Abstract
The detection, stabilization, and repair of stress-induced damage are essential requirements for cellular life. All cells respond to osmotic stress-induced water loss with increased expression of genes that mediate accumulation of organic osmolytes, solutes that function as chemical chaperones and restore osmotic homeostasis. The signals and signaling mechanisms that regulate osmoprotective gene expression in animal cells are poorly understood. Here, we show that gpdh-1 and gpdh-2, genes that mediate the accumulation of the organic osmolyte glycerol, are essential for survival of the nematode Caenorhabditis elegans during osmotic stress. Expression of GFP driven by the gpdh-1 promoter (Pgpdh-1::GFP) is detected only during hypertonic stress but is not induced by other stressors. Using Pgpdh-1::GFP expression as a phenotype, we screened ≈16,000 genes by RNAi feeding and identified 122 that cause constitutive activation of gpdh-1 expression and glycerol accumulation. Many of these genes function to regulate protein translation and cotranslational protein folding and to target and degrade denatured proteins, suggesting that the accumulation of misfolded proteins functions as a signal to activate osmoprotective gene expression and organic osmolyte accumulation in animal cells. Consistent with this hypothesis, 73% of these protein-homeostasis genes have been shown to slow age-dependent protein aggregation in C. elegans. Because diverse environmental stressors and numerous disease states result in protein misfolding, mechanisms must exist that discriminate between osmotically induced and other forms of stress-induced protein damage. Our findings provide a foundation for understanding how these damage-selectivity mechanisms function.
Keywords: Caenorhabditis elegans, functional genomics, organic osmolytes, osmotic stress
The ability to detect, repair, and stabilize cellular and molecular damage induced by environmental stress is essential for cell survival and function. All organisms respond to environmental stress with increased expression of stress-protective genes. For example, heat shock causes protein denaturation and induces the expression of molecular chaperones that function to refold denatured proteins (1). Oxidative stress results in lipid and protein damage and activates the expression of antioxidant enzymes that detoxify free radicals (2).
Hypertonic stress causes cellular water loss, cell shrinkage, elevated cytoplasmic ionic strength, and increased expression of genes that mediate organic osmolyte accumulation. Organic osmolytes function as chemical chaperones and can be accumulated by cells to concentrations of hundreds of millimolar without adverse effects (3). Replacement of inorganic ions with organic osmolytes allows cells to maintain normal cytoplasmic ionic strength and survive in hypertonic environments (4, 5).
The effector mechanisms that mediate organic osmolyte accumulation are generally well defined. For example, in Escherichia coli, ProP is activated by hypertonicity and mediates uptake of compatible solutes such as proline, glycine betaine, and ectoine (6). Plants accumulate proline through hypertonicity-induced expression of biosynthetic enzymes (7). In yeast, hypertonicity induces expression of the glycerol biosynthesis enzyme glycerol 3-phosphate dehydrogenase (GPD), which mediates accumulation of the organic osmolyte glycerol (8). Cells of the renal inner medulla accumulate sorbitol and myo-inositol through increased expression of the sorbitol biosynthetic enzyme aldose reductase (9) and the sodium-coupled myo-inositol transporter SMIT (10).
The signals and signaling mechanisms that activate organic osmolyte accumulation in bacteria, plants, and yeast have been studied extensively (6, 7, 11). For example, genetic and molecular characterization of yeast has identified a MAP kinase signaling cascade that regulates hypertonicity-induced GPD expression (11). Activation of MAP kinase signaling is mediated by interaction of components of the cascade with the membrane protein Sho1. In animal cells, both the hypertonic stress signals and the signaling cascades that mediate osmosensitive gene expression are unknown. Sho1 homologs are absent from animal genomes, and, although animal homologs of yeast hypertonicity-activated kinases exist, none of them have been consistently shown to play a significant role in regulating osmosensitive gene expression (12, 13).
We have undertaken studies of the hypertonic stress response in Caenorhabditis elegans with the goal of exploiting this animal's genetic and molecular tractability to define animal cell osmosensing mechanisms. Recently, we demonstrated that C. elegans adapts to hypertonic stress by accumulating the organic osmolyte glycerol and that hypertonicity induces expression of glycerol biosynthetic enzymes (14). Here, we show that glycerol biosynthesis is essential for survival in hypertonic environments. Based on these findings, we developed an in vivo GFP reporter that reflects the activation state of signaling pathways controlling osmosensitive gene expression. Using genome-wide RNAi screening, we identified 122 gene inactivations that cause constitutive activation of this osmosensitive GFP reporter in the absence of hypertonic stress. The majority of these genes function normally to prevent the accumulation of damaged and denatured proteins in the cell cytoplasm. The results of our studies suggest the hypothesis that signaling pathways controlling osmosensitive gene expression in animal cells are activated, at least in part, by increased levels of hypertonicity-induced protein damage, specifically damage to proteins undergoing de novo synthesis.
