Abstract
Active nucleocytoplasmic transport of protein and RNA in eukaryotes depends on the Ran-GTPase system to regulate cargo–receptor interactions. Several viruses, including the RNA picornaviruses, encode factors that alter nuclear transport with the aim of suppressing synthesis of antiviral factors and promoting viral replication. Picornaviruses in the cardiovirus genus express a unique 67-aa Leader protein (L), known to alter the subcellular distribution of IFN regulatory proteins targeted to the nucleus. We report here that L binds directly to Ran and blocks nuclear export of new mRNAs. In Xenopus egg extracts, recombinant L also inhibits mitotic spindle assembly, a RanGTP function crucial to cell-cycle progression. We propose that L inhibits nucleocytoplasmic transport during infection by disrupting the RanGDP/GTP gradient. This inhibition triggers an efflux of nuclear proteins necessary for viral replication and causes IFN suppression. To our knowledge, L is the first viral picornaviral protein to interact directly with Ran and modulate the Ran-dependent nucleocytoplasmic pathway.
Keywords: encephalomyocarditis virus, Leader protein, traffic inhibition
The eukaryotic nucleus provides a unique environment to separate DNA replication, transcription, and RNA processing events from protein synthesis in the cytoplasm. Communication between the nucleus and the cytoplasm occurs through massive nuclear pore complexes (NPCs) that span the nuclear envelope (NE). Protein or RNA transport in either direction is signal-dependent and requires interaction with an extended family of importin β-related receptors to chaperone the traffic (1). Cargo association with these receptors is regulated by the type of guanine nucleotide (GTP or GDP) bound to the small GTPase, Ran (2, 3). Cytoplasmic Ran is GDP-bound, whereas the GTP form predominates in the nucleus. There is a steep concentration gradient of the respective pools across the NPC. During interphase of the cell cycle, the gradient is maintained by Ran-associated factors separated by the NE (2). Guanine nucleotide exchange factor RCC1 is exclusively nuclear and promotes RanGDP/GTP exchange. In the cytoplasm, RanGAP and RanBP1 accelerate the intrinsic slow rate of RanGTP hydrolysis, cycling the complex back to the GDP form (4). Cytoplasmic RanGDP has a low affinity for importin β, allowing the free form of the receptor to bind cargo and traffic through the NPC (5). Inside the nucleus, higher-affinity RanGTP releases the cargo, then escorts the receptor back to the cytoplasm for another cycle (6). If GTP hydrolysis is inhibited, importin β remains bound to RanGTP and unavailable for active protein import. Without import, export of mRNA and ribosomal subunits are also inhibited because these processes require continuous nuclear reuptake of proteins that comprise export-competent ribonucleoproteins (RNPs) (2). Therefore, if the Ran gradient fails, only proteins small enough to diffuse through the NPC (<40–60 kDa) can exchange.
The metabolic bottleneck posed by nuclear trafficking makes the NPC and associated processes vulnerable to attack by viruses intent on impeding signal transduction to the nucleus. NPC abrogation can prevent up-regulation of antiviral genes and the export of detrimental cellular mRNAs or enhance the redistribution of nuclear proteins required for viral replication (7). The family of RNA picornavirus includes a variety of pathogenic agents. Among the better known members are poliovirus, rhinovirus, and foot-and-mouth disease virus. Others include the Coxsackie viruses, hepatitis A, swine vesicular disease, and cardioviruses like Mengo virus, encephalomyocarditis virus (EMCV), and Theiler's murine encephalomyelitis virus (8). These positive-sense RNA genomes direct a life cycle that is predominantly cytoplasmic and, indeed, can be recapitulated in a test tube in the absence of nuclei (9). During infection, however, picornaviruses are adept at subverting innate cellular immunity traps, crippling the capacity of the cell to mount a defense. Within 2–3 h, infection brings to a halt all cellular mRNA transcription, cap-dependent mRNA translation, antiviral signal transduction, and active protein/RNA exchange between the nucleus and cytoplasm. The shutoff is profound. The viruses replicate with fecundity, and the cell dies before it ever triggers an alarm. Among the molecular processes involved in shutoff, poliovirus and rhinovirus encode a protease, 2Apro, which attacks nucleoporins within the NPC (7, 10, 11). When visualized by electron microscopy, a “bar-like” structure spanning normal NPC channel is found to be missing (7), and its absence correlates with an onset of unregulated efflux of small proteins from the nucleus into the cytoplasm.
