Abstract
Herbicides that act by inhibiting protoporphyrinogen oxidase (PPO) are widely used to control weeds in a variety of crops. The first weed to evolve resistance to PPO-inhibiting herbicides was Amaranthus tuberculatus, a problematic weed in the midwestern United States that previously had evolved multiple resistances to herbicides inhibiting two other target sites. Evaluation of a PPO-inhibitor-resistant A. tuberculatus biotype revealed that resistance was a (incompletely) dominant trait conferred by a single, nuclear gene. Three genes predicted to encode PPO were identified in A. tuberculatus. One gene from the resistant biotype, designated PPX2L, contained a codon deletion that was shown to confer resistance by complementation of a hemG mutant strain of Escherichia coli grown in the presence and absence of the PPO inhibitor lactofen. PPX2L is predicted to encode both plastid- and mitochondria-targeted PPO isoforms, allowing a mutation in a single gene to confer resistance to two herbicide target sites. Unique aspects of the resistance mechanism include an amino acid deletion, rather than a substitution, and the dual-targeting nature of the gene, which may explain why resistance to PPO inhibitors has been rare.
Keywords: Amaranthus, evolution, waterhemp, weed resistance, herbicide resistance
A major concern with the use of herbicides for weed control is the selection of resistant populations. To date, over 300 different herbicide-resistant weed biotypes have been identified worldwide (www.weedscience.com). Numerous factors influence the likelihood of herbicide-resistance evolution in a weed population, and certain herbicides are more prone to resistance evolution than are others. For example, populations of 95 weed species have been reported with resistance to herbicides that inhibit acetolactate synthase, whereas evolved resistance to herbicides that inhibit protoporphyrinogen oxidase (PPO) has been reported for only three weeds (www.weedscience.com), even though these herbicides were first commercialized in the 1960s (1). The first weed to evolve resistance to PPO inhibitors was Amaranthus tuberculatus (waterhemp), an increasingly problematic weed of agronomic production systems throughout the midwestern United States (2–4).
PPO is the last common enzyme in the tetrapyrrole biosynthetic pathway that produces heme and chlorophyll (5). In plants, chlorophyll biosynthesis takes place exclusively in plastids, whereas heme is produced in both plastids and mitochondria (6, 7). In both organelles, PPO converts protoporphyrinogen IX (protogen IX) to protoporphyrin IX (proto IX) (8). Two different nuclear genes, PPX1 and PPX2, encode plastid and mitochondrial PPO isozymes, respectively (9, 10). When susceptible plants are treated with PPO inhibitors, the substrate of PPO, protogen IX, accumulates and is exported from the organelles into the cytoplasm (11) where herbicide-insensitive peroxidase-like enzymes in the plasma membrane convert it to proto IX (12). Proto IX accumulates in the cytoplasm and, in the presence of light, induces the formation of singlet oxygen that is damaging to cell membranes (13).
Although PPO-inhibitor-resistant plants have been generated through genetic engineering (14–20), A. tuberculatus populations have evolved resistance from the repeated use of these herbicides in agronomic production systems. The consequence of A. tuberculatus evolving resistance to PPO inhibitors, combined with its already widespread resistance to acetolactate synthase-inhibiting herbicides, is that the only remaining chemical option for its control following emergence in Glycine max (soybean) production systems is glyphosate, which requires the planting of glyphosate-resistant varieties (4). Although the molecular mechanisms of evolved resistance to many herbicides have been identified, such a mechanism has not yet been elucidated for resistance to PPO inhibitors.
Results
Inheritance of PPO Inhibitor Resistance.
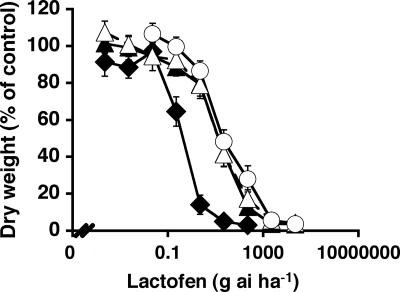
To characterize the resistance mechanism, plants from a PPO-inhibitor-resistant (R) A. tuberculatus biotype were reciprocally crossed with wild-type [herbicide-susceptible (S)] plants to create F1 lines, followed by subsequent crossing to generate F2 and backcross (BC) lines. In response to the PPO inhibitor lactofen, the resultant A. tuberculatus lines segregated for resistance in ratios similar to those expected for a single genetic unit of inheritance (Table 1). Furthermore, plants from lines that were homozygous or heterozygous for resistance survived 53-fold or 31-fold higher doses of lactofen, respectively, when compared with S plants (Fig. 1 and Table 2 and Fig. 6, which are published as supporting information on the PNAS web site). Thus, resistance to lactofen was inherited as a single, incompletely dominant gene.
Table 1.
