Abstract
Despite recent progress in understanding lamellipodia extension, the molecular mechanisms regulating filopodia formation remain largely unknown. Myo10 is a MyTH4-FERM myosin that localizes to the tips of filopodia and is hypothesized to function in filopodia formation. To determine whether endogenous Myo10 is required for filopodia formation, we have used scanning EM to assay the numerous filopodia normally present on the dorsal surfaces of HeLa cells. We show here that siRNA-mediated knockdown of Myo10 in HeLa cells leads to a dramatic loss of dorsal filopodia. Overexpressing the coiled coil region from Myo10 as a dominant- negative also leads to a loss of dorsal filopodia, thus providing independent evidence that Myo10 functions in filopodia formation. We also show that expressing Myo10 in COS-7 cells, a cell line that normally lacks dorsal filopodia, leads to a massive induction of dorsal filopodia. Because the dorsal filopodia induced by Myo10 are not attached to the substrate, Myo10 can promote filopodia by a mechanism that is independent of substrate attachment. Consistent with this observation, a Myo10 construct that lacks the FERM domain, the region that binds to integrin, retains the ability to induce dorsal filopodia. Deletion of the MyTH4-FERM region, however, completely abolishes Myo10's filopodia-promoting activity, as does deletion of the motor domain. Additional experiments on the mechanism of Myo10 action indicate that it acts downstream of Cdc42 and can promote filopodia in the absence of VASP proteins. Together, these data demonstrate that Myo10 is a molecular motor that functions in filopodia formation.
Keywords: actin, cytoskeleton, Myo10, microvilli, cell spreading
The finger-like cellular extensions known as filopodia play important roles in numerous biological processes including growth cone guidance (1), wound-healing (2), angiogenesis (3), and cell–cell signaling (4). Despite these important roles, the molecular mechanisms underlying the formation of filopodia and related structures, such as intestinal microvilli and inner ear stereocilia, are not yet understood (5, 6). Filopodia are known to contain a core of parallel-bundled actin filaments whose barbed ends are located at the filopodial tip, and filopodial growth requires actin polymerization at these barbed ends. The GTPase Cdc42 is a master regulator of filopodia formation (7) and can interact with proteins such as N-WASP (Wiskott-Aldrich Syndrome Protein) to activate Arp2/3 and nucleate new actin filaments (5). Although the branched actin array generated by activated Arp2/3 may be important for initiating filopodia (8), Arp2/3 is not present on mature filopodial actin bundles (9). Thus, other proteins such as VASP and formins are likely to regulate actin polymerization at the tips of filopodia. VASP family proteins, for example, are present at the tips of filopodia and can stimulate filopodia formation, presumably because of their anticapping activity (10, 11). Filopodia formation also involves actin bundling, and fascin appears to serve as a major actin bundling protein in filopodia (8).
Myo10 is a vertebrate-specific MyTH4-FERM myosin that is expressed in most tissues, exhibits a striking localization at the tips of filopodia, and can undergo movements known as intrafilopodial motility within filopodia (12). The Myo10 heavy chain contains a myosin head domain responsible for motor activity (13, 14), a neck domain consisting of 3 IQ motifs that bind to calmodulin light chains, and a unique tail (15). The tail includes a region predicted to form a coiled coil that can form dimers (16), 3 PH domains implicated in phosphatidylinositol 3-kinase signaling, a MyTH4 domain that can bind microtubules (17), and a FERM domain that can bind β-integrins (18). Overexpressing full-length Myo10 increased substrate-attached filopodia by 4-fold (19), which suggests that Myo10 plays an important role in filopodia formation. Myo10 could promote filopodia indirectly by transporting or anchoring integrins at the filopodial tip (18), therefore stabilizing filopodia by enhancing substrate attachment. Myo10 also could induce filopodia more directly, e.g., by functioning as part of a filopodial tip complex or by transporting molecules required for filopodia formation. We show here that Myo10 is a potent inducer of dorsal filopodia and, thus, can induce filopodia independently of effects on substrate attachment. Furthermore, we show that endogenous Myo10 is required for formation of normal levels of dorsal filopodia and acts downstream of Cdc42, thus demonstrating that this MyTH4-FERM myosin functions in filopodia formation.