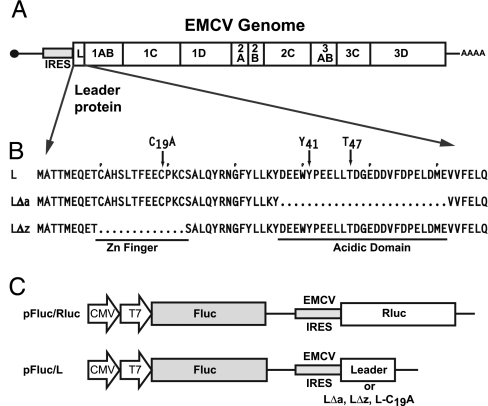
Cardioviruses like EMCV also abrogate nucleocytoplasmic trafficking, but their genomes lack a 2A protease and instead encode another protein, the Leader (L), at the amino terminus of the viral polyprotein (Fig. 1A). The L protein is 67 aa long, with a novel CHCC zinc-finger motif, a highly acidic carboxyl domain (protein pI: 3.8), and no known homologs (Fig. 1). EMCV with L deletions are viable but have attenuated growth phenotypes (12). They are inefficient at shutting off host protein synthesis (13), and they stimulate an increased antiviral IFN activity (14). For Theiler's murine encephalomyelitis virus the related L likewise affects nuclear trafficking. This slightly larger L (76 aa) stimulates IFN transcriptional activator IRF-3, normally a cytoplasmic entity, to redistribute aberrantly between the nucleus and cytoplasm. Simultaneously, polypyrimidine tract-binding protein, a nuclear component of premRNA splicing complexes, redistributes to the cytoplasm and is usurped into viral replication machinery. As a result, viral replication is enhanced by the cytoplasmic availability of polypyrimidine tract binding protein, and the cell is unable to mount a viable IFN-dependent antiviral response to the infection. The antitrafficking activities of EMCV and Theiler's murine encephalomyelitis virus are compromised if either virus harbors a mutation in the zinc-finger region of the L protein (15). It has been proposed that the cardiovirus L, like the poliovirus/rhinovirus 2Apro, disrupts the integrity of the NPC, leading to the leakage of nuclear proteins by passive diffusion (16). But it is difficult to envision how a small protein, lacking any known enzymatic activity, could effectively attack the massive NPC with the scant copy number presented to an infected cell. We hypothesized that an alternative target might be the nuclear transport system composed of Ran-GTPase or the cofactors required for maintaining the RanGTP gradient. We now describe experiments showing that the EMCV L binds directly to Ran and disrupts nucleocytoplasmic trafficking.
Fig. 1.
Virus, Leader, and plasmid maps. (A) A genome map shows that EMCV L is derived from the amino terminus of the polyprotein. (B) The L, L(Δa), and L(Δz) sequences highlight the zinc-finger motif (amino acids 10–23) and acidic domain (amino acids 37–61). (C) Bicistronic cDNAs drive Fluc expression from a capped mRNA and the coexpression Rluc or the L proteins from an internal EMCV IRES.
Results and Discussion
L Alone Is Active.
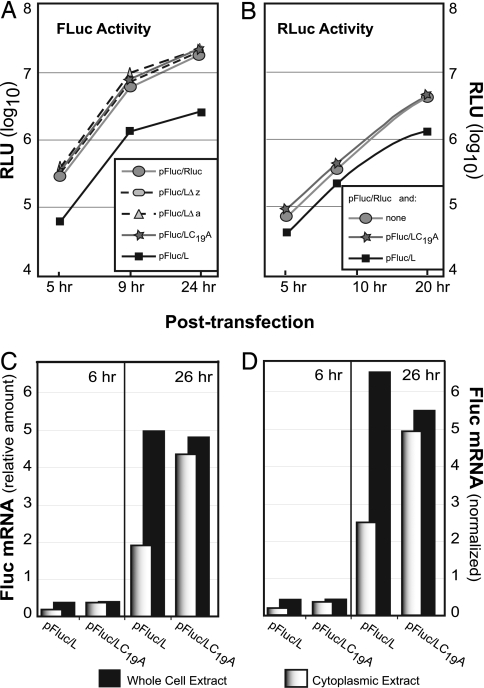
Infection of cells with EMCV blocks active nucleocytoplasmic transport. Mutant viruses with L deletions do not effect the same response (16). Given the potential for synergy with other viral proteins, it has never been clear whether L alone was sufficient for this activity. We reasoned that if L were responsible for virus-induced shutoff, its activity should be maintained as an independent protein. Bicistronic cDNAs encoding EMCV L derivatives and Renilla luciferase (Rluc) or firefly luciferase (Fluc) reporter combinations were created to test this idea. Upon transfection, the cDNAs enter the nucleus and are transcribed from an internal CMV promoter. RNA transcripts are exported back to the cytoplasm, where translation of the 5′ cistron is cap-dependent. Translation of the 3′ cistron is driven by a native EMCV internal ribosome entry site (IRES). Compared to a control cDNA (pFluc/Rluc), Fluc activity from the 5′ cistron was reduced 5- to 8-fold when L was expressed from the 3′ cistron (pFluc/L) (Fig. 2A). Truncated L constructs, missing the zinc-finger domain (Δz) or the acidic domain (Δa), or with a point mutation (C19A) that prevented zinc binding, were not inhibitory. L effects on 3′ cistron expression