Inheritance of resistance to the PPO inhibitor lactofen in A. tuberculatus
| Male parent | Female parent | N | No. observed |
Expected ratio (R:S) | χ2 | P value | |
|---|---|---|---|---|---|---|---|
| R | S | ||||||
| F1(R) | F1(R) | 400 | 297 | 103 | 3:1 | 0.120 | 0.7290 |
| S | 200 | 98 | 102 | 1:1 | 0.080 | 0.7772 | |
| R | 200 | 200 | 0 | 1:0 | 0 | 1 | |
| F1(S) | F1(S) | 400 | 304 | 96 | 3:1 | 0.213 | 0.6441 |
| S | 200 | 109 | 91 | 1:1 | 1.620 | 0.2030 | |
| R | 200 | 200 | 0 | 1:0 | 0 | 1 | |
F1 plants were obtained from reciprocal crosses between a resistant (R) and a sensitive (S) biotype [F1(R), female parent was R; F1(S), female parent was S]. Plants from the F2 and BC lines were treated with lactofen at 110 g ai·ha−1 plus 1% (vol/vol) COC and scored as R or S 15 days after treatment. The expected segregation ratio of R to S responses assumes a single genetic unit of inheritance.
Fig. 1.
Dose–response curves of different A. tuberculatus lines to the PPO inhibitor lactofen. Lactofen was foliar-applied to greenhouse-grown plants from the S (filled diamonds), R (open circles), F1(R) (open triangles), or F1(S) (filled triangles) A. tuberculatus lines. Data were collected 15 days after treatment. Vertical bars represent ± SEM (n = 12).
Molecular Characterization of PPX Genes.
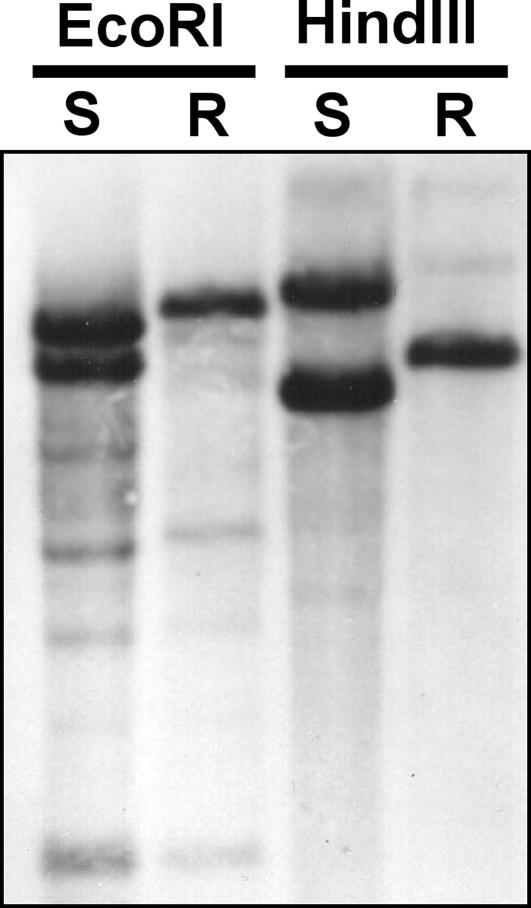
cDNA sequences that encode PPO isozymes were obtained from R and S A. tuberculatus plants, but with unexpected results. From S plants, cDNA sequences for PPX1, PPX2, and a longer version of PPX2, PPX2L, were identified (GenBank accession nos. DQ386112, DQ386113, and DQ386114, respectively) (Fig. 7, which is published as supporting information on the PNAS web site). Comparison of translated sequences of PPX2 and PPX2L indicated that they shared 98% amino acid identity, with the exception of a 30-aa extension in the 5′ end that was unique to PPX2L. This extension is predicted to encode a signaling sequence for plastid import (21). Thus, the PPX2L gene isolated from A. tuberculatus likely encodes both plastid- and mitochondria-targeted PPO isoforms due to the presence of alternate in-frame initiation codons, a phenomenon that was reported previously for Spinacia oleracea (spinach) PPX2 (10). In comparison, PPX1 shared 26% and 25% amino acid identity with PPX2 and PPX2L, respectively, and thus is an evolutionarily distinct isozyme. From R plants, only PPX1 and PPX2L genes (GenBank accession nos. DQ386115 and DQ386116, respectively) were identified on the basis of cDNA sequencing. To confirm the lack of PPX2 identification in R plants, Southern blot analysis was performed by using genomic DNA (gDNA) probed with a fragment of PPX2L. Probing with the fragment of PPX2L identified two major bands (presumably PPX2 and PPX2L loci) from S plants but only a single major band (presumably the PPX2L locus) from R plants, thus confirming the results obtained from sequencing efforts (Fig. 2).
Fig. 2.
Southern blot of A. tuberculatus gDNA probed with a fragment of PPX2L. DNA was isolated from plants that were derived from the S or R biotype and digested with EcoRI or HindIII.