Results and Discussion
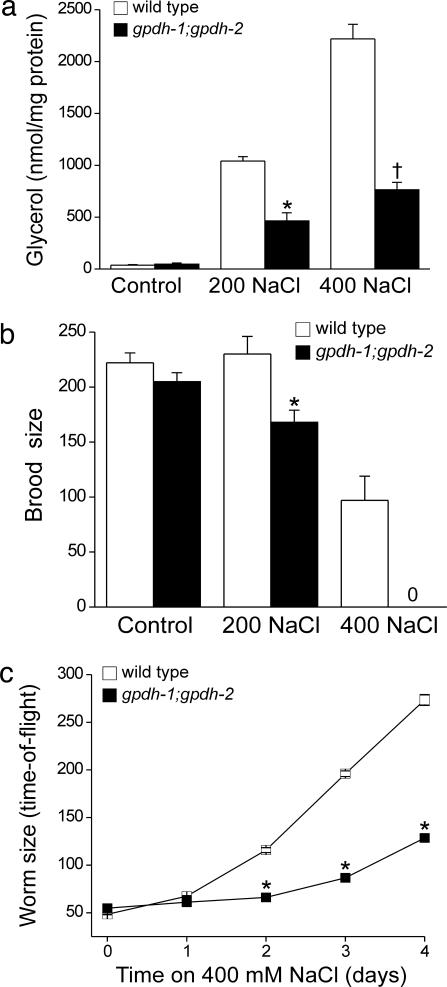
When exposed to hypertonic stress, C. elegans accumulates the organic osmolyte glycerol by de novo synthesis (14). The worm genome contains two genes encoding glycerol-3-phosphate-dehydrogenase (gpdh), which catalyzes the rate-limiting step of glycerol biosynthesis. Microarray (data not shown) and Northern blot analyses (14) demonstrated that gpdh-1 exhibits a strong and sustained transcriptional up-regulation during hypertonic stress, whereas gpdh-2 is weakly and transiently up-regulated. To test the physiological role of glycerol biosynthesis in the hypertonic stress response, we used the deletion alleles of gpdh-1 and gpdh-2, ok1558, and kb33, respectively. Although the rate of hypertonicity-induced glycerol accumulation was slowed in the gpdh-1-deletion mutant, steady-state glycerol levels under control and hypertonic conditions were similar to those in wild-type animals (data not shown). We therefore crossed ok1558 and kb33 worms to generate gpdh-1;gpdh-2 double mutants. Glycerol levels (Fig. 1a), fertility (Fig. 1b), and growth rates (data not shown) of gpdh-1(ok1558);gpdh-2(kb33) worms were indistinguishable from wild-type animals under normal culture conditions. However, when exposed to hypertonic stress, gpdh-1(ok1558);gpdh-2(kb33) worms exhibited greatly reduced glycerol accumulation (Fig. 1a) and fertility (Fig. 1b) and grew considerably more slowly than wild-type animals (Fig. 1c). Therefore, GPDH-1- and GPDH-2-mediated glycerol accumulation is essential for survival in hypertonic environments.
Fig. 1.
gpdh-1 and gpdh-2 are required for adaptation to hypertonic stress. (a) Whole-animal glycerol levels in the wild type and gpdh-1;gpdh-2 double mutants. L1 larvae were grown on control or 200 mM NaCl agar plates for 3–4 days before glycerol measurements. For measurements at 400 mM NaCl, L1 larvae were grown on 200 mM NaCl until the L4 stage of development and then shifted to 400 mM NaCl plates for 24 h. Values are means ± SE (n = 3). ∗, P < 0.01 compared with wild type; †, P < 0.001 compared with wild type. (b) Brood size in wild-type and gpdh-1;gpdh-2 double-mutant worms. Values are means ± SE (n = 9–21). ∗, P < 0.01 compared with wild type. (c) Rate of growth in wild-type and gpdh-1;gpdh-2 double-mutant worms exposed to 400 mM NaCl. Worm size was quantified as time-of-flight by using a COPAS Biosort. Values are means ± SE (n > 300). ∗, P < 0.01 compared with wild type.
