative image with higher magnification shows that mPer1-expressing cells in the DMH are neurons. (Scale bars: a and b, 1 mm; c and d, 0.1 mm; e–g, 0.2 mm; h, 0.05 mm.)
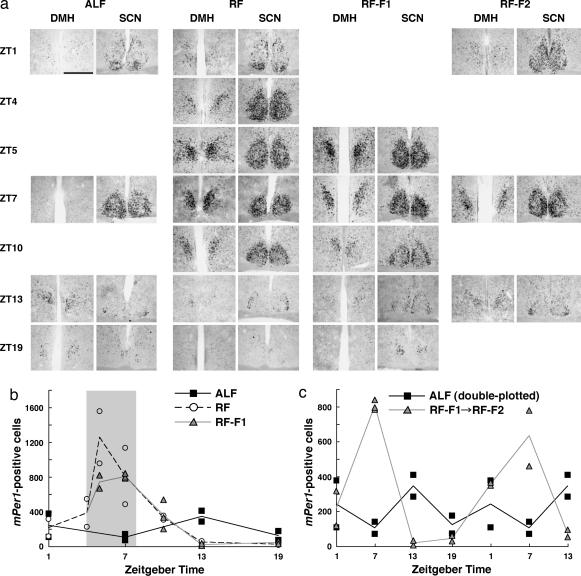
We quantified temporal patterns of mPer1 expression in the DMH around the period of food availability by counting the number of mPer1-positive cells in the ventromedial part of the DMHc (Fig. 4b and c) (note that the intensity of labeling in each positive cell was not considered in this procedure). Under ALF, mPer1 expression in the DMHc was relatively low and had relatively little, if any, oscillation, was higher at the onset of dark phase (ZT13), and was lowest in the middle of light phase (ZT7); these results may reflect the nocturnal feeding pattern of mice (Fig. 4 a and b). On the seventh day under RF, robust circadian oscillation of mPer1 expression was observed in the DMHc; expression started to increase immediately before food availability (ZT4), peaked during the period of food availability (ZT5 and ZT7), rapidly decreased after the food availability (ZT10), and reached nadir during the dark phase (ZT13 and ZT19). To exclude the possibility that this pattern of mPer1 expression was simply a reflection of food availability, we examined mPer1 expression when mice were given no food during the expected feeding period on the seventh day under RF (RF-F1). The temporal pattern of mPer1 expression under this condition was similar to that with food (RF). Furthermore, circadian oscillation of mPer1 in the DMHc persisted under prolonged fasting for an additional day, although peak expression level during the period of expected food availability tended to be slightly reduced (RF-F2, Fig. 4 a and c). These results indicate that circadian expression of mPer1 in the DMHc is entrained by feeding and free-runs at least for two cycles without further entrainment.
Fig. 4.
Detailed temporal patterns of circadian mPer1 expression in the DMH under RF. (a) Coronal brain sections, prepared from mice under ALF, RF, or RF followed by fast (RF-F1) on day 7 or under prolonged fasting on day 8 after RF-F (RF-F2) at time points shown in ZT, were hybridized in situ to a digoxigenin-labeled mPer1 antisense probe. Representative sections from duplicate mice at the level of the DMH and SCN are shown. Note that samples at ZT1 and ZT4 under RF are shared by the RF-F1 condition. (Scale bar: 0.2 mm.) (b and c) Numbers of mPer1-expressing cells in the ventromedial part of the DMHc. Values from two mice per data point (three mice for ZT7 under RF-F1) are dot-plotted; averages of duplicate counts are line-plotted. (b) Comparison of cell numbers under ALF, RF, and RF-F1. The period of food availability is shaded. (c) Food-entrained circadian oscillation of mPer1 expression in the DMHc sustains for at least 2 days without food. The preceding four time points (ZT1, ZT7, ZT13, and ZT19) are the same as those of RF-F1 in b, followed by three time points (ZT1, ZT7, and ZT13) on the second day of prolonged fasting (RF-F2). At the end of the experiments, mice under RF-F2 fasted for 53 h since the last food availability. For comparison, values under ALF, the same as those in b, are double-plotted.