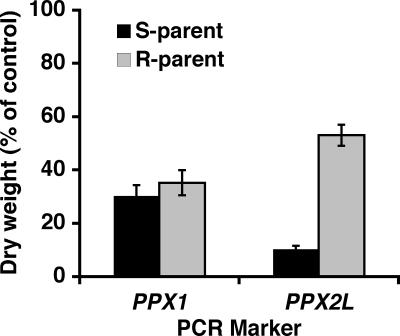
To determine whether PPX1 or PPX2L mediated PPO inhibitor resistance, PCR-based molecular markers were used to follow the inheritance of alleles of these two genes in A. tuberculatus lines segregating 1:1 for R or S responses to lactofen. The molecular marker for PPX2L was significantly correlated with lactofen responses (P < 0.0001), whereas the marker for PPX1 was not (P = 0.4278) (Fig. 3). In other words, plants were resistant to lactofen only if they inherited the PPX2L allele from the R parent. Results of molecular marker studies focused further efforts toward differences among PPX2L alleles.
Fig. 3.
PCR-based molecular marker analysis of PPX1 or PPX2L alleles. A. tuberculatus plants used in the study were derived from F1 hybrids backcrossed to the S parent (BCS). Markers were used to determine whether the F1-derived pollen carried an S or R parental allele. BCS plants were treated with lactofen at 110 g ai·ha−1 plus 1% (vol/vol) COC and harvested 15 days after treatment. Vertical bars represent ± SEM (PPX1, n = 42 or 40 for S or R parental alleles, respectively; PPX2L, n = 39 or 49 for S or R parental alleles, respectively).
Inspection of the inferred amino acid sequences of PPX2L among S and R plants revealed two amino acid polymorphisms that were correlated with resistance. In an attempt to identify only a single amino acid polymorphism, additional R and S plants were sequenced from independently identified A. tuberculatus biotypes (GenBank accession nos. DQ386117 and DQ386118). Sequencing results and subsequent comparisons identified three additional amino acid polymorphisms (five total); however, only one, a glycine deletion at position 210 (ΔG210), was consistently polymorphic between all R and S plants analyzed (Fig. 8, which is published as supporting information on the PNAS web site). PPX2L also was sequenced by using gDNA as a template (GenBank accession nos. DQ394875 and DQ394876 for S and R plants, respectively) to further confirm the existence of the 3-bp deletion corresponding to the G210 codon. Alignment of gDNA and cDNA sequences of PPX2L identified the codon corresponding to the G210 residue in the ninth exon when starting from the 5′ end (Fig. 9, which is published as supporting information on the PNAS web site). The 3-bp deletion was also identified in PPX2L gDNA sequences of R plants, indicating that the ΔG210 mutation in PPO2L was not the result of an error introduced during mRNA processing.
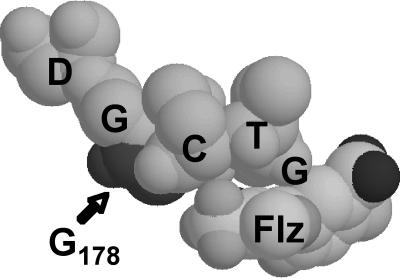
The ΔG210 mutation was also assessed by using the resolved protein structure of PPO2 from Nicotiana tabacum (tobacco) as a reference (22, 23). The equivalent amino acid to G210 of A. tuberculatus PPO2L (G178 of N. tabacum PPO) was located near the herbicide-binding site, thus supporting the prediction that the G210 deletion was responsible for herbicide resistance (Fig. 4).
Fig. 4.
Selected amino acid residues of N. tabacum PPO2 in proximity to the herbicide-binding site. A. tuberculatus plants resistant to PPO inhibitors are missing a glycine residue equivalent to G178 of N. tabacum. This amino acid deletion is predicted to hinder PPO inhibitor binding. Amino acid residues: D, aspartic acid; G, glycine; C, cysteine; and T, threonine. PPO-inhibiting herbicide: Flz, fluazolate.
Functional Complementation.
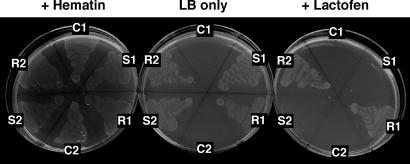
Complementation assays used a hemG (PPO) mutant strain of Escherichia coli, SASX38, (24) to assess the effect of the G210 deletion toward herbicide responses. The SASX38 strain grows very slowly unless supplied with exogenous heme or rescued with an alternative source of PPO. Furthermore, because wild-type E. coli is naturally tolerant to PPO inhibitors, use of the SASX38 strain enabled a relatively direct assay for herbicide sensitivity of the S and putative R PPO2Ls from A. tuberculatus (25, 26). The SASX38 E. coli strain was transformed with plasmid constructs encoding PPO2L proteins that differed only in the presence/absence of G210. Both constructs were able to rescue growth of the SASX38 E. coli strain, thus indicating that both PPX2L genes encoded functional proteins (Fig. 5). However, supplementation of the growth medium with lactofen dramatically inhibited growth of E. coli transformed with the wild-type PPX2L but not E. coli transformed with the ΔG210 PPX2L (Fig. 5). Thus, the 3-bp deletion in PPX2L resulting in deletion of a glycine at position 210 of PPO2L was sufficient to confer resistance to lactofen.