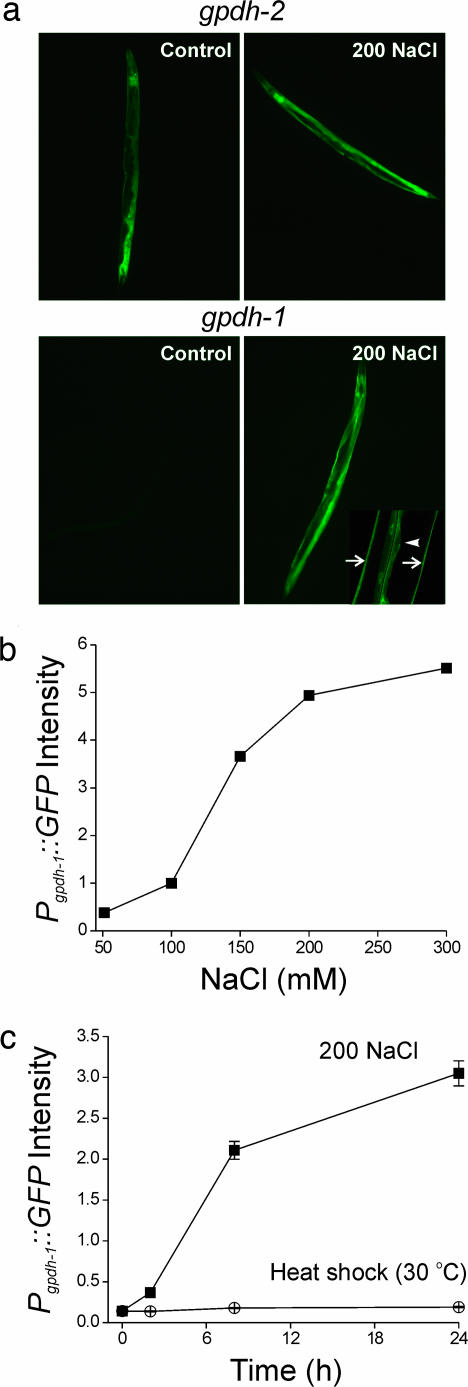
GFP transcriptional reporters demonstrated constitutive expression of gpdh-2 in the intestine, hypodermis, and excretory cell (Fig. 2a). In contrast, Pgpdh-1::GFP expression was virtually undetectable under control conditions but showed dramatic up-regulation in the intestine and hypodermis in worms exposed to hypertonicity (Fig. 2a). Pgpdh-1-GFP expression was proportional to the level of hypertonic stress (Fig. 2b), and the time course of expression (Fig. 2c) correlated well with that of glycerol accumulation (14). Heat shock (30°C; Fig. 2c), endoplasmic reticulum stress (10 μg/ml tunicamycin), cold (4°C), and oxidative stress (230 μM juglone and 100% O2; data not shown) failed to activate Pgpdh-1::GFP. These data demonstrate that Pgpdh-1::GFP specifically reports activation of signaling pathways required for glycerol synthesis and adaptation of C. elegans to hypertonic stress.
Fig. 2.
gpdh transcriptional GFP reporters are activated by hypertonic stress. (a Upper) Pgpdh-2::GFP is constitutively expressed in the hypodermis, intestine, and excretory cell under control conditions. (a Lower) Pgpdh-1::GFP expression is undetectable under isotonic conditions and is dramatically elevated after exposure to 200 mM NaCl. Pgpdh-1::GFP is expressed in the intestine (Lower Right, arrowhead) and hypodermis (Lower Right, arrows). (b) Effect of NaCl concentration on Pgpdh-1::GFP expression in young adult worms. Animals were placed on agar plates containing the indicated amounts of NaCl for 24 h. Values are means ± SE (n > 1,000). Pgpdh-1::GFP expression was also induced by hypertonic KCl, sucrose, or sorbitol (data not shown), indicating that gpdh-1 expression is regulated specifically by hypertonicity-induced water loss rather than elevated Na+ or Cl− levels. (c) Changes in Pgpdh-1::GFP expression in young adult worms during hypertonic stress or heat shock (30°C). Values are means ± SE (n > 150). GFP expression in b and c was quantified by using a COPAS BioSort and normalized to time-of-flight (i.e., GFP fluorescence divided by time-of-flight).
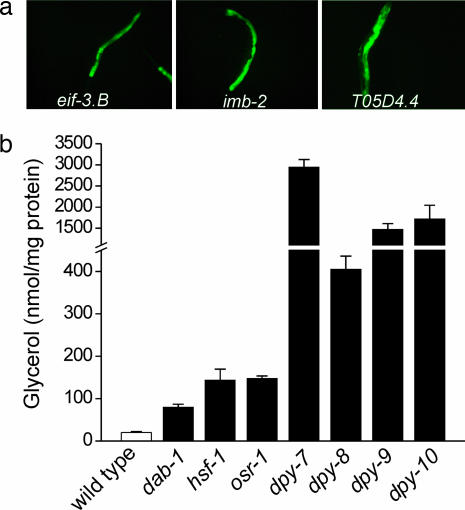
To identify signals and signaling mechanisms that regulate osmoprotective gene expression in animal cells, we performed a genome-wide RNAi feeding screen based on Pgpdh-1::GFP expression. The Pgpdh-1::GFP reporter strain was used in this screen, because gpdh-1 undergoes striking and sustained up-regulation in response to hypertonicity (ref. 14 and data not shown), and because Pgpdh-1::GFP expression was undetectable in the absence of hypertonic stress (Fig. 2a). L1-stage larvae on 20 mM NaCl growth plates were fed dsRNA-producing bacteria, and animals were visually scored for GFP expression after 3 days. We identified 106 gene inactivations that consistently activated Pgpdh-1::GFP expression in the absence of hypertonic stress (e.g., Fig. 3a). These genes are termed regulators of glycerol-3-phosphate-dehydrogenase expression (rgpd; Table 1; and see Table 2, which is published as supporting information on the PNAS web site).