Results
Expressing Myo10 Induces a Massive Increase in Dorsal Filopodia.
To test whether Myo10 can directly induce filopodia, we used SEM as a high-resolution assay to image the filopodia on the dorsal surfaces of cultured cells (Fig. 1 A–D). Control COS-7 cells transfected with GFP had virtually no filopodia (1 ± 0.4 dorsal filopodia per cell), whereas COS-7 cells transfected with GFP-Myo10 exhibited a massive increase in dorsal filopodia (553 ± 88). COS-7 cells that expressed higher levels of GFP-Myo10 elaborated more dorsal filopodia (Fig. 1). These data also show that the substrate-attached filopodia visible along the edges of cells that we (19) and most other researchers have focused on previously by using light microscopy sometimes constitute only a tiny subset of the total filopodia (Fig. 1 C and D). Because dorsal filopodia are not attached to the substrate, analyzing them also simplifies interpretation and avoids the difficulties that arise if substrate-attached filopodia are confounded with retraction fibers.
Fig. 1.
Expressing Myo10 leads to a massive increase in dorsal filopodia. (A and B) The dorsal surfaces of control COS-7 cells transfected with GFP alone are smooth and lack dorsal filopodia. (C and D) Cells transfected with GFP-Myo10, however, exhibit a massive increase in dorsal filopodia. A few substrate-attached filopodia also are visible. (E–G) Fluorescence microscopy demonstrates that the dorsal structures induced by GFP-Myo10 contain known markers of filopodia, including F-actin (E) and fascin (F), which label filopodial shafts, and VASP (G), which is present in filopodial tips. Note that GFP-Myo10 is visible at the tips of the numerous dorsal filopodia induced in these cells and, in some cases, forms streaks extending into the filopodial shaft. Although the SEM images in this figure were obtained from an experiment where FACS was used to isolate transfected cells before SEM, similar results were obtained when fluorescence-correlative SEM was used to image transfected cells (Figs. 6 A–C and 7A, which are published as supporting information on the PNAS web site).
To verify that the Myo10-induced extensions are indeed filopodia, COS-7 cells expressing GFP-Myo10 were stained for filopodial marker proteins. As expected for filopodia, the extensions induced by GFP-Myo10 contain F-actin and fascin along their length and have VASP at their tips (Fig. 1 E–G). To further verify our assay, COS-7 cells were transfected with proteins previously reported to induce filopodia and imaged by SEM. GFP-VASP, GFP-fascin, and constitutively active Cdc42 each induced the formation of numerous dorsal filopodia morphologically similar to those induced by GFP-Myo10 (Fig. 2 A–C). Similar experiments showed that GFP-Myo10 also induces dorsal filopodia in many other cell types (data not shown) including HEK-293 (human embryonic kidney), HUVEC (human umbilical vein endothelial cells), and CAD cells (a mouse neuronal cell line). Myo10 therefore promotes formation of dorsal filopodia in many different cell types and appears to be as effective as known filopodia inducers such as Cdc42 and fascin.
Fig. 2.
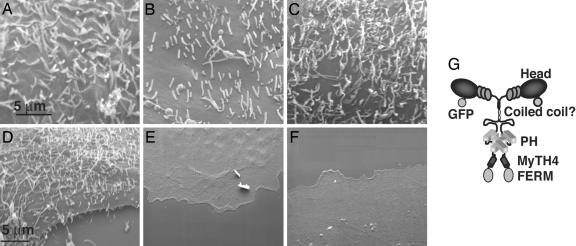
Expressing VASP, fascin, Cdc42, or a Myo10 construct lacking the FERM domain also lead to massive increases in dorsal filopodia. (A–C) Transfecting known inducers of filopodia such as GFP-VASP (A), GFP-fascin (B), and constitutively active GFP-Cdc42(61L) (C) in COS-7 cells all lead to massive increases in dorsal filopodia. Note that in addition to filopodia, the VASP and Cdc42 constructs sometimes also induced small ruffle-like structures. (D–F) Domain-mapping experiments show that GFP-Myo10 lacking the FERM domain retains the ability to induce dorsal filopodia when expressed in COS-7 cells, whereas GFP-Myo10 lacking the MyTH4 (E) and FERM (D) domains fails to induce filopodia. The GFP-Myo10-heavy meromyosin construct, which lacks the PH, MyTH4, and FERM (F) domains, also fails to induce filopodia. (G) A hypothetical model of Myo10 as a dimer illustrating its major domains. The SEM images illustrated here and in all subsequent figures were obtained by using fluorescence-correlative SEM.