were measured after simultaneous addition of different effector and reporter cDNAs (Fig. 2B). In this case, with a transfection efficiency of 60–80%, the IRES-driven Rluc activity (pFluc/Rluc) was diminished 3- to 6-fold whenever pFluc/L, but not pFluc/L(C19A), was included in the mix. The results show that L alone, in the absence of other viral proteins, was a potent down-regulator of new protein synthesis from both cistrons. Because cell-free translation systems are not inhibited by the presence of L (12, 17), it is unlikely the measured inhibition was due to a cytoplasmic activity common to both IRES and cap-dependent translation. Rather, the results suggest that L affected some step in the mutual pathway used by all mRNAs, namely, a process inherent to nuclear mRNA synthesis, maturation, or export.
Fig. 2.
Luciferase expression in the presence of Leader. (A) Plasmid cDNAs were transfected into HeLa cells, and Fluc activity [relative light units (RLU)] was assayed at the indicated times. Values are averages for duplicate samples (<10 RLU variance). (B) Plasmid pFluc/Rluc (1 μg per plate) was augmented with pFluc/L (1 μg per plate) or pFluc/L(C19A) (1 μg per plate) and then cotransfected into HeLa cells. Rluc activity (RLU) was assayed at the indicated times. The values are averages for duplicate samples (<10 RLU variance). (C) RNA from transfected cells prepared as in A were isolated from whole cells or cytoplasmic extracts. The relative Fluc RNA titers were assessed by quantitative PCR by using a standard curve derived for this sequence. (D) The β-actin mRNA titers were determined in parallel to C. The Fluc titers (from C) were normalized to the β-actin mRNA content observed at 6 h or 26 h in the pFluc/L(C19A) noninhibitory samples. This plot controls for changes in mRNA content from 6 to 26 h due to cell growth rather than transcription.
To test this hypothesis, the distribution of Fluc and β-actin mRNAs was measured in extracts derived from whole cells or fractionated cytoplasm after transfection with pFluc/L or pFluc/L(C19A) cDNAs. Appropriate sample preparation ensured that all signals were reverse transcriptase-dependent. Whether the Fluc mRNA signals were plotted relative to each other (Fig. 2C) or normalized to the β-actin content (Fig. 2D), the mutant and wild-type cDNAs each synthesized similar total levels of transcripts, which increased equivalently from 6 to 26 h. Therefore, L was not a direct inhibitor of mRNA transcription. Instead, when the total levels were compared to the cytoplasmic levels, it was obvious that L had an effect on mRNA distribution. In samples directed by pFluc/L(C19A), >91% of the whole cell signal was represented by the cytoplasmic signal, as expected for cells undergoing normal transcription and mRNA export. But for pFluc/L, less than half of the total signal was cytoplasmic at either time point. The remainder was in the nucleus, implicating L in a direct inhibition of mRNA export. Therefore, the decrease in Fluc/Rluc enzyme activities (Fig. 2 A and B) can be explained in part as a direct consequence of the smaller pools of cytoplasmic mRNAs available for translation. To be sure, an L-dependent down-regulation of its own mRNA export and expression, over the length of these assays, had the potential to trigger additional cell responses detrimental to Fluc/Rluc translation, including, e.g., PKR phosphorylation of initiation factor eIF2b (18) or the nuclear sequestration (or turnover) of polypyrimidine tract-binding protein, poly(A) binding protein, or other ribosome factors. It will require a careful kinetic study with measured gene doses to parse such the translational responsibilities along the feedback loop of bicistronic expression, but, certainly, mRNA export was among the key pathways impinged by L, and L alone.