Fig. 3.
Loss of gene function by RNAi or mutation causes constitutive expression of Pgpdh-1::GFP and glycerol accumulation in the absence of hypertonic stress. (a) Examples of genes identified in genome-wide RNAi screening that activate Pgpdh-1::GFP expression when silenced. (b) Glycerol content in worms harboring loss-of-function mutations in genes identified by RNAi screening. Values are means ± SE (n = 3). All glycerol levels in mutant worms were significantly (P < 0.02) different from wild type.
Because our screen was based on activation of a GFP reporter, the genes we identified could encode regulators of transgene expression rather than components of signaling pathways controlling glycerol accumulation. To determine the role of rgpd genes in hypertonic stress resistance, we measured glycerol levels for a subset of rgpd genes in which viable and fertile loss-of-function mutants were available. Glycerol levels were significantly elevated 4- to 144-fold over control animals in worm strains harboring loss-of-function mutations in the genes tested (Fig. 3b).
Genome-wide RNAi screening results in significant numbers of false negatives (15). To identify additional rgpd genes, we queried the C. elegans Interactome, a genome-wide protein–protein interaction map comprising 3,228 genes and 5,685 yeast two-hybrid interactions (16). Forty-eight rgpd genes were present in the Interactome and showed few direct interactions. However, we identified 148 genes that interact with the rgpd genes (see Fig. 5, which is published as supporting information on the PNAS web site). RNAi constructs for 124 of these genes were present in our library. We rescreened these interacting genes (see Methods) and identified an additional 16 rgpd genes (see Fig. 5). The Interactome screen increased the rgpd genes by 15% to 122. This increase is consistent with false-negative rates of 10–30% that have been estimated for C. elegans genome-wide RNAi screens (15).
Many of the rgpd genes encode conserved proteins, and 72 of them have human homologs (Table 2). Expression patterns have been determined by using GFP reporters for 35 of the genes, and 30 are coexpressed with gpdh-1 in the hypodermis and/or intestine (Table 2) (17). rgpd gene functions fell into six defined cellular processes and a group of genes with unassigned functions (Tables 1 and 2). The defined processes include protein homeostasis, extracellular matrix, signaling, metabolism, protein trafficking, and transcriptional regulation.
Table 1.
Summary of gene knockdowns that cause constitutive gpdh-1 expression
| Process (no. of genes) | Molecular function (no. of genes) | Gene example | Description | Human homolog |
|---|---|---|---|---|
| Protein homeostasis (54) | ||||
| Protein synthesis (38) | Translation initiation (10) | B0511.10/eif-3.E | Translation initiation factor 3, subunit e | EIF3S6 |
| tRNA synthetase (10) | F22D6.3/nrs-1 | Asparaginyl-tRNA synthetase | NARS | |
| RNA processing (18) | W08E3.1/snr-2 | U1 snRNP component | SNRPN | |
| Protein folding (7) | Chaperonins (4) | K01C8.10/cct-4 | Chaperonin complex, δ-subunit | CCT4 |
| Hsp70 (1) | F26D10.3/hsp-1 | Molecular chaperones HSP70 superfamily | HSPA8 | |
| Protein degradation (9) | 26S proteasome (4) | C23G10.4/rpn-2 | 26S proteasome-regulatory complex | PSMD1 |
| Extracellular matrix (5) | Collagen (4) | T14B4.7/dpy-10 | Collagens (types IV and XIII) | Collagen α IV |
| Signaling (15) | Scaffolding (2) | ZK849.2 | PDZ domain | GOPC |
| Transduction (6) | F38H4.9/let-92 | Ser/thr protein phosphatase 2A, catalytic subunit | PPP2CB | |
| Metabolism (9) | Trehalose synthesis (1) | H13N06.3/gob-1 | Trehalose 6-phosphate synthase | None |
| Protein trafficking (6) | Nuclear import (1) | R06A4.4/imb-2 | Nuclear transport receptor | TNPO2 |
| Transcriptional regulation (9) | CCR4/NOT complex (2) | F57B9.2/ntl-1 | Negative regulator of transcription | CNOT1 |
| Unassigned function (24) | Secreted protein (10) | C32E12.3/osr-1 | Osmotic stress-resistance protein | None |
| Unknown function (14) | B0035.11 | Uncharacterized conserved protein | LEO1 |
Four of the extracellular matrix genes encode the collagens DPY-7, DPY-8, DPY-9, and DPY-10. Loss-of-function mutations in these genes increased Pgpdh-1::GFP expression (data not shown) and glycerol accumulation (Fig. 3b). The DPY collagens are secreted proteins that likely function extracellularly. Interestingly, we also identified 10 genes with unassigned functions that are predicted to encode secreted proteins (Tables 1 and 2). Secreted proteins have been shown to play important roles in mechanotransduction. For example, MEC-1 and MEC-9 are secreted by C. elegans touch neurons and genetically interact with the collagen MEC-5 (18, 19). These three proteins are essential components of the C. elegans touch neuron mechanosensory complex (20). Similarly, DPY collagens and other secreted proteins could function to detect hypertonic stress-induced mechanical signals. In vertebrates, collagens and integrins function in cellular mechanotransduction (21, 22) and osmotic stress-induced signaling processes (23–26).