To determine which domains of Myo10 are required to induce filopodia, we tested a series of Myo10 deletion constructs for their ability to induce filopodia in COS-7 cells. Importantly, a construct lacking the FERM domain, the region that binds to integrins, was able to induce dorsal filopodia (Fig. 2D). Constructs lacking both the MyTH4 and FERM domains, however, were unable to induce dorsal filopodia (Fig. 2E). Consistent with this observation, a heavy meromyosin-like Myo10 construct that consists of only the head, neck, and coiled coil region (ΔPH-MyTH4-FERM), also failed to induce dorsal filopodia (Fig. 2F). Because all three deletion constructs localize to the tips of substrate-attached filopodia (Fig. 6), the failure of the ΔMyTH4-FERM and the ΔPH-MyTH4-FERM constructs to induce filopodia is not due to an inability to localize to filopodia, but instead is due to deletion of regions in the Myo10 tail required for filopodia formation.
Knockdown of Myo10 Leads to Loss of Dorsal Filopodia.
We next asked whether endogenous Myo10 is necessary for filopodia formation. We thus used siRNA to knockdown Myo10 in HeLa cells, a cell type that elaborates numerous dorsal filopodia under our standard culture conditions. Densitometry of immunoblots demonstrated that the Myo10 siRNA specifically knocked down ≈90% of Myo10 protein by 60–72 h (Fig. 3A). SEM revealed that HeLa cells treated with control siRNA have numerous dorsal filopodia per cell (861 ± 37), whereas cells treated with the Myo10 siRNA exhibit decreased dorsal filopodia (207 ± 23) (Figs. 3 B and C and 7). These data demonstrate that Myo10 is required for formation of normal numbers of dorsal filopodia. It is important to note, however, that the loss of dorsal filopodia was not complete and that live cell imaging showed that knockdown cells still were able to extend occasional filopodia (Movies 1 and 2, which are published as supporting information on the PNAS web site). These results may reflect incomplete knockdown of Myo10, but it could also indicate that there are Myo10-independent pathways for filopodia formation.
Fig. 3.
Inhibiting Myo10 suppresses dorsal filopodia. (A) Immunoblot of HeLa cells treated with control or Myo10 siRNA showing specific knockdown of Myo10. Samples were run with a 4–20% SDS/PAGE, transferred to nitrocellulose, and stained with Ponceau to reveal total protein and then blotted with anti-Myo10 to confirm knockdown. (B) SEM of a HeLa cell from the same experiment treated with control siRNA showing the numerous dorsal filopodia normally present on these cells. (C) SEM of a HeLa cell from the same experiment treated with Myo10 siRNA showing the loss of dorsal filopodia induced by Myo10 siRNA. (D) SEM of a HeLa cell illustrating the loss of dorsal filopodia observed in HeLa cells expressing the dominant-negative GFP-Myo10 coiled-coil construct.
Expressing a Dominant-Negative GFP-Myo10 Coiled-Coil Construct Also Leads to Loss of Dorsal Filopodia.