Fig. 5.
PPO expression in a hemG mutant strain of E. coli. E. coli were grown on LB medium alone or supplemented with hematin (20 μg·ml−1) or lactofen (100 nM). E. coli isolates were as follows: C1 and C2, nontransformed controls; S1 and S2, transformed with a vector encoding A. tuberculatus-derived PPO2L with glycine at position 210; and R1 and R2, transformed with a vector encoding identical PPO2L with the exception of a deletion of glycine at position 210.
Discussion
Seven different mechanisms of PPO inhibitor resistance have been proposed for plants (27). Two of these mechanisms include either enhanced metabolic degradation of the herbicide or an alteration of the herbicide target site, which together constitute the majority of mechanisms for herbicide resistance in weed species. Of these, an altered herbicide target enzyme (PPO) was investigated, based on previous characterization of R A. tuberculatus plants (4). It was later determined, in an independently identified PPO-inhibitor-resistant A. tuberculatus population, that enhanced metabolism was not responsible for resistance (28).
The mechanism of PPO inhibitor resistance that was selected within natural populations of A. tuberculatus was a codon deletion in a gene encoding PPO. Although alterations of herbicide target proteins are common mechanisms for conferring resistance, several characteristics of this specific mechanism merit highlighting. First, PPO inhibitors have two herbicide target sites in plants [i.e., in plastids and in mitochondria (8)]; therefore, in order for target-site resistance to occur, two altered genes would need to be selected. However, A. tuberculatus plants have overcome this obstacle by means of mutation in a single gene (PPX2L) that is predicted to encode both plastidic and mitochondrial PPO isoforms. Second, the specific alteration of PPO2L that confers resistance to PPO-inhibiting herbicides is an amino acid deletion, rather than a substitution, resulting from a 3-bp deletion in the gDNA. Although intentional selection for resistance to PPO inhibitors identified amino acid substitutions that conferred resistance (20, 29), the codon-deletion approach revealed by A. tuberculatus is instructive of an alternative approach to achieve resistance. Third, the R biotype was found to be resistant to multiple chemical families of PPO inhibitors, albeit at different levels (4), indicating that the ΔG210 mutation confers resistance to all PPO inhibitors. Finally, the fact that R A. tuberculatus plants lacked one of the PPO genes (PPX2) found in plants from the S biotype is curious and requires further research. However, the absence of PPX2 in the R biotype likely is not related to the resistance phenotype, inasmuch as (i) resistance was (incompletely) dominant and exhibited single-locus inheritance, (ii) PPX2L cosegregated with resistance, and (iii) the ΔG210 mutation was sufficient to confer lactofen insensitivity.
Although the origin of the G210 codon deletion of PPX2L identified in the R A. tuberculatus biotype is uncertain, nucleotide length polymorphisms are not uncommon in this plant species. Codon insertion/deletions (indels) among populations of A. tuberculatus were previously identified in other genes encoding herbicide target proteins [e.g., ALS (acetolactate synthase) and EPSPS (5-enolpyruvylshikimate-3-phosphate synthase)] (W.L.P and P.J.T., unpublished results). Furthermore, other indels in addition to the G210 indel were found among PPX genes in this study. In PPX1 (GenBank accession nos. DQ386112 and DQ386115), there were two additional, adjacent proline codons in the nucleotide sequence from R plants relative to S plants. A codon indel was also identified when PPX2 was compared with PPX2L from S plants (GenBank accession nos. DQ386113 and DQ386114). This indel, like the G210 polymorphism between R and S PPX2Ls, also resulted in a glycine amino acid indel but it was located at a different position (128 nucleotides downstream of the G210 codon). The codon indels observed in A. tuberculatus typically are associated with short, simple sequence repeats (SSRs). The G210 indel in PPX2L is part of a bi-GTG repeat (or a bi-TGG repeat); the PPX2/PPX2L indel is part of a tri-GGA repeat; and the PPX1 indel is part of a hexa-CCT repeat. Because of their high mutability, SSRs are recognized as a means for providing adaptive genetic variation for evolutionary processes (30). Although the numbers of repeats associated with some of the PPX indels are fewer than typically recognized for SSRs, that the indels are found within repeated nucleotides suggests a means for their evolutionary origin.