L Binds Ran-GTPase.
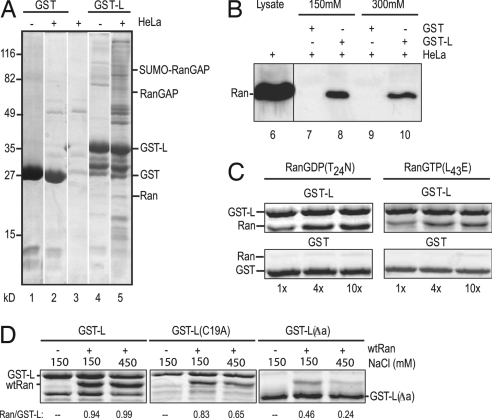
The EMCV L has no known enzymatic function, and its sequence lacks recognizable catalytic motifs. Reasonably, it must bind or sequester some component(s) essential to trafficking. The residue compositions of the zinc-finger and acidic domains evoke a weak comparison to the binding regions of RanGAP and RanBP1, respectively, that interact with Ran-GTPase (19). We speculated that if L bound to Ran or its associated factors, active transport of mRNA or protein through the NPC might be inhibited. To explore this idea, recombinant L fused to GST was prepared. When bound to glutathione beads and incubated with cell lysates, GST-L, but not GST alone, extracted several bands of cellular proteins that could be visualized by staining (Fig. 3A). One of these bands was identified as Ran by immunoreactivity (Fig. 3B). The binding contacts between GST-L:Ran, or with the complex that contained Ran, were strong enough to resist disruption by 300 mM salt (Fig. 3B).
Fig. 3.
Leader–Ran interactions. (A) Recombinant GST or GST-L, bound to glutathione beads, was reacted with HeLa extracts (lanes 2 and 5) or with buffer (lanes 1 and 4). Bound protein was fractionated by SDS/PAGE and then visualized after staining. Control beads without recombinant proteins were treated in parallel (lane 3). (B) GST or GST-L beads were incubated with HeLa extracts as in A. Before boiling, the beads were washed with 150 mM NaCl (lanes 7 and 8) or 300 mM NaCl (lanes 9 and 10). After SDS/PAGE, the bands were visualized by Western assay by using anti-Ran polyclonal antibodies. Unfractionated cell extract (lane 6) was included as a Ran marker. (C) GST or GST-L beads were incubated with the indicated molar equivalents of recombinant Ran(T24N) or Ran(L43E). Extracted protein was fractionated by SDS/PAGE and then visualized by staining. (D) GST-L, GST-L(C19A), or GST-L(Δa) beads were incubated with (wild-type) Ran (1:4) as in C. The beads were washed with 150 mM or 450 mM NaCl as in B before the bound proteins were extracted, fractionated by SDS/PAGE, and visualized by staining. Relative band intensities were captured (ImageQuant) and are presented as the ratio of Ran/GST-L (or its derivatives) on the gels.
RanBP1 and RCC1 are not present, but Western assays (data not shown) have detected RanGAP and SUMO-RanGAP, a modified derivative. Several additional bands are in reasonable agreement with NTF2, importin β, and Crm1, members of the large family of known Ran contact proteins (20). The cohort of cellular proteins bound to any particular Ran is intrinsically dependent on the status of the current nucleotide (GTP, GDP, or “empty”). As Ran cycles within cells or extracts, these populations can fluctuate dramatically. Therefore, the exact contacts that initiate pull-down in such experiments are difficult to assign, because any of the Ran nucleotide formats has the potential to react differently with the beads, or with different forms of L, or through different auxiliary proteins.
To determine whether the L extraction of Ran had been direct or required mediation by other proteins, GST beads and GST-L beads were incubated with recombinant Ran(L43E) or Ran(T24N). These forms are nucleotide-independent structural mimics of RanGTP and RanGDP, respectively, and they do not undergo GDP/GTP exchange or hydrolysis (21). When the retained complexes were visualized by staining, all samples showed (approximately) 1:1 molar ratios of GST-L:Ran, regardless of the input concentrations (Fig. 3C). The assay showed no apparent specificity for either Ran conformation. Both forms bound equivalently in a L-dependent manner, and neither bound to GST alone. Therefore, Ran itself is a binding partner for L, regardless of its nucleotide-dependent conformation. Furthermore, when the mutant L sequences were configured as recombinant proteins and tested in similar assays (Fig. 3D), neither GST-L(C19A) nor GST-L(Δa) bound as effectively to (wild-type) Ran. Beads with the mutant proteins retained 10–50% less Ran than GST-L at 150 mM salt, and the binding was further reduced (30–70%) at 450 mM salt. This weakened stability is consistent with an impaired L:Ran interaction in the observed mutant phenotypes.