As an independent strategy to confirm that Myo10 functions in filopodia formation, we also developed a dominant-negative approach to inhibit Myo10. Because we had found that a head-neck construct fails to localize to filopodial tips whereas a longer construct that includes the coiled coil does localize to filopodial tips (19), we reasoned that Myo10 is likely to function as a dimer and that overexpression of the coiled-coil region therefore might act as a dominant-negative. We thus generated a GFP-tagged construct from the putative coiled-coil region of human Myo10. Like Myo10 knockdown cells, HeLa cells transfected with the GFP-Myo10 coiled-coil exhibit decreased dorsal filopodia (Fig. 3D). Quantitative experiments revealed that HeLa cells transfected with GFP had 851 ± 51 dorsal filopodia per cell, whereas cells transfected with GFP-Myo10 coiled coil had only 331 ± 62 (Fig. 7). It should be noted that the GFP-Myo10 coiled coil did not localize to filopodial tips and that expressing a “control” coiled coil from chicken Myo5a did not reduce dorsal filopodia (data not shown). The dominant-negative experiments thus provide independent confirmation that Myo10 is required for formation of normal numbers of dorsal filopodia.
Inhibiting Myo10 Increases Cell Spreading.
In addition to decreased dorsal filopodia, the most obvious phenotype of HeLa cells treated with siRNA to Myo10 and replated overnight was an ≈4-fold increase in cell spreading (Fig. 8, which is published as supporting information on the PNAS web site). A similar increase in spread area was observed in HeLa cells transfected with GFP coiled coil and replated overnight. Because these results raised the possibility that decreases in dorsal filopodia are associated with increases in cell spreading, we tested whether increases in dorsal filopodia are associated with decreases in cell spreading. We thus transfected COS-7 cells with GFP-Myo10 to induce dorsal filopodia and replated them for ≈12 h. These cells exhibited a ≈3-fold decrease in their spread area. Although the precise basis of this effect is not yet clear, it is not due to changes in cell volume, and expressing GFP-VASP or GFP-fascin led to similar decreases (Figs. 7 and 8). It thus is likely that the decreased cell spreading is a consequence of the massive increase in dorsal filopodia induced by all three constructs rather than a specific effect of Myo10 expression.
Myo10 Acts Downstream of Cdc42.
To dissect the molecular mechanisms by which Myo10 induces filopodia, we next investigated the relationship between Myo10 and Cdc42, a master regulator of filopodia formation. We first verified that constitutively active Cdc42 induces dorsal filopodia and that dominant-negative Cdc42 suppresses dorsal filopodia (Fig. 2C and data not shown). To determine whether Myo10 function requires Cdc42, COS-7 cells were cotransfected with cyan fluorescent protein-Myo10 to induce dorsal filopodia and dominant-negative GFP-Cdc42 to inhibit Cdc42. These cells elaborated numerous dorsal filopodia, indicating that Myo10 acts either independently or downstream of Cdc42 (Fig. 9, which is published as supporting information on the PNAS web site). To test whether Myo10 acts downstream of Cdc42, HeLa cells were treated with siRNA to deplete Myo10 and then transfected with constitutively active Cdc42. Constitutively active Cdc42 was unable to induce filopodia in the absence of Myo10 (Fig. 4 A and B), suggesting that Myo10 functions downstream of Cdc42. Importantly, the loss of dorsal filopodia in HeLa cells treated with siRNA to Myo10 could be rescued by transfection with the bovine GFP-Myo10 construct (Fig. 4 C and D), which also provides additional evidence for the specificity of the Myo10 siRNA. Although GFP-fascin also was able to induce numerous filopodia in Myo10 siRNA cells, results with GFP-VASP were less clear (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
Myo10 acts downstream of Cdc42. (A) Like untreated HeLa cells, HeLa cells treated with a control siRNA and then transfected with constitutively active GFP-Cdc42 exhibit numerous dorsal filopodia. (B) Parallel samples treated with the Myo10 siRNA, however, have very few dorsal filopodia, even in the presence of constitutively active GFP-Cdc42, indicating that Myo10 functions downstream of Cdc42. (C and D) The loss of dorsal filopodia induced by the siRNA to human Myo10 (C) can be rescued by transfection with bovine GFP-Myo10 (D). Note that the constitutively active GFP-Cdc42(61L) construct used here led to a massive induction of filopodia in other situations (Fig. 2C).