In regard to PPO-inhibitor-resistant A. tuberculatus in agroecosystems, resistance can be transmitted both maternally and paternally and therefore is able to spread through seed dispersal or, more rapidly, by wind dispersal of pollen. Because A. tuberculatus is a dioecious plant, it is forced to outcross. This obligate outcrossing, combined with a significant level of resistance that is expressed in the heterozygous state (Fig. 1), will make pollen a very effective means for dissemination of the resistance. In addition to dissemination from a single “source” population, resistance to PPO inhibitors could become more widespread in A. tuberculatus populations by independent selection events. In fact, it seems likely that PPO-inhibitor-resistant populations already have evolved independently, given the distinct locations where they have been identified (2–4) and the different PPX2L alleles containing the ΔG210 mutation identified in this study (Fig. 8).
A. tuberculatus is one of the most problematic weeds in agronomic fields throughout the midwestern United States. In particular, the propensity of A. tuberculatus to rapidly evolve herbicide resistance makes its management difficult (4). The herbicide resistance mechanism reported herein illustrates the sophisticated means by which A. tuberculatus can adapt and evolve in response to weed control efforts. With the loss of PPO inhibitors as an effective A. tuberculatus management tool in soybean production, farmers will become even more reliant on glyphosate.
Materials and Methods
Detailed procedures for the generation and analysis of A. tuberculatus lines and for herbicide dose–response experiments and degree-of-dominance calculations are given in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
A. tuberculatus Biotypes.
The R biotype used in this study was derived from an A. tuberculatus population originally collected in Adams County, Illinois, and confirmed resistant to PPO-, acetolactate synthase-, and photosystem II-inhibiting herbicides (4). The S biotype was collected in Wayne County, Illinois, and was identified in previous experiments to be susceptible to all herbicides tested (31). A. tuberculatus plants derived from the original Adams County population that were PPO-inhibitor-susceptible (S-BioAC), and those from a PPO-inhibitor-resistant biotype collected in Clinton County, Illinois (R-BioCC), were used for sequencing of PPX2L alleles only.
Plant Culture.
A. tuberculatus seeds for each experiment were sown in flats (surface area = 930 cm2) containing a 1:1:1 mixture of soil:peat:sand. Seedlings for each experiment were transplanted into square plastic pots containing 800 ml of soil plus 0.2% (vol/vol) 14-14-14 Nutricote (Agrivert, New York, NY) when the plants were ≈1 cm in height. Plants were grown in a greenhouse maintained at 28/22°C day/night, with supplemental light (minimum of 800 μmol·m−2·s−1 photon flux at the plant canopy) provided by mercury halide and sodium vapor lamps programmed for a 16-hr photoperiod.
Herbicide Applications.
Herbicide treatments were applied with a compressed-air, moving-nozzle laboratory sprayer equipped with an 80° flat fan nozzle (Teejet; Spraying Systems Co., Wheaton, IL) delivering 187 liters·ha−1 of water at 207 kPa. The nozzle was maintained ≈45 cm above the plant canopy. Plants were returned to the greenhouse immediately after herbicide treatment. All foliar-applied herbicide treatments were made when the A. tuberculatus plants were 10–12 cm in height.
Generation of F1, F2, and BC Lines.
To create F1 lines, A. tuberculatus plants from the R biotype were crossed with plants from the S biotype. F1 lines were created in which the maternal parent was either S [F1(S)] or R [F1(R)]. After maturity, seeds were harvested from each female individually as full-sib lines. F1 male plants were crossed with female plants from the S biotype, R biotype, or F1 full-sibs to create BCS, BCR, or F2 lines, respectively. Separate crosses were conducted using males from F1(S) or F1(R) lines. Each genetic combination was conducted twice with new A. tuberculatus plants, thus constituting a complete replication of the experiment. Crosses were conducted in growth chambers maintained at 28/22°C day/night with fluorescent and incandescent bulbs providing 400 μmol·m−2·s−1 photon flux at the plant canopy, programmed for a 16-hr photoperiod.
Evaluation of F2 and BC Lines.
Inheritance of PPO inhibitor resistance was determined by evaluating R or S responses of plants from F2 and BC lines 15 days after treatment with lactofen at 110 grams of active ingredient per hectare (g ai·ha−1) plus 1% (vol/vol) crop oil concentrate (COC). From each F2 or BC line, 50 plants from each cross (including replicated crosses) were assessed in a completely randomized design. The entire experiment was conducted twice, with a total of 100 plants being assessed from each cross. Responses for each cross were subjected to χ2 analysis to determine whether the responses were due to a single genetic unit of inheritance. Because we observed no differences among replications of the same cross, data obtained from similar crosses were combined.
cDNA Sequencing.