L Localizes Near the NE.
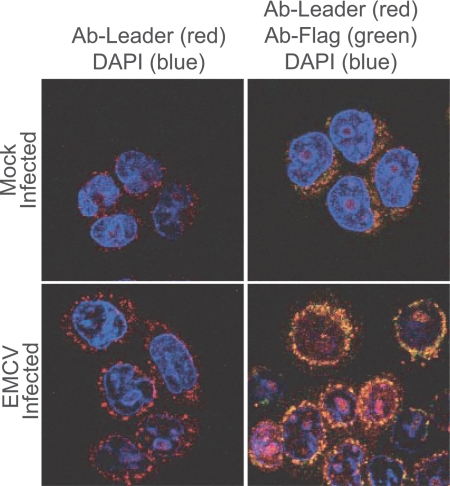
Ideally, in pull-down experiments there is reciprocity in reagents. It would be interesting to know, for example, whether recombinant Ran or its antibodies could extract L from infected cells. Unfortunately, our Ran antibodies are ineffective reagents for pull-down experiments even with uninfected cells, and, as an added complication, native L is very small and highly charged. In the absence of a tag like GST, L is not retained on membranes for Western assays. Furthermore, antibodies against recombinant L are only weakly reactive to the native protein. Still, when used to probe infected cells by confocal microscopy, these antibodies formed a pattern of dots, like a necklace of beads, around the outer regions of NE (Fig. 4). To better resolve this signal, a Flag tag was fused to the amino terminus of L in a recombinant virus context. After infection the Flag signal overlapped with that of L and augmented the necklace effect near the NE. Although preliminary, these localizations are at least indicative of a potential L locale near the cytoplasmic side of the NPC.
Fig. 4.
Leader localization. HeLa cells were mock-infected or infected with vEC9-Flag. At 5 h after infection, the cells were fixed and stained with primary antibodies against L (polyclonal rat serum) or Flag (polyclonal mouse serum); DAPI (blue) was included as a DNA marker. After reaction with appropriate secondary antibodies (red is anti-rat, and green is anti-mouse), image capture was as described in Materials and Methods.
L Inhibits Ran-Dependent Mitotic Spindle Assembly.
During mitosis in normal cells the NPCs and NE are dismantled. A natural Ran gradient persists nonetheless because RanGTP can be generated locally by chromatin-bound RCC1, whereas RanGAP and RanBP1 ensure that the remainder is hydrolyzed to the GDP form throughout the cytoplasm (22, 23). As a consequence, “cargo” remains inexorably bound to importin β everywhere except near the chromatin. Among the favored mitotic cargos of importin β are protein factors that initiate mitotic spindle assembly (24–26). Consequently, spindles form preferentially near chromatin, where the RanGTP concentrations are high enough to dissociate these factors from importin β (27). Cell-free extracts that allow RanGTP to sequester importin β are commonly used to examine Ran activities. For example, in Xenopus egg extracts, tubulin polymerization into asters and spindle structures requires an obligate prior conversion of endogenous RanGDP into RanGTP, as catalyzed by RCC1 associated with added sperm chromatin (28).
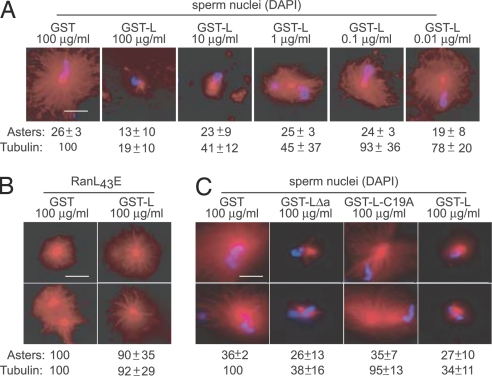
Consistent with a role for L in the abrogation of Ran function, we found that recombinant GST-L, but not GST alone, was a potent inhibitor of sperm-induced aster formation (Fig. 5A). In repeated experiments, the addition of GST-L markedly reduced the number of observed asters relative to GST alone. The representative images also capture the dose-dependent reduction in rhodamine-labeled tubulin per aster, characteristically observed with GST-L. Typically, the count of chromatin units (blue) that generated asters was reduced by half, and the size of the asters that did form (red) was only 20% that of the controls.