dpy-7 and dpy-10 have been shown to suppress temperature-sensitive mutations in several unrelated genes (27, 28). Phenotypes associated with temperature-sensitive mutations are thought to be due to misfolding of the mutated protein that is reduced at low temperatures (29, 30). Because organic osmolytes such as glycerol aid in the refolding of denatured proteins (31), the ability of the dpy mutants to suppress temperature-sensitive gene mutations might be a general property of all rgpd genes that activate gpdh-1 expression and cause glycerol accumulation (Fig. 3b).
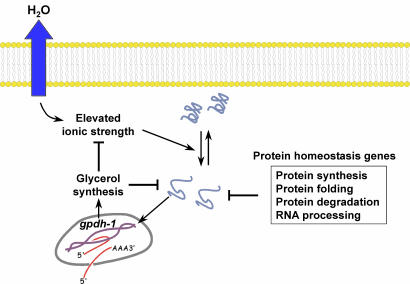
Surprisingly, the majority (44%, or 54 of 122) of rgpd genes fell into a category defined as protein homeostasis (Table 1). These genes encode proteins required for RNA processing, protein synthesis, protein folding, and protein degradation. Protein homeostasis genes function to maintain levels of properly folded and functioning cellular proteins. Inhibition of protein homeostasis genes is expected to increase the levels of damaged cellular proteins. Recent studies by Nollen et al. (32) support this idea. Wild-type GFP expressed in C. elegans muscle cells is distributed uniformly in the cytoplasm. However, modified GFPs containing repeats of glutamine undergo age-dependent aggregation (33). Genome-wide RNAi screening identified 187 genes that function to slow aging-induced protein aggregation (32). We found that 34 of the 122 rgpd genes overlapped with this 187-gene data set (Table 2), a 24-fold greater overlap than expected by chance alone (P < 0.001). Strikingly, 25 of the 34 overlapping genes are predicted to function in RNA processing, protein synthesis, protein folding, and protein degradation. Thus, genes that function to prevent protein aggregation also function to inhibit gpdh-1 expression. When the function of these genes is disrupted, damaged and denatured proteins accumulate in cells, and gpdh-1 expression is increased, leading to glycerol accumulation. Our results are consistent with a model in which increased levels of damaged or denatured proteins act as a signal that triggers osmoprotective gene expression and organic osmolyte accumulation (Fig. 4). Accumulation of organic osmolytes is expected to stabilize protein structure and decrease protein misfolding (31), which, in turn, would serve to autoregulate pathway activity.
Fig. 4.
Model for regulation of osmosensitive gene expression by disruption of protein homeostasis. Hypertonic stress-induced water loss causes elevated cytoplasmic ionic strength, which, in turn, causes protein damage. Damaged proteins function as a signal that activates gpdh-1 expression and glycerol synthesis. Glycerol replaces inorganic ions in the cytoplasm and functions as a chemical chaperone that aids in the refolding of misfolded proteins. Loss of function of protein homeostasis genes also causes accumulation of damaged proteins and activation of gpdh-1 expression.
Interestingly, protein damage induced by numerous stressors including heat shock (Fig. 2c) does not activate Pgpdh-1::GFP. Thus, osmotic stress must cause a type of protein damage that selectively activates gpdh-1 expression. Recent studies in eukaryotic cells suggest a mechanism for how osmotic stress-induced damage may be discriminated from other forms of protein damage. Albanese et al. (34) demonstrated that eukaryotes have at least two distinct systems for detecting and repairing protein misfolding. Canonical heat-shock proteins (HSPs) function to refold stress-denatured proteins (35). In contrast, chaperones that are linked to protein synthesis (CLIPS) function to regulate de novo protein folding (34).