Total RNA was isolated by using young leaf tissue from a single plant from each of the R and S biotypes (32), followed by purification of mRNA (Promega, Madison, WI). Purified mRNA was used to obtain full-length sequences of PPX1 or PPX2 by using 5′ and 3′ RACE (Invitrogen, Carlsbad, CA). Primers were designed based on conserved regions of nucleotide sequences of PPX1 or PPX2 from numerous plant species (9, 10, 33). Sequencing of the resultant fragments facilitated the design of gene-specific primers for A. tuberculatus PPX1 and PPX2 that were used to obtain their full-length sequences. Total RNA was individually isolated from three A. tuberculatus plants each of the R or S biotypes and used to create cDNA in reactions with reverse transcriptase (Invitrogen). PCR was used to amplify PPX1, PPX2, or PPX2L with the following primers: PPX1, forward 5′-GAGAGAGTGCGAGAGAGATGAG-3′ and reverse 5′-CAAGATGCTGGAGCCCTATTGAC-3′; PPX2, forward 5′-GCCATCGCCATTGTCAGTTTAC-3′ and reverse 5′-GAATTACGCGGTCTTCTCATCCAT-3′; and PPX2L, forward 5′-GACAAAATTGGATTCAGAATTTAGC-3′ and reverse 5′-GAATTACGCGGTCTTCTCATCCAT-3′. PCRs contained 1 μl of cDNA; 400 nM each of forward and reverse primers; 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 1.5 mM MgCl2; and 1.0 unit of high-fidelity Taq polymerase (Roche Molecular Biochemicals, Indianapolis, IN) with a 1× concentration of supplied buffer in a final volume of 25 μl. The reactions were subjected to a 3-min incubation at 95°C; 35 cycles of 0.5 min at 95°C, 1 min at 58°C, and 1.5 min at 72°C; and then 5 min at 72°C. Resultant PCR products were isolated by gel electrophoresis, sequenced (34), and compared by using both Sequencher 4.1 and online software (35). Because sequences among plants from the same biotypes were similar, only a single sequence is presented for each gene/biotype combination.
Southern Blot Analysis.
gDNA was isolated from young leaves of A. tuberculatus plants from the S or R biotypes (36). PPO inhibitor responses of each plant were confirmed by treatment with lactofen at 175 g ai·ha−1 plus 1% (vol/vol) COC. Samples were prepared by digesting 7.5 μg of gDNA with 100 units of either EcoRI or HindIII to completion, followed by separation in a 1% (wt/wt) agarose gel, and then were transferred to a nylon membrane (Roche Molecular Biochemicals). The membrane was probed with a digoxigenin-labeled (Roche Molecular Biochemicals) PCR fragment of PPX2L amplified from gDNA isolated from a single S plant. Hybridization and probe detection were performed in accordance with the manufacturer’s instructions.
PCR-Based Molecular Markers.
Inheritance of PPX1 and PPX2L alleles in BCS progeny was studied by treating plants with lactofen at 110 g ai·ha−1 plus 1% (vol/vol) COC. Prior to lactofen applications, tissue samples were obtained from each plant to isolate DNA (37). PCR-based molecular markers were used to identify the parental origin (R or S) of the PPX alleles contributed by the F1 male to the BCS progeny. To differentiate R or S PPX1 alleles, a fragment of genomic PPX1 was amplified by PCR using the forward primer 5′-TGATAAGTCGCTCAATGGAGA-3′ and reverse primer 5′-AGATTTGTAGCACCTCCAATG-3′, followed by BspDI digestion to identify S alleles (i.e., S PPX1 alleles contain a recognition sequence for BspDI, whereas R alleles do not). To identify parent-specific PPX2L alleles, a fragment of genomic PPX2L was amplified by PCR using the forward primer 5′-AAGAGACCTCTTGAGGGCTTC-3′ and the reverse primer 5′-GAATTACGCGGTCTTCTCATCCAT-3′, followed by TfiI digestion to identify S alleles (i.e., S PPX2L alleles contain a recognition sequence for TfiI, whereas R alleles do not). PCRs contained 40 ng of total DNA; 400 nM each of forward and reverse primers; 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 2.0 mM MgCl2; and 1 unit of Taq polymerase (Invitrogen) with a 1× concentration of supplied buffer in a final volume of 20 μl. The reactions were subjected to a 3-min incubation at 95°C; 40 cycles of 0.5 min at 95°C, 1 min at 60°C or 64°C (for reactions with PPX1 or PPX2L primers, respectively), and 1.5 min at 72°C; and then 5 min at 72°C. After PCR amplification, a mixture containing 0.5 unit of the appropriate restriction enzyme with a 1× concentration of supplied buffer in a final volume of 10 μl was added to each reaction. Digests with BspDI were incubated at 37°C for 4 hr; digests with TfiI were incubated at 65°C for 2 hr. PCR products were fractionated in a 1% (wt/wt) agarose gel containing 0.5 μg·ml−1 ethidium bromide and were visualized with UV light.