To understand how feeding entrains rhythmic expression of mPer1 in the DMHc, we next examined acute effects of feeding on its expression in this area (Fig. 5). On the first day under RF, scheduled feeding after 20-h fasting rapidly induced mPer1 expression in DMHc within 1 h (RF1). This expression continued at least for 3 h and then returned to the baseline level within 6 h from the onset of feeding. Fasting by itself did not cause such a robust mPer1 expression (F in Fig. 5). Interestingly, the basal mPer1 expression in the DMHc under fasting was higher than that under ALF during the late dark phase and the light phase (ZT19, ZT1, and ZT7; collectively, P < 0.05, unpaired t test); a small number of DMH neurons might sense negative energy balance to up-regulate mPer1 expression. Although we have not performed systematic quantification of expression, mPer2 expression detected by in situ hybridization using digoxigenin-labeled riboprobe also demonstrated spatial and temporal patterns similar to those of mPer1 under various feeding conditions (data no shown).
Fig. 5.
Rapid induction of mPer1 in the DMHc by feeding. (a) Coronal brain sections, prepared from mice on the first day under RF (RF1) or fasting (F) at time points shown in ZT, were hybridized in situ to a digoxigenin-labeled mPer1 antisense probe. Representative sections from duplicate mice at the level of the DMH and SCN are shown. Note that samples at ZT1 and ZT4 under RF1 are shared by the fasting condition. (Scale bar: 0.2 mm.) (b) Numbers of mPer1-expressing cells in the ventromedial part of the DMHc. Values from two mice per data point (one mouse for ZT13 under RF1) are dot-plotted; averages of duplicate counts are line-plotted. Food was removed at ZT8 preceding time points shown in the graph, and returned, only under the RF1 condition, during the period shaded.
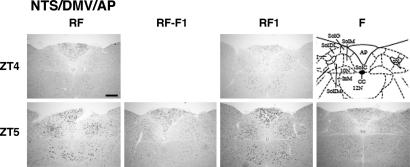
Consistent with the results of our in situ hybridization experiments using 35S-labeled riboprobe for mPer2, we observed rhythmic mPer1 expression in the NTS and AP under RF by in situ hybridization using digoxigenin-labeled riboprobe for mPer1 (Fig. 6). Expression peaked during the food availability, although the maximum levels were blunted compared with those in the DMHc. Expression in the NTS was highest at the anatomical level of the AP; most mPer1-expressing cells were restricted within the medial nucleus of the NTS and the dorsal motor nucleus of vagus (DMV). In contrast to the aforementioned situation in the DMHc, however, rhythmic mPer1 expression in the NTS/DMV/AP area under RF depends on periodical feeding per se; when mice were given no food during the expected feeding period on the seventh day under RF, little induction of mPer1, if any, was observed at ZT5 compared with that of fed mice under RF. On the first day under RF (RF1), like in the DMHc, feeding rapidly induced mPer1 expression in the NTS/DMV/AP area. These results suggest that the NTS/DMV/AP area increases mPer1 expression in acute response to feeding, but it does not contain an autonomous circadian oscillator entrained by feeding.
Fig. 6.
Feeding-dependent up-regulation of mPer1 in the NTS, DMV, and AP. Coronal brain sections, prepared from mice at ZT4 and ZT5 under RF or RF followed by fast (RF-F1) on day 7, or RF (RF1) or fasting (F) on day 1, were hybridized in situ to a digoxigenin-labeled mPer1 antisense probe. Representative sections from duplicate mice are shown, together with a coronal diagram from the mouse brain atlas (39). Note that samples at ZT4 are shared between RF and RF-F1 and between RF1 and F. (Scale bar: 0.2 mm.)