The results of our screen suggest that de novo protein folding associated with protein synthesis plays a critical role in the hypertonic stress response. Canonical HSPs were not detected in our screen. Instead, the four T complex chaperones and hsp-1/F26D10.3 are predicted CLIPS (Tables 1 and 2). Furthermore, RNAi silencing of 38 genes involved in RNA processing and protein translation activates Pgpdh-1::GFP expression (Tables 1 and 2). In both cases, inhibition of these genes is predicted to disrupt protein synthesis.
Previous studies have shown that hypertonic stress, but not heat or oxidative stress, inhibits protein synthesis in yeast. Inhibition is transient, and recovery occurs via a Hog1p-dependent process that likely requires glycerol accumulation (36). Importantly, the initiation and elongation steps of protein synthesis are inhibited in vitro by increases in salt concentration of as little as 10 mM, and salt-induced inhibition is fully reversed by organic osmolytes (37). Inhibition of elongation causes disengagement of actively translating ribosomes from mRNAs and premature termination of protein translation, resulting in accumulation of incomplete and aberrantly folded polypeptides. Because cell shrinkage increases cytoplasmic salt concentration, these observations are consistent with a model in which gpdh-1 expression is specifically activated by osmotically induced disruption of new protein synthesis and cotranslational folding rather than by denaturation of existing proteins. Such a mechanism would allow cells to discriminate between osmotically induced protein damage and other forms of stress-induced damage. Our proposed model is analogous to the unfolded protein response, which is an intracellular signaling and transcriptional/translational program activated by the accumulation of unfolded proteins in the endoplasmic reitculum (ER) lumen that functions to restore ER protein homeostasis (38).
The response of a multicellular organism such as C. elegans to hypertonic stress likely involves the integration of a number of hypertonicity-induced signals and signal-transduction pathways. However, our RNAi studies indicate that disruption of protein homeostasis alone is sufficient to activate cellular osmoprotective pathways. Given that a wide range of environmental stressors induces protein damage, mechanisms must exist that discriminate between osmotically induced protein damage and other forms of stress-induced protein damage. Our findings provide a new foundation for understanding how these damage-selectivity mechanisms function and for defining the signaling pathways by which animal cells respond to osmotic stress. Because accumulation of misfolded proteins is a hallmark of diseases such as Alzheimer's and Parkinson's (39), understanding the molecular events underlying the detection of osmotically induced protein damage will provide an important paradigm for defining the cellular response to disease-induced protein damage.
Methods
C. elegans Strains.
All strains used in this study were derived from the N2 Bristol wild-type strain. Unless otherwise noted, worms were cultured at 20°C. The following strains were used: LGI-RB1032 [osr-1(ok959)], PS3551 [hsf-1(sy441)], LGII-VC616 [dab-1(gk216)], CB128 [dpy-10(e128)], LGIV-CB12 [dpy-9(e12)], LGX-CB88 [dpy-7(e88)], and CB130 [dpy-8(e130)].
gpdh-1(ok1558) was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis). We generated gpdh-2(kb33) by PCR-based screening. Both mutants were outcrossed to wild-type animals three times and exhibited no gross phenotypic defects as homozygotes. ok1558 encodes a 1,227-bp deletion in F47G4.3 (flanking sequences, tgcaactgat/ctagaaacca), and kb33 encodes a 758-bp deletion in K11H3.1 (flanking sequences, tttattcctc/aagaactgtt). gpdh-1(ok1558)I;gpdh-2(kb33)III animals were generated by crossing and PCR genotyping.
Plasmid Construction.
To create Pgpdh-1::GFP, a 3.3-kb PCR fragment, containing 3 kb of sequence 5′ to the start ATG and the first six amino acids from exon 2, was cloned into BamHI and SphI sites of pPD95.75. To create Pgpdh-2::GFP, a 4.4-kb PCR fragment, containing 2.3 kb of sequence 5′ to the start ATG and the first eight amino acids of exon 2 of the K11H3.1a gene, was cloned into SphI sites of pPD95.75. Both constructs were confirmed by sequencing. Primer sequences are available upon request.
Transgenics.
Wild-type animals were injected with 75 ng/μl rol-6 marker pRF4 and 25 ng/μl GFP construct by using standard methods (40). Three of the Pgpdh-1::GFP and two of the Pgpdh-2::GFP lines generated showed identical patterns of GFP expression under control and hypertonic growth conditions. Array kbEx144 was used to generate the integrated strain kbIs5. Integration was carried out by exposing ≈50 kbEx144 L4 animals to 30,000 μJ/cm2 generated from a UV cross-linker (Hoeffer Instruments, San Francisco, CA). A single integrated line segregating 100% rol-6 animals was isolated and outcrossed three times to wild-type animals to generate kbIs5 [Pgpdh-1::GFP;rol-6(su1006)], which was used in all subsequent studies.