PPX2L gDNA Sequencing.
gDNA was isolated from leaf tissue of S or R plants (37) to sequence a portion of genomic PPX2L. Primers were designed that flanked the G210 codon of PP02L; then, subsequent sequencing of amplified fragments facilitated the design of new primers until the exon containing the G210 codon was identified. Primer sets (A–D, forward and reverse), starting with the largest fragment, were as follows: A, 5′-GCCATCGCCATTGTCAGTTTAC-3′ and 5′-GGAGCAGTGACAACCACAGCATCA-3′; B, 5′-ATCGATGATCTTGGGCTTCGTG-3′ and 5′-AATGGTAAGGAGTCGCACCAAC-3′; C, 5′-CTTCAAATCCCGCTGCACTA-3′ and 5′-TACTTCTGGAAATGTATGG-3′; and D, 5′-GAGAAAACACAATGCTACTGAA-3′ and 5′-ACAGCCTCCAGAAAATGTTG-3′. PCR amplification, sequencing, and analysis were performed similar to the method used for cDNA sequencing of PPX genes.
Functional Complementation.
A shortened version of PPX2L from the S A. tuberculatus biotype was cloned into a pBAD-TOPO expression vector (Invitrogen) so that translation began at the second ATG start codon (+91). PPX2L cDNA was PCR-amplified by using the forward primer 5′-CAGGAATAAGTAATGGGCAACATTTCTGAG-3′, containing both a ribosome binding site (AGGA) and an ATG start codon, and the reverse primer 5′-GAAGAATTACGCGGTCTTCTCATC-3′ containing a stop codon. To create putative PPO-inhibitor R and S plasmids that would encode proteins differing only in the presence/absence of G210, PPX2L was PCR-amplified from multiple cDNA samples and a region of the gene encompassing an ≈500-bp XhoI/DraIII fragment was sequenced. The 3-bp polymorphism corresponding to the ΔG210 mutation was within this XhoI/DraIII fragment. Two XhoI/DraIII fragments were identified that were identical except for the presence/absence of the G210 codon and a C/T nucleotide polymorphism that was in the third position of a serine codon (and therefore did not alter the encoded protein). These two fragments were each used to replace the corresponding fragment in the pBAD-TOPO PPX2L construct. The region encompassing the replaced fragment was sequenced from the two resulting constructs to confirm the existence of the 3-bp polymorphism and that no other polymorphisms were created during the cloning process. Susceptible and putative R PPO plasmids were used to transform a hemG mutant strain of E. coli, SASX38 (24), kindly provided by Harry Dailey (University of Georgia, Athens, GA). The SASX38 E. coli strain was maintained on LB medium supplemented with 20 μg·ml−1 hematin. Transformation-competent E. coli were prepared by using CaCl2 (38). Transformed colonies of SASX38, and nontransformed controls, were tested for their ability to grow on LB medium alone or supplemented with 20 μg·ml−1 hematin or with the PPO inhibitor lactofen ranging from 0.01 to 100 μM, and incubated at 37°C for 14 hr.
Supplementary Material
Acknowledgments
We thank Danman Zheng for assistance in making the E. coli expression vectors. This work was supported by the Illinois Soybean Program Operating Board.
Glossary
Abbreviations
- PPO
protoporphyrinogen oxidase
- BC
backcross
- gDNA
genomic DNA
- ai
active ingredient
- COC
crop oil concentrate.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ386112–DQ386118, DQ394875, and DQ394876).
See Commentary on page 12215.