Myo10 Can Induce Dorsal Filopodia Independently of VASP Proteins.
Finally, because VASP is present at the tips of filopodia and can induce dorsal filopodia, we also tested whether Myo10 could induce filopodia in MVD7 cells, a cell line engineered to lack all three members of the VASP family (10). Like COS-7 cells, control MVD7 cells transfected with GFP alone virtually had no dorsal filopodia (Fig. 5A). MVD7 cells expressing GFP-Myo10, however, elaborated numerous dorsal filopodia (Fig. 5B). This result clearly demonstrates that Myo10 can induce dorsal filopodia in the absence of VASP proteins.
Fig. 5.
Myo10 can induce dorsal filopodia in the absence of VASP family proteins. (A) Ena/VASP null cells (MVD7) transfected with GFP alone exhibit very few dorsal filopodia. (B) Ena/VASP null cells transfected with GFP-Myo10, however, exhibit numerous dorsal filopodia, thus demonstrating that Myo10 does not require VASP proteins for its filopodia promoting activity.
Discussion
The SEM data presented here demonstrate that Myo10 is a remarkably potent promoter of dorsal filopodia. These data also demonstrate that Myo10 can promote filopodia via a mechanism that does not involve stabilization of substrate-attached filopodia. Consistent with this mechanism, deletion of Myo10's FERM domain (the region that binds to integrins) did not impair Myo10's ability to induce filopodia. Thus, although the FERM domain and integrin binding may facilitate the formation of substrate-attached filopodia (18), Myo10 also can induce dorsal filopodia independently of substrate attachment. Deletion of both the MyTH4 domain and the FERM domain of Myo10, however, led to a complete loss of Myo10's ability to induce dorsal filopodia, even though this construct was able to localize to the tips of substrate-attached filopodia. This result suggests that the MyTH4 domain, one of the defining features of the MyTH4-FERM myosins, plays an important role in filopodia formation. Because deletion constructs that lack the Myo10 motor domain fail to localize to filopodial tips and do not induce filopodia (19), our results support a model for Myo10 function where the motor domain is required to properly localize the tail domain, and a properly localized tail is required for filopodia formation.
We also show here that endogenous Myo10 is necessary for formation of normal numbers of dorsal filopodia by using two independent strategies, Myo10 siRNA and a dominant-negative construct. These results, together with the data showing that Myo10 is a potent inducer of filopodia, demonstrates that Myo10 functions as a molecular motor for filopodia formation. Our work also raises the question of how Myo10 acts to promote filopodia. Our data indicate that Myo10 acts downstream of Cdc42, a regulator of filopodia that acts upstream of actin nucleators such ARP2/3 (20) and formins (21) and also stimulates actin bundling (6). Because Myo10 is an actin-based motor and its motor is necessary for filopodia induction, Myo10 may act on actin generated downstream of Cdc42 action. In the convergent elongation model of filopodia formation, branched actin filaments nucleated by Arp2/3 associate via their tips to form “Λ-precursors” proposed to initiate filopodia (8). Although VASP is present at the tips of Λ-precursors (8), the protein(s) that lead the tips of actin filaments to associate with one another in Λ-precursors remain unknown. Like VASP, Myo10 is present at the tips of nascent filopodia as soon as they can be detected (19). As expected for a component of Λ-precursors, puncta of GFP-Myo10 recently have been observed to move laterally along the leading edge, where they collide and fuse (22). Because our data show that Myo10 can induce dorsal filopodia in the absence of VASP proteins, Myo10 may be a component of Λ-precursors and, thus, function in filopodial initiation. If Myo10 promotes filopodia by “focusing” the barbed ends of actin filaments into Λ-precursors, our results show that Myo10's heads are not sufficient for this activity because the heavy meromyosin-like construct consisting only of the head, neck, and coiled coil is unable to induce dorsal filopodia (Fig. 2F). It also should be noted that although convergent elongation provides a useful conceptual model for filopodia formation, the nature of the actin network underlying dorsal filopodia is not yet clear and it may differ from the dendritic array observed at the leading edge. It thus will be important to consider other mechanisms by which Myo10 could promote filopodia, such as by delivering/localizing materials required for polymerization to the filopodial tip, interacting with formins, or by pushing the plasma membrane away from the ends of growing actin filaments to facilitate monomer insertion.