Genome-Wide RNAi Screening.
Genome-wide RNAi screening was carried out by using a commercially available RNAi feeding library (MRC Geneservice, Cambridge, U.K.). Single colonies were inoculated into 100 μl of LB media containing 25 μg/ml carbenicillin and grown for 6–8 h at 37°C. Twenty microliters of each culture was spotted onto individual wells of 24-well nematode growth medium (NGM) agar plates containing 20 mM NaCl, 1 mM isopropyl β-d-thiogalactoside (IPTG), and 25 μg/ml carbenicillin. After overnight induction of dsRNA, 30–40 L1-stage animals were dispensed into each well. GFP expression was monitored after 72 h at 16°C by using an M2Bio fluorescence microscope (Zeiss, Thornwood, NY). Clones were scored as positive if at least five animals exhibited visible GFP fluorescence. These clones were rescreened in quadruplicate, and those that induced GFP expression in at least three of four trials were considered bona fide regulators of gpdh-1 expression, or rgpd genes.
rgpd genes present in the C. elegans Interactome (16), and their interacting proteins were rescreened for the rgpd phenotype by RNAi. Bacterial strains were grown as described above. Forty microliters of each bacterial culture was spotted onto individual wells of 24-well NGM RNAi plates and left at room temperature overnight. Five to 10 L1-stage animals were dispensed into each well and allowed to develop into gravid adults at 16°C. After laying >20 eggs, the adults were removed from the plate. F1 RNAi-treated animals were examined for GFP expression 3–4 days after hatching. The screen was repeated twice, and positive clones were assigned a score based on the level of GFP induction from 1 (weak) to 3 (strong). Scores from each screen were summed, and clones that had a total score of >2 were considered to confer an rgpd phenotype. Both the genome-wide and Interactome screens were performed blind to the identity of the genes being screened.
Measurement of Brood Size and Rate of Growth.
L1-stage worms were placed on NGM plates containing 51, 200, or 400 mM NaCl. After 2–4 days, single L4-stage hermaphrodites were placed into individual wells of 24-well plates. Worms were transferred to new wells every day until egg-laying ceased (3–4 days). Brood size was determined by summing the number of viable progeny in each well.
Growth rate was estimated by transferring L1-stage worms onto NGM plates containing 400 mM NaCl. Worms were washed off plates with M9 solution containing 400 mM NaCl at 24 h intervals, and animal size was quantified as time-of-flight, which was measured using a COPAS Biosort (Union Biometrica, Somerville, MA). Time-of-flight is a direct measure of worm axial length (41).
Glycerol Measurements.
Hypochlorite-synchronized L1-stage larvae were placed onto enriched peptone plates streaked with NA22 bacteria. After 3–4 days at 20°C, gravid worms were washed off plates and processed for glycerol measurements as described (14).
Statistical Analyses.
Data are presented as means ± SE. Statistical significance was determined by using Student's two-tailed t test for unpaired means. Significance of the overlap between two sets of genes was determined by calculating the representation factor, which is the total number of overlapping genes divided by the expected number of overlapping genes. Calculation of the representation factor and P value were carried out as described (42). P values of <0.05 were considered to indicate significance.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants DK61168 and DK64743 (to K.S.) T.L. was supported by a fellowship from the National Kidney Foundation. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Young J. C., Agashe V. R., Siegers K., Hartl F. U. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 2.Liu H., Colavitti R., Rovira I. I., Finkel T. Circ. Res. 2005;97:967–974. doi: 10.1161/01.RES.0000188210.72062.10. [DOI] [PubMed] [Google Scholar]
- 3.Yancey P. H. J. Exp. Biol. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- 4.Burg M. B. Am. J. Physiol. 1995;268:F983–F996. doi: 10.1152/ajprenal.1995.268.6.F983. [DOI] [PubMed] [Google Scholar]
- 5.Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 6.Wood J. M., Bremer E., Csonka L. N., Kraemer R., Poolman B., van der Heide T., Smith L. T. Comp. Biochem. Physiol. A. 2001;130:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa P. M., Bressan R. A., Zhu J. K., Bohnert H. J. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 8.Hohmann S. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smardo F. L., Jr., Burg M. B., Garcia-Perez A. Am. J. Physiol. 1992;262:C776–C782. doi: 10.1152/ajpcell.1992.262.3.C776. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi A., Nakanishi T., Takamitsu Y., Sugita M., Imai E., Noguchi T., Fujiwara Y., Kamada T., Ueda N. J. Am. Soc. Nephrol. 1994;5:62–67. doi: 10.1681/ASN.V5162. [DOI] [PubMed] [Google Scholar]