Discussion
Our observations suggest that the DMHc contains a self-sustained, autonomous circadian oscillator entrained by feeding, although additional lines of evidence such as demonstration of sustained rhythmicity in explants of the DMHc in vitro will be required to definitively prove this idea. One of the salient characteristics of the DMHc that differentiates it from other peripheral circadian oscillators is that the amplitude of Per1/Per2 oscillation was much higher under RF than under ALF; whereas RF does shift the phase of other extra-SCN oscillators, their amplitudes do not differ overtly between ALF and RF (9, 10, 12). Notably, the DMHc was found to be the only brain locus other than the SCN that contains densely clustered neurons exhibiting robust mPer1 and mPer2 oscillation under RF, whereas mPer genes are diffusely expressed in nearly all CNS and peripheral tissues.
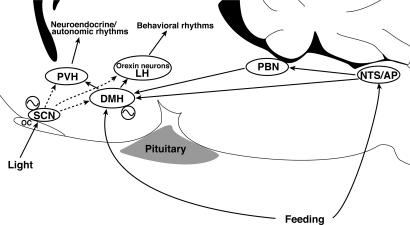
Accumulating lines of evidence suggest that the DMH is a critical locus for integration of circadian information from the SCN and feeding information from the peripheral organs to modulate a wide variety of behaviors and physiology (Fig. 7) (17). The DMH receives direct and indirect projections from the SCN (18, 19) and senses metabolic and feeding cues such as leptin (20). Furthermore, the DMH sends projections to regions critical for vigilance state regulation, such as the ventrolateral preoptic area (GABAergic projections) (19, 21), the locus coeruleus (22), and the orexin neurons (glutamatergic projections) (19, 23). The DMH also projects to the paraventricular hypothalamic nucleus (20), which regulates hypothalamic outflow to the autonomic nervous systems and endocrine systems. Indeed, studies have implicated the DMH in the SCN-entrainable circadian regulation of feeding, drinking, corticosterone secretion, locomotor activity, and vigilance states (19, 24). In the present study, we also found that the food-entrained oscillation of mPer2 in the DMHc and NTS/AP under RF was unaffected in the orexin/ataxin-3 transgenic mice (13) in which orexin neurons are genetically ablated (data not shown). This result further supports the idea that orexin neurons are located downstream of the DMH within the neural pathway govering food-entrained circadian behaviors.
Fig. 7.
The DMH as a food-entrainable circadian oscillator. When food is freely available (ALF), the light-entrainable oscillator in the SCN predominates in orchestrating circadian behavioral and physiological rhythms according to the environmental photic cycle. The DMH mediates circadian information from the SCN and also receives feeding information directly and indirectly via humoral and neural pathways. When food availability is restricted to a period scheduled at a fixed time of the day (RF), a FEO in the DMH entrains to feeding information, uncouples from the SCN, and starts regulating food-entrainable circadian rhythms. Dotted lines from the SCN designate that the predominance of SCN-derived signals is contingent on food availability. LH, lateral hypothalamus; OC, optic chiasm; PBN, parabrachial nucleus; PVH, paraventricular hypothalamic nucleus.
Collectively, the DMH seems well suited to be a central locus to regulate behaviors and physiology under such situations in which the SCN and peripheral clocks are phase-uncoupled, as under RF (Fig. 7). Recent reports showed that c-Fos immunoreactivity (an indicator of neuronal activity) is increased in the DMH before and during food availability under RF (15, 25). Most importantly, during the course of our independent study, Gooley et al. (15) demonstrated that lesions in DMH neurons in rats blocked food entrainment of wakefulness, locomotor activity, and core body temperature. However, although they demonstrated a critical role of the DMH in the expression of food-entrainable circadian rhythms, their findings did not discriminate among the possibilities: (i) that the DMH itself acts as the primary FEO (in other words, a pacemaker under RF), (ii) that the DMH is a key relay site from a still-unknown primary FEO (output pathway), and (iii) that it is a key relay site on the entrainment pathway to an FEO (input pathway). The present study provides support for the first possibility (Fig. 7). Interestingly, Gooley et al. showed food-anticipatory increases in c-Fos expression throughout the DMH under RF, whereas we demonstrated a robust oscillation of mPer1 mostly in a subregion of the DMH with its peak during the food availability. This finding may not necessarily be a contradiction, because c-Fos is an indicator of cumulative neuronal activity, whereas Per1 is a core component of neuronal circadian oscillators, which may influence the activity of these and surrounding cells quite indirectly. Thus, it would be important to delineate local networks surrounding the mPer1-oscillating cells within the DMH.