Conserved Functions of MyTH4-FERM Myosins in Filopodia and Related Structures.
Like Myo10, other MyTH4-FERM myosins may have similar functions in the formation of filopodia and related structures such as microvilli and stereocilia. Experiments with human Myo15, a MyTH4-FERM myosin that is expressed in the inner ear, show that it localizes to the tips of stereocilia (23, 24), is necessary for proper stereociliary elongation, and exhibits movements within filopodia strikingly similar to the intrafilopodial motility of Myo10 (25). Loss of a Drosophila MyTH4-FERM myosin, myosin-VIIa, is the basis of crinkled, a usually lethal mutation where escapers exhibit defects in bristles and other structures derived from actin bundles (26). Another MyTH4-FERM myosin, myosin-VIIb, localizes to the microvilli of secretory epithelial cells (27). Finally, deletion of Dictyostelium myosin-VII leads to a 90% loss of filopodia and dramatic defects in adhesion and phagocytosis (28, 29). Together with the work on Myo10 presented here, these data provide strong evidence that MyTH4-FERM myosins have ancient and highly conserved functions in membrane–cytoskeleton interactions underlying the formation of filopodia and related structures.
Materials and Methods
Constructs.
Human GFP-fascin in pEGFP-C1 (30), human GFP-VASP in pEGFP-N1 (8), untagged bovine Myo10 in pcDNA3.1, and the bovine GFP-Myo10, GFP-Myo10-heavy meromyosin, GFP-Myo10 tail, and GFP-Myo10 headless constructs in pEGFP-C2 have been described in ref. 19. The bovine GFP-Myo10 construct was converted to cyan fluorescent protein-Myo10 by replacing GFP with cyan fluorescent protein. The bovine GFP-Myo10 ΔFERM construct in pEGFP-C2 includes amino acids 1–1916, and the GFP-Myo10 ΔMyTH4-FERM construct in pEGFP-C2 includes amino acids 1–1523. The GFP-Myo10 coiled-coil construct in pEGFP-C2 includes amino acids 812–946 and was generated by PCR from human Myo10 (31), and the control GFP-Myo5a coiled-coil construct includes amino acids 913-1116 of chicken Myo5a. The dominant-negative human GFP-Cdc42(15A) (32) and the constitutively active GFP-Cdc42(61L) in pEGFP-C3 were generous gifts of Keith Burridge (University of North Carolina, Chapel Hill). Because the GFP-Cdc42(15A) construct appeared to be more effective than a Myc-Cdc42(N17) construct in suppressing dorsal filopodia in HeLa cells, the 15A construct was used here.
Transfection.
All cells were transfected with cDNA constructs by using Polyfect (Qiagen, Valencia, CA) except for MVD7 cells, which were transfected by using the Amaxa nucleoporation protocol for mouse embryonic fibroblasts. After the overnight transfections, cells were trypsinized and replated onto glass coverslips for 12 h in the presence of serum before fixation. For fluorescence-correlative SEM, the cells were plated onto 12-mm gridded round glass coverslips, (Eppendorf-USA, Westbury, NY), whereas cells sorted by FACS were plated onto 12-mm round glass coverslips. Although COS-7 cells normally lack dorsal filopodia, during mitosis, they round up and elaborate numerous dorsal filopodia, so mitotic cells were excluded from our analysis (≈3% of cells). Quantitative immunoblotting with anti-Myo10 indicated that COS-7 cells express ≈65% as much endogenous Myo10 as HeLa cells and that the average level of overexpression with GFP-Myo10 in COS-7 cells was ≈100-fold. Similar blots indicated that the GFP-Myo10 coiled coil was overexpressed at least ≈30 fold, and blots with anti-GFP indicated that the GFP-Myo5a coiled coil was expressed at approximately the same level.
Knockdown of Myo10 by Using siRNA.