Fig. 5.
Microtubule assembly assay. (A) Xenopus egg extracts were induced to form microtubule asters by the addition of sperm nuclei as described in Materials and Methods. The assays contained GST or GST-L at the indicated concentrations. Representative images were captured by fluorescence microscopy of rhodamine-labeled microtubules (red) or DNA (Hoechst dye, blue). In three separate experiments, the number of asters relative to GST alone was quantitated for 50 randomly selected microscope fields. Aster count and SD are indicated, along with the observed rhodamine florescence (tubulin) per aster (and SD) relative to the average intensity from control samples. (B) Assay conditions were similar to A except recombinant Ran(L43E) replaced sperm nuclei. The data are averages (and SD) from six independent experiments. (C) Assay conditions were identical to A, except GST-L(Δa) and GST-L(C19A) were tested in parallel to GST and GST-L. (Scale bars: 25 μm.)
To rule out nonspecific effects of L on this assay and determine whether the observed aster inhibition was indeed Ran-dependent, GST and GST-L were retested in a modified Xenopus egg system, where Ran(L43E), the RanGTP mimic, was added in place of sperm nuclei. Ran(L43E) can bind perpetually to importin β, circumventing the need for GTP hydrolysis or RCC1-facilitiated nucleotide exchange. The asters that now formed in the presence of GST-L were indistinguishable in morphology and quantity from those of the control (Fig. 5B). Therefore, GST-L was not toxic to the generation of microtubule structures per se and did not interfere with the binding of Ran(L43E) to importin β. Rather, the presence of GST-L in the first experiment (Fig. 5A) must have prevented aster formation by inhibiting the generation of RanGTP.
Models for L Activity in Cells.
In contrast to cell-free assays, cellular forms of Ran are partitioned across the NE according to the activity and segregation of the Ran-associated factors. RCC1 is on chromatin. RanGAP and RanBP1 are predominantly cytoplasmic. Any activity that disrupts GDP/GTP exchange, or Ran cycling across the NE, has the potential to perturb the required cellular gradient and bring active trafficking to a halt. We do not yet know which specific step in the Ran pathway is impeded by L during EMCV infection. The combined data could be explained (i) if L prevented RanGTP hydrolysis or (ii) if L inhibited RanGDP/GTP exchange by RCC1. It is also possible that L works to accelerate RanGTP hydrolysis, because, in theory, a significant boost in GTPase activity could upset the endogenous gradient. But, given the rapidity with which L acts in cells during infection and the already high turnover of GTP during normal hydrolysis (6), we consider this model to be less probable. More logically, model i predicts that L binding to Ran might simply trap the GTP form on the cytoplasmic side of the NE, making it unavailable for further cycling. Indeed, this is where our antibodies localize L. Our mutants (C19A and Δa) with demonstrably lower binding affinities for Ran are doubtlessly defective because they are less efficient at this sequestration. Model ii predicts that L bound to RanGDP could become trapped in a cargo-bound form on the nuclear side of the membrane. In the absence of precise measurements of GDP/GTP exchange in cells and cell-free extracts, we cannot yet distinguish these scenarios. But either mechanism would presumably trigger the same effect in cells, namely, a potent disruption of the RanGDP/GTP gradient across the NE and a rapid, significant, probably irreversible block to active nucleocytoplasmic transport of protein (in) and cellular mRNA and protein (out). Either model might also explain other reported phenotypes currently attributed to L, including host protein synthesis inhibition and adverse IFN responses. Most likely, the L phenomenon is not IFN-specific but, rather, a general consequence of L-dependent destruction of the Ran gradient during infection.
Format of the Active L.