References
- 1.Matsunaka S. In: Herbicides: Chemistry, Degradation, and Mode of Action. Kearney P. C., Kaufman D. D., editors. Vol. 2. New York: Marcel Dekker; 1976. pp. 709–739. [Google Scholar]
- 2.Shoup D. E., Al-Khatib K., Peterson D. E. Weed Sci. 2003;51:145–150. [Google Scholar]
- 3.Li J., Smeda R. J., Nelson K. A., Dayan F. E. Weed Sci. 2004;52:333–338. [Google Scholar]
- 4.Patzoldt W. L., Tranel P. J., Hager A. G. Weed Sci. 2005;53:30–36. [Google Scholar]
- 5.Beale S. I., Weinstein J. D. In: Tetrapyrrole Metabolism in Photosynthetic Organisms. Dailey H. A., editor. New York: McGraw–Hill; 1990. pp. 287–391. [Google Scholar]
- 6.Smith A. G., Marsh O., Elder G. H. Biochem. J. 1993;292:503–508. doi: 10.1042/bj2920503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow K. S., Singh D. P., Roper J. M., Smith A. G. J. Biol. Chem. 1997;272:27565–27571. doi: 10.1074/jbc.272.44.27565. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs J. M., Jacobs N. J. Arch. Biochem. Biophys. 1984;229:312–319. doi: 10.1016/0003-9861(84)90157-7. [DOI] [PubMed] [Google Scholar]
- 9.Lermontova I., Kruse E., Mock H. P., Grimm B. Proc. Natl. Acad. Sci. USA. 1997;94:8895–8900. doi: 10.1073/pnas.94.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe N., Che F. S., Iwano M., Takayama S., Yoshida S. J. Biol. Chem. 2001;276:20474–20481. doi: 10.1074/jbc.M101140200. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs J. M., Jacobs N. J. Plant Physiol. 1993;101:1181–1187. doi: 10.1104/pp.101.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H. J., Duke S. O. J. Agric. Food Chem. 1994;42:2610–2618. [Google Scholar]
- 13.Duke S. O., Lydon J., Becerril J. M., Sherman T. D., Lehnen L. P., Matsumoto H. Weed Sci. 1991;39:465–473. [Google Scholar]
- 14.Choi K. W., Han O., Lee H. J., Yun Y. C., Moon Y. H., Kim M., Kuk Y. I., Han S. U., Guh J. O. Biosci. Biotechnol. Biochem. 1998;62:558–560. doi: 10.1271/bbb.62.558. [DOI] [PubMed] [Google Scholar]
- 15.Lee H. J., Lee S. B., Chung J. S., Han S. U., Han O., Guh J. O., Jeon J. S., An G., Back K. Plant Cell Physiol. 2000;41:743–749. doi: 10.1093/pcp/41.6.743. [DOI] [PubMed] [Google Scholar]
- 16.Lermontova I., Grimm B. Plant Physiol. 2000;122:75–83. doi: 10.1104/pp.122.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha S. B., Lee S. B., Lee Y., Yang K., Lee N., Jang S. M., Chung J. S., Jung S., Kim Y. S., Wi S. G., Back K. Plant Cell Environ. 2004;27:79–88. [Google Scholar]
- 18.Jung S., Lee Y., Yang K., Lee S. B., Jang S. M., Ha S. B., Back K. Plant Cell Environ. 2004;27:1436–1446. [Google Scholar]
- 19.Lee Y., Jung S., Back K. Pestic. Biochem. Physiol. 2004;80:65–74. [Google Scholar]
- 20.Li X., Nicholl D. Pest Manage. Sci. 2005;61:277–285. doi: 10.1002/ps.1011. [DOI] [PubMed] [Google Scholar]
- 21.Emanuelsson O., Nielsen H., Brunak S., Heijne G. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 22.Koch M., Breithaupt C., Kiefersauer R., Freigang J., Huber R., Messerschmidt A. EMBO J. 2004;23:1720–1728. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martz E. Trends Biochem. Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
- 24.Sasarman A., Chartrand P., Lavoie M., Tardif D., Proschek R., Lapointe C. J. Gen. Microbiol. 1979;113:297–303. doi: 10.1099/00221287-113-2-297. [DOI] [PubMed] [Google Scholar]
- 25.Li X., Volrath S. L., Nicholl D. B. G., Chilcott C. E., Johnson M. A., Ward E. R., Law M. D. Plant Physiol. 2003;133:736–747. doi: 10.1104/pp.103.026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasarman A., Letowski J., Czaika G., Ramirez V., Nead M. A., Jacobs J. M., Morais R. Can. J. Microbiol. 1993;39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- 27.Dayan F. E., Duke S. O. In: Herbicide Activity: Toxicology, Biochemistry and Molecular Biology. Roe R. M., Burton J. D., Kuhr R. J., editors. Amsterdam: IOS Press; 1997. pp. 11–36. [Google Scholar]
- 28.Shoup D. E., Al-Khatib K. Weed Sci. 2005;53:284–289. [Google Scholar]
- 29.Volrath S. L., Johnson M. A., Ward E. R., Heifetz P. B., inventors. 5,939,602. U.S. Patent. 1999
- 30.Kashi Y., King D. G. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Patzoldt W. L., Tranel P. J., Hager A. G. Crop Prot. 2002;21:707–712. [Google Scholar]
- 32.McCarty D. R. Maize Genet. Coop. Newslett. 1986;60:61. [Google Scholar]
- 33.Narita S., Tanaka R., Ito T., Okada K., Inokuchi H. Gene. 1996;182:169–175. doi: 10.1016/s0378-1119(96)00545-8. [DOI] [PubMed] [Google Scholar]
- 34.Patzoldt W. L., Tranel P. J., Alexander A. L., Schmitzer P. R. Weed Sci. 2001;49:485–490. [Google Scholar]
- 35.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4690. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Short Protocols in Molecular Biology. 4th Ed. New York: Wiley; 1999. pp. 2.11–2.12. [Google Scholar]
- 37.Doyle J. J., Doyle J. L. Focus (Rochester, N.Y.) 1990;12:13–15. [Google Scholar]
- 38.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Woodbury, NY: Cold Spring Habor Lab. Press; 1989. pp. 1.82–1.84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.