A synthetic siRNA targeting human Myo10 (5′-AAGTGCGAACGGCAAAAGAGA-3′) that differs at 7 positions from bovine Myo10 and a control siRNA (5′-AATTCTCCGAACGTGTCACGT-3′) were obtained from Qiagen with fluorescein tags at their 3′ ends. A day before siRNA treatment, ≈100,000 cells per well were plated onto six-well plates at 50–60% confluency and incubated at 37°C for 12 h. Cells were then treated with a final concentration of 110–150 nM siRNA by using RNAifect (Qiagen) following the manufacturer's instructions. The media was replaced ≈16 h after transfection, and fluorescence microscopy was used to verify that ≈100% of the cells had taken up the siRNA. At ≈48 h, the cells from each well were replated, typically into 18 wells of a 24-well plate containing 12-mm glass coverslips. At ≈60–72 h, cells were processed for scanning EM or light microscopy, and parallel samples were assayed by immunoblotting to verify knockdown. For siRNA experiments that also involved transfection with an expression plasmid, siRNA-treated cells were transfected with GFP-Myo10 or constitutively active GFP-Cdc42 constructs at ≈48 h. Cells were allowed to grow overnight after transfection, replated onto glass coverslips for ≈12 h, and then fixed and processed for fluorescence-correlative SEM.
SEM Experiments.
SEM experiments to test the filopodia promoting activity of GFP-Myo10 and other constructs were performed by using three different approaches (1). For preliminary comparative experiments, cells were transfected overnight with Polyfect and then replated for ≈12 h onto 12-mm coverslips. The transfection efficiency for each construct was assessed by fluorescence microscopy of one coverslip and compared with the fraction of cells exhibiting dorsal filopodia on a duplicate coverslip prepared for SEM. Because control COS-7 cells normally lack dorsal filopodia, this approach provided a simple and rapid screen for filopodia induction, especially when the transfection efficiency exceeded ≈30%. This method also allowed us to determine that the untagged bovine construct is a potent inducer of dorsal filopodia (2). For the FACS approach, cells in six-well dishes were transfected overnight by using Polyfect, trypsinized, and subjected to FACS. The transfected cells were replated onto 12-mm coverslips for ≈12 h and then prepared for SEM. This approach provided the advantage for scanning EM that 100% of the cells on a given coverslip were transfected (3). For the fluorescence-correlative SEM approach, cells were transfected overnight and then replated for ≈12 h on gridded coverslips. Coverslips were rinsed briefly with PBS, prefixed 3.7% paraformaldehyde for 5 min at room temperature, and then rinsed three times with PBS. To identify and record the positions of individual transfected cells, the coverslips then were placed cell side up in a glass bottom dish (Willco, Amsterdam, The Netherlands) and imaged with fluorescence by using an inverted microscope and a 20 × 0.75 NA dry lens (Nikon, Melville, NY). Coverslips then were prepared for SEM by using standard procedures, and SEM images of the transfected cells were collected. Because this correlative approach worked well even when transfection efficiencies were low and it provided the additional internal control of untransfected cells on each coverslip, it was used for most experiments.
Supplementary Material
Acknowledgments
We thank Keith Burridge and Krister Wennerberg (University of North Carolina) for supplying Cdc42 constructs; Frank Gertler for GFP-VASP and MVD7 cells (Massachusetts Institute of Technology, Boston, MA); Josephine Adams for GFP-fascin (Cleveland Clinic, Cleveland, OH); Aurea Sousa for assistance with siRNA; Hal Mekeel and Robert Bagnell for assistance with EM; and Jonathan Berg, Omar Quintero, Damon Jacobs, and Julian Miller for help and suggestions. A.B.B. was supported by a predoctoral fellowship from the American Heart Association. This research was supported by National Institues of Health Grant R01-DC03299 (to R.E.C.).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Supporting Information.
Supporting Materials and Methods, which is published as supporting information on the PNAS web site, provides additional details on the constructs, cell culture conditions, and microscopy procedures used here.