Many previous descriptions of L phenotypes have relied on L(Δa) and L(Δz) mutations to remove large portions of the protein (12, 16). Other mutations have suggested that phosphorylation at Tyr-41 (12) or Thr-47 (14) may influence functionality. Given the efficiency of the spindle assembly assays as a test for Ran activity, we prepared two additional recombinant variants of L and tested them for aster inhibition, carefully quantitating (×3) the number of asters per sperm (50 nuclei), and the relative tubulin content per aster (Fig. 5C). As before (Fig. 5A), GST-L showed a marked decrease (25%) in the number of asters per field and a reduction (66%) in the amount of tubulin per aster when compared with GST alone. The GST-L(C19A) was not inhibitory. It allowed about the same number of asters, with the same amount of tubulin, as the GST control. The results suggest that the zinc-finger motif within L is a key element for observation of Ran inhibition. Reinforcing this idea, the second recombinant protein, GST-L(Δa), was a strong inhibitor of aster formation. This fragment represents only a 42-aa piece of L. It lacks the acidic domain and both potential phosphorylation sites, yet its activity was equivalent to full-length L in this assay. This finding is of special interest, because the identical L(Δa) sequence expressed in cells from a bicistronic cDNA behaved like zinc-finger mutations L(Δz) and L(C19A) and was not inhibitory to Fluc expression (Fig. 2A). This finding suggests a special requirement for the acidic region in cells, but not in cell-free assays. Perhaps, as suggested by recombinant Ran-binding assays (Fig. 3D), the acidic domain and its potential phosphorylation sites are responsible for additional Ran impedance activities or increased binding affinities evident only in cells. Nevertheless, in cell-free extracts, we propose that this very limited fragment might be a very useful tool for further investigations into Ran functions during nuclear transport and mitotic spindle assembly. Recently, the NMR structure of the amino-terminal portion (32 residues) of the Mengoviral L was solved in a collaborative effort with the Center for Eukaryotic Structural Genomics (University of Wisconsin, Madison, WI). The coordinates (Protein Data Bank ID code 2BAI) show limited structural flexibility in the zinc-finger region, consistent with the idea that this unique domain could be involved as a primary Ran contact (C. Cornilescu, personal communication).
Materials and Methods
Bicistronic Plasmids.
Bicistronic vector pFluc/− was a generous gift from R. Groppo (University of Wisconsin, Madison, WI). The plasmid has an immediate–early cytomegalovirus promoter (PCMVIE) that drives transcription of capped, polyadenylated mRNA after transfection into cells. The 5′ cistron (cap-dependent translation) of the mRNA encodes a full-length Fluc gene flanked on the 3′ side by a wild-type EMCV IRES (29). In plasmid pFluc/Rluc, a full-length Rluc gene was linked to the start codon of the IRES. In plasmids pFluc/L, pFluc/L(Δz), and pFluc/L(Δa), the EMCV L, L(Δz), or L(Δa) sequences (12) replaced the Rluc gene. For pFluc/L(C19A), two-step PCRs changed the wild-type Cys-19 to Ala (UGC to GGC) in the L gene. The identity of all cDNAs was confirmed by restriction analysis and sequencing. (All primer sequences used for cloning and PCR are available upon request.) HeLa cell monolayers were transfected with plasmid DNAs by using liposomes (30). Fluc and Rluc activities were assayed in cell lysates at appropriate times after transfection by using a dual luciferase assay system (Promega, Madison, WI) according to the manufacturer's instructions.
Quantitative PCR.
Primer sequences for the detection of Fluc and human β-actin mRNAs were designed by using Primer Express software (ABI, Foster City, CA). Total or cytoplasmic RNAs were isolated from transfected HeLa cells (duplicate plates) by using RNeasy mini kits (Qiagen). DNA contamination was reduced by treatment with RQ1 DNase (Promega) followed by an RNeasy step. The process was repeated a second time. Treated RNA (10 μl) was reacted (in 20 μl) with Moloney murine leukemia virus reverse transcriptase (M-MLV; Invitrogen, Carlsbad, CA) in the presence of appropriate primers (2 pg of primers flanking Fluc or β-actin). The cDNAs were diluted (10×) and then used to program duplicate (25 μl each), real-time PCR amplifications by using SYBR Green PCR master mix (ABI) and flanking primers as appropriate for each gene. The Fluc signal was normalized to the β-actin signal (Sequence Detection System software). Controls (2×) for each primer pair, in which water or an RNA sample without an RT-MLV step was replaced for template cDNA, were used in each experiment.
Recombinant L.
The EMCV L segment from pEC9 (31) was amplified by PCR and then ligated into pGEX-P2 (GE Bioscience, Piscataway, NJ) to form plasmid pGST-L. Similar procedures linked the L(Δa) and L(C19A) segments into analogous plasmids. Escherichia coli (BL-21) was transformed with these plasmids, amplified, and induced with isopropyl β-d-thiogalactoside. Four hours later the cells were lysed, and the extracts were reacted with glutathione Sepharose HP resin (GE Bioscience). The bound proteins GST, GST-L, GST-L(Δa), and GST-L(C19A) were eluted (50 mM Tris, pH 8/10 mM reduced glutathione) and then analyzed for content and purity by SDS/PAGE.