- 11.O'Rourke S. M., Herskowitz I., O'Shea E. K. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- 12.Kwon H. M., Itoh T., Rim J. S., Handler J. S. Biochem. Biophys. Res. Commun. 1995;213:975–979. doi: 10.1006/bbrc.1995.2224. [DOI] [PubMed] [Google Scholar]
- 13.Kultz D., Garcia-Perez A., Ferraris J. D., Burg M. B. J. Biol. Chem. 1997;272:13165–13170. doi: 10.1074/jbc.272.20.13165. [DOI] [PubMed] [Google Scholar]
- 14.Lamitina S. T., Morrison R., Moeckel G. W., Strange K. Am. J. Physiol. 2004;286:C785–C791. doi: 10.1152/ajpcell.00381.2003. [DOI] [PubMed] [Google Scholar]
- 15.Simmer F., Moorman C., Van Der Linden A. M., Kuijk E., Van Den Berghe P. V., Kamath R., Fraser A. G., Ahringer J., Plasterk R. H. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., et al. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKay S. J., Johnsen R., Khattra J., Asano J., Baillie D. L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. Cold Spring Harbor Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 18.Gu G., Caldwell G. A., Chalfie M. Proc. Natl. Acad. Sci. USA. 1996;93:6577–6582. doi: 10.1073/pnas.93.13.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du H., Gu G., William C. M., Chalfie M. Neuron. 1996;16:183–194. doi: 10.1016/s0896-6273(00)80035-5. [DOI] [PubMed] [Google Scholar]
- 20.Emtage L., Gu G., Hartwieg E., Chalfie M. Neuron. 2004;44:795–807. doi: 10.1016/j.neuron.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 21.DeMali K. A., Wennerberg K., Burridge K. Curr. Opin. Cell Biol. 2003;15:572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 22.ffrench-Constant C., Colognato H. Trends Cell Biol. 2004;14:678–686. doi: 10.1016/j.tcb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Vom D. S., Schliess F., Reissmann R., Gorg B., Weiergraber O., Kocalkova M., Dombrowski F., Haussinger D. J. Biol. Chem. 2003;278:27088–27095. doi: 10.1074/jbc.M210699200. [DOI] [PubMed] [Google Scholar]
- 24.Sheikh-Hamad D., Youker K., Truong L. D., Nielsen S., Entman M. L. Am. J. Physiol. 2000;279:C136–C146. doi: 10.1152/ajpcell.2000.279.1.C136. [DOI] [PubMed] [Google Scholar]
- 25.Browe D. M., Baumgarten C. M. J. Gen. Physiol. 2003;122:689–702. doi: 10.1085/jgp.200308899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moeckel G. W., Zhang L., Chen X., Rossini M., Zent R., Pozzi A. Am. J. Physiol. 2006;290:F223–F231. doi: 10.1152/ajprenal.00371.2004. [DOI] [PubMed] [Google Scholar]
- 27.Nishiwaki K., Miwa J. Mol. Gen. Genet. 1998;259:2–12. doi: 10.1007/s004380050782. [DOI] [PubMed] [Google Scholar]
- 28.Levy A. D., Yang J., Kramer J. M. Mol. Biol. Cell. 1993;4:803–817. doi: 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gidalevitz T., Ben Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. Science. 2006;311:1385–1386. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 30.Van Dyk T. K., Gatenby A. A., LaRossa R. A. Nature. 1989;342:451–453. doi: 10.1038/342451a0. [DOI] [PubMed] [Google Scholar]
- 31.Auton M., Bolen D. W. Proc. Natl. Acad. Sci. USA. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nollen E. A., Garcia S. M., van Haaften G., Kim S., Chavez A., Morimoto R. I., Plasterk R. H. Proc. Natl. Acad. Sci. USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morley J. F., Brignull H. R., Weyers J. J., Morimoto R. I. Proc. Natl. Acad. Sci. USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albanese V., Yam A. Y., Baughman J., Parnot C., Frydman J. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 35.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uesono Y., Toh E. J. Biol. Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- 37.Brigotti M., Petronini P. G., Carnicelli D., Alfieri R. R., Bonelli M. A., Borghetti A. F., Wheeler K. P. Biochem. J. 2003;369:369–374. doi: 10.1042/BJ20021056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder M., Kaufman R. J. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 39.Selkoe D. J. Nat. Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 40.Mello C. C., Fire A. In: Caenorhabditis elegans; Modern Biological Analysis of an Organism. Epstein H. F., Shakes D. C., editors. San Diego: Academic; 1995. pp. 452–482. [Google Scholar]
- 41.Pulak R. C. elegans: Methods and Applications. Totowa, NJ: Humana Press, Inc; 2006. in press. [Google Scholar]
- 42.Kim S. K., Lund J., Kiraly M., Duke K., Jiang M., Stuart J. M., Eizinger A., Wylie B. N., Davidson G. S. Science. Vol. 293. 2001. pp. 2087–2092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.