References
- 1.O’Connor T. P., Duerr J. S., Bentley D. J. Neurosci. 1990;10:3935–3946. doi: 10.1523/JNEUROSCI.10-12-03935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood W., Jacinto A., Grose R., Woolner S., Gale J., Wilson C., Martin P. Nat. Cell Biol. 2002;4:907–912. doi: 10.1038/ncb875. [DOI] [PubMed] [Google Scholar]
- 3.Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., Betsholtz C. J. Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez-Weber F. A., Kornberg T. B. Cell. 2000;103:189–192. doi: 10.1016/s0092-8674(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 5.Pollard T. D., Borisy G. G. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 6.Faix J., Rottner K. Curr. Opin. Cell Biol. 2005;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Nobes C. D., Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 8.Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svitkina T. M., Borisy G. G. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bear J. E., Loureiro J. J., Libova I., Fassler R., Wehland J., Gertler F. B. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 11.Lebrand C., Dent E. W., Strasser G. A., Lanier L. M., Krause M., Svitkina T. M., Borisy G. G., Gertler F. B. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 12.Sousa A. D., Cheney R. E. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Kovacs M., Wang F., Sellers J. R. J. Biol. Chem. 2005;280:15071–15083. doi: 10.1074/jbc.M500616200. [DOI] [PubMed] [Google Scholar]
- 14.Homma K., Ikebe M. J. Biol. Chem. 2005;280:29381–29391. doi: 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 15.Berg J. S., Derfler B. H., Pennisi C. M., Corey D. P., Cheney R. E. J. Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 16.Knight P. J., Thirumurugan K., Yu Y., Wang F., Kalverda A. P., Stafford W. F., III, Sellers J. R., Peckham M. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 17.Weber K. L., Sokac A. M., Berg J. S., Cheney R. E., Bement W. M. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., Berg J. S., Li Z., Wang Y., Lang P., Sousa A. D., Bhaskar A., Cheney R. E., Stromblad S. Nat. Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 19.Berg J. S., Cheney R. E. Nat. Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 20.Higgs H. N., Pollard T. D. J. Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. Curr. Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- 22.Sousa A. D., Berg J. S., Robertson B. W., Meeker R. B., Cheney R. E. J. Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 23.Rzadzinska A. K., Schneider M. E., Davies C., Riordan G. P., Kachar B. J. Cell Biol. 2004;164:887–897. doi: 10.1083/jcb.200310055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belyantseva I. A., Boger E. T., Friedman T. B. Proc. Natl. Acad. Sci. USA. 2003;100:13958–13963. doi: 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belyantseva I. A., Boger E. T., Naz S., Frolenkov G. I., Sellers J. R., Ahmed Z. M., Griffith A. J., Friedman T. B. Nat. Cell Biol. 2005;7:148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- 26.Kiehart D. P., Franke J. D., Chee M. K., Montague R. A., Chen T. L., Roote J., Ashburner M. Genetics. 2004;168:1337–1352. doi: 10.1534/genetics.104.026369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z.-Y., Hasson T., Zhang D.-S., Schwender B. J., Derfler B. H., Mooseker M. S., Corey D. P. Genomics. 2001;72:285–296. doi: 10.1006/geno.2000.6456. [DOI] [PubMed] [Google Scholar]
- 28.Tuxworth R. I., Weber I., Wessels D., Addicks G. C., Soll D. R., Gerisch G., Titus M. A. Curr. Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 29.Titus M. A. Curr. Biol. 1999;9:1297–1303. doi: 10.1016/s0960-9822(00)80051-2. [DOI] [PubMed] [Google Scholar]
- 30.Adams J. C., Schwartz M. A. J. Cell Biol. 2000;150:807–822. doi: 10.1083/jcb.150.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers M. S., Strehler E. E. J. Biol. Chem. 2001;276:12182–12189. doi: 10.1074/jbc.M010056200. [DOI] [PubMed] [Google Scholar]
- 32.Reuther G. W., Lambert Q. T., Booden M. A., Wennerberg K., Becknell B., Marcucci G., Sondek J., Caligiuri M. A., Der C. J. J. Biol. Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.