In the SCN core, phase-shifting light pulses rapidly induce expression of Per1 and Per2 in the light-responsive cells, and this induction has been suggested as a critical step for photic entrainment (26). Our observations that refeeding after fasting rapidly induces mPer1 (and mPer2 as well; data not shown) expression in the DMHc suggest that an analogous mechanism exists in the DMH for feeding-triggered entrainment of Per oscillation. Thus, this step may play a key role in resetting circadian rhythms by feeding. The SCN is a heterogeneous network of functionally and phenotypically differentiated neurons; cells directly responsive to the light are shown to represent a population distinct from those with robust circadian oscillation (26). It is currently unclear whether cells in the DMHc that acutely responded to feeding by increasing mPer1 expression are the same ones as those with robust mPer1 rhythmicity under RF.
Rhythmic expression of mPer1 and mPer2 in the NTS/DMV/AP area of the brainstem under RF is consistent with a recent report showing a feeding-dependent up-regulation of c-Fos expression under RF in the NTS, DMV, and AP (27). The NTS is a relay nucleus for visceral and gustatory sensory afferents, and it has also been suggested as a site of integration for visceral autonomic and sensory responses (28). In the rat, vagal afferents responding to gastric and duodenal distension tend to map to the medial and commissural divisions of the NTS (29), the former found to be the major area of mPer1 expression in the present study. The motor nucleus of the vagus, located just ventral to the NTS, is the primary site of motor efferents to the gut and is densely innervated by fibers from the NTS. The AP lacks the blood–brain barrier, resulting in its ability to sense circulating factors directly, and it is known to be involved in the regulation of autonomic functions and feeding (30). The NTS and AP also have reciprocal projections (28). Notably, the DMH receives direct projections from the NTS and the parabrachial nucleus (18), which in turn receives dense projections from the NTS/AP.
Not only the locus of FEO but also its molecular components have been elusive. It has been demonstrated that FAA is sustained in arrhythmic Clock mutant mice (31). However, each component of the molecular clock has multiple paralogs whose expression patterns are distinct but overlapping throughout the brain and peripheral tissues: Clock–Npas2, BMAL1–BMAL2, Cry1–Cry2, Per1–Per2–Per3, etc. (2). Thus, tissue-specific combinations of clock genes may be forming molecular clocks outside of the SCN (1, 32). In accordance with this idea, mice deficient for Npas2, which is expressed mainly in the forebrain but not in the SCN, has been demonstrated to have defects in food-anticipatory behaviors (33). Furthermore, mCry1−/−;mCry2−/− double knockout mice demonstrate markedly attenuated FAA (34). The molecular substrate of the DMH oscillator may consist of partly overlapping paralogous members of the same gene families that comprise the core clock in the SCN; in particular, the Cry and Per genes may be shared by both oscillators.
Disturbed eating patterns are often associated with human obesity (35). Dissociation of the circadian control of feeding relative to sleep/wake has been suggested in night eating syndrome, which is frequently seen in obese individuals (36). Moreover, Clock mutant mice were recently reported to demonstrate not only a defect in diurnal feeding rhythm but also hyperphagia, obesity, and metabolic syndrome, for which altered Clock expression in CNS feeding centers and/or peripheral tissues involved in metabolism may be a causal factor (37). Understanding the neural mechanism of reciprocal interactions between biological clocks and feeding centers is of potential practical importance, and the DMH might play a key role in these interactions as a putative FEO.
Methods
The methods we used are described in detail in Supporting Methods, which is published as supporting information on the PNAS web site. RF protocols were essentially the same as described (13). Tissues were collected in duplicate (two mice per data point, except for a data point in triplicate shown in figure legends). Coronal brain sections (30 μm) were collected in four series. In situ hybridization was performed as described (13, 38) with modifications. The numbers of stained cells were counted bilaterally in four coronal sections that were spaced 90 μm apart through the ventromedial part of the DMHc (39). We estimated the number of mPer1-expressing cells in the vetromedial DMHc per mouse.
Supplementary Material
Acknowledgments
We thank the Kazusa DNA Research Institute (Chiba, Japan) for full-length mPer1 and mPer2 cDNAs; S. A. Dixon, R. O. Floyd, and M. Thornton for technical support; and T. Miyashita for advice concerning nonradioisotope in situ hybridization methods. M.Y. is an Investigator of the Howard Hughes Medical Institute. M.M. is a former long-term fellow of the Human Frontier Science Program. This work was supported in part by research funds from the Keck Foundation, the Perot Family Foundation, Exploratory Research for Advanced Technology/Japan Science and Technology Agency, the Narishige Kagaku Foundation, the Ichiro Kanehara Foundation, and the Suzuken Memorial Foundation.
Abbreviations
- FEO
food-entrainable oscillator
- RF
restricted feeding
- FAA
food-anticipatory activity
- ALF
ad libitum feeding
- ZT
zeitgeber time
- SCN
suprachiasmatic nucleus
- DMH
dorsomedial hypothalamic nucleus
- DMHc
compact part of DMH
- NTS
nucleus of the solitary tract
- AP
area postrema
- DMV
dorsal motor nucleus of vagus.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hastings M. H., Reddy A. B., Maywood E. S. Nat. Rev. Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 2.Lowrey P. L., Takahashi J. S. Annu. Rev. Genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mistlberger R. E. Neurosci. Biobehav. Rev. 1994;18:171–195. doi: 10.1016/0149-7634(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 4.Stephan F. K. J. Biol. Rhythms. 2002;17:284–292. doi: 10.1177/074873040201700402. [DOI] [PubMed] [Google Scholar]
- 5.Abe M., Herzog E. D., Yamazaki S., Straume M., Tei H., Sakaki Y., Menaker M., Block G. D. J. Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkovsky P., Veisenberger E., LeSauter J., Yan L., Johnson M., Zhang D. Q., McMahon D., Silver R. J. Neurosci. 2003;23:7670–7676. doi: 10.1523/JNEUROSCI.23-20-07670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amir S., Lamont E. W., Robinson B., Stewart J. J. Neurosci. 2004;24:781–790. doi: 10.1523/JNEUROSCI.4488-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamont E. W., Robinson B., Stewart J., Amir S. Proc. Natl. Acad. Sci. USA. 2005;102:4180–4184. doi: 10.1073/pnas.0500901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara R., Wan K., Wakamatsu H., Aida R., Moriya T., Akiyama M., Shibata S. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 11.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., Menaker M. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 12.Wakamatsu H., Yoshinobu Y., Aida R., Moriya T., Akiyama M., Shibata S. Eur. J. Neurosci. 2001;13:1190–1196. doi: 10.1046/j.0953-816x.2001.01483.x. [DOI] [PubMed] [Google Scholar]
- 13.Mieda M., Williams S. C., Sinton C. M., Richardson J. A., Sakurai T., Yanagisawa M. J. Neurosci. 2004;24:10493–10501. doi: 10.1523/JNEUROSCI.3171-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akiyama M., Yuasa T., Hayasaka N., Horikawa K., Sakurai T., Shibata S. Eur. J. Neurosci. 2004;20:3054–3062. doi: 10.1111/j.1460-9568.2004.03749.x. [DOI] [PubMed] [Google Scholar]
- 15.Gooley J. J., Schomer A., Saper C. B. Nat. Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- 16.Landry G. J., Simon M. M., Webb I. C., Mistlberger R. E. Am. J. Physiol. 2006;290:R1527–R1534. doi: 10.1152/ajpregu.00874.2005. [DOI] [PubMed] [Google Scholar]
- 17.Saper C. B., Lu J., Chou T. C., Gooley J. Trends Neurosci. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Thompson R. H., Swanson L. W. Brain Res. Brain Res. Rev. 1998;27:89–118. doi: 10.1016/s0165-0173(98)00010-1. [DOI] [PubMed] [Google Scholar]
- 19.Chou T. C., Scammell T. E., Gooley J. J., Gaus S. E., Saper C. B., Lu J. J. Neurosci. 2003;23:10691–10702. doi: 10.1523/JNEUROSCI.23-33-10691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmquist J. K., Ahima R. S., Elias C. F., Flier J. S., Saper C. B. Proc. Natl. Acad. Sci. USA. 1998;95:741–746. doi: 10.1073/pnas.95.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou T. C., Bjorkum A. A., Gaus S. E., Lu J., Scammell T. E., Saper C. B. J. Neurosci. 2002;22:977–990. doi: 10.1523/JNEUROSCI.22-03-00977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aston-Jones G., Chen S., Zhu Y., Oshinsky M. L. Nat. Neurosci. 2001;4:732–738. doi: 10.1038/89522. [DOI] [PubMed] [Google Scholar]
- 23.Sakurai T., Nagata R., Yamanaka A., Kawamura H., Tsujino N., Muraki Y., Kageyama H., Kunita S., Takahashi S., Goto K., et al. Neuron. 2005;46:297–308. doi: 10.1016/j.neuron.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Bernardis L. L., Bellinger L. L. Proc. Soc. Exp. Biol. Med.; 1998. pp. 284–306. [DOI] [PubMed] [Google Scholar]
- 25.Angeles-Castellanos M., Aguilar-Roblero R., Escobar C. Am. J. Physiol. 2004;286:R158–R165. doi: 10.1152/ajpregu.00216.2003. [DOI] [PubMed] [Google Scholar]
- 26.Antle M. C., Silver R. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Angeles-Castellanos M., Mendoza J., Diaz-Munoz M., Escobar C. Am. J. Physiol. 2005;288:R678–R684. doi: 10.1152/ajpregu.00590.2004. [DOI] [PubMed] [Google Scholar]
- 28.Luckman S. M., Lawrence C. B. Trends Endocrinol. Metab. 2003;14:60–65. doi: 10.1016/s1043-2760(02)00033-4. [DOI] [PubMed] [Google Scholar]
- 29.Cone R. D. Nat. Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- 30.Edwards G. L., Ritter R. C. Brain Res. 1981;216:265–276. doi: 10.1016/0006-8993(81)90129-3. [DOI] [PubMed] [Google Scholar]
- 31.Pitts S., Perone E., Silver R. Am. J. Physiol. 2003;285:R57–R67. doi: 10.1152/ajpregu.00023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reick M., Garcia J. A., Dudley C., McKnight S. L. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 33.Dudley C. A., Erbel-Sieler C., Estill S. J., Reick M., Franken P., Pitts S., McKnight S. L. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 34.Iijima M., Yamaguchi S., van der Horst G. T., Bonnefont X., Okamura H., Shibata S. Neurosci. Res. 2005;52:166–173. doi: 10.1016/j.neures.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 35.de Zwaan M., Burgard M. A., Schenck C. H., Mitchell J. E. Eur. Eating Disorders Rev. 2003;11:7–24. [Google Scholar]
- 36.O’Reardon J. P., Ringel B. L., Dinges D. F., Allison K. C., Rogers N. L., Martino N. S., Stunkard A. J. Obes. Res. 2004;12:1789–1796. doi: 10.1038/oby.2004.222. [DOI] [PubMed] [Google Scholar]
- 37.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., et al. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyashita T., Nishimura-Akiyoshi S., Itohara S., Rockland K. S. Neuroscience. 2005;136:487–496. doi: 10.1016/j.neuroscience.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Paxinos G., Franklin K. B. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.