For binding studies with cell extracts, confluent HeLa cell monolayers (6 × 106 cells) were washed with PBS and then lysed (0.5 ml of 50 mM Hepes, pH 7.4/150 mM NaCl/2 mM DTT/1 mM PMSF/0.5% vol/vol IPEGAL CA-630). After clarification, the extracts were added to glutathione Sepharose 4B-beads (GE Bioscience) prereacted with GST, GST-L, GST-L(C19A), or GST-L(Δa). Incubation was for 1 h at 22°C, and then the beads were washed twice with buffer (50 mM Hepes/0.5% vol/vol IPEGAL CA-630) containing NaCl (150 mM and 300 mM). Protein bound to the beads was eluted with SDS (boiling), fractionated by SDS/PAGE (12%), and then visualized by Western blot analysis using a primary antibody against Ran (goat polyclonal IgG, C-20; Santa Cruz Biotechnology, Santa Cruz, CA). For binding assays with recombinant Ran, the GST- or GST-L-bound Sepharose beads were reacted with Ran, RanGTP(L43E), or RanGDP(T24N) (32) at molar ratios of 1:1, 4:1, or 10:1 in phosphate buffer (20 mM, pH 7.6) for 1 h. The beads were washed with buffer containing NaCl (150 mM or 450 mM) before bound proteins were eluted with SDS (boiling), fractionated by SDS/PAGE, and then visualized by staining with Coomassie R-250.
Confocal Microscopy.
Recombinant virus vEC9 was modified to encode a Flag tag (DYKDDDK) at the start of the polyprotein (MA-DYKDDDK-MATT). HeLa monolayers were infected at a multiplicity of infection of 20 (33). At 4 h after infection, the cells were washed, fixed, permeabilized, and then reacted with appropriate primary and secondary antibodies or DAPI stain, as described previously (29). The images were visualized by laser confocal microscope. For L protein detection, the primary antibody was anti-L polyclonal rat sera (511-2) precleared by incubation with HeLa cells. The secondary antibody was TRITC-conjugated rabbit, anti-rat IgG (T-4280; Sigma, St. Louis, MO). The Flag tag was detected with a murine monoclonal (F3165, Sigma). The secondary antibody was FITC-conjugated goat anti-mouse IgG (F5387; Sigma).
Egg Extract Preparation and Aster Assembly.
CSF-arrested Xenopus egg extracts and demembranated sperm chromatin were prepared as described (34). Microtubule structures were assembled by adding 25 μM Ran(L43E) (32) or chromatin (150 sperm per microliter) to an egg extract supplemented with rhodamine-labeled tubulin (0.2 mg/ml) and incubating (at 22–25°C for 15 min) as described (35). GST or GST-L was diluted in CSF-XB (10 mM K-Hepes, pH 7.6/100 mM KCl/2 mM MgCl2/0.1 mM CaCl2/50 mM sucrose/5 mM EGTA) and added to the extract at final concentrations of 0.01–100 μg/ml. For quantitation of tubulin fluorescence, the images were captured at the same camera setting for all samples. Images were quantified by using MetaMorph software. Briefly, fluorescence intensity of each aster was measured, and then a background value was subtracted by using an equal-sized area on the same image. At least 16 asters were measured per sample. For Ran(L43E)-induced structures, the number of asters in 50 randomly chosen microscope fields was recorded.
Acknowledgments
We thank Rachel Groppo for plasmid pFluc/−. This work was supported by National Institutes of Health Grant AI-17331 (to A.C.P.), American Heart Association Grant 0330388N (to C.W.), National Science Foundation Grant MCB 0344723 (to C.W.), and a Steenbock Predoctoral Fellowship from the Department of Biochemistry of the University of Wisconsin (to A.J.A.).
Glossary
Abbreviations
- EMCV
encephalomyocarditis virus
- NPC
nuclear pore complex
- NE
nuclear envelope
- IRES
internal ribosome entry site
- Fluc
firefly luciferase
- Rluc
Renilla luciferase
- RLU
relative light unit
- L
Leader protein
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Stewart M. L., Baker R. P., Bayless R., Clayton L., Grant R. P., Littlewood T., Matsuura Y. FEBS Lett. 2001;498:145–149. doi: 10.1016/s0014-5793(01)02489-9. [DOI] [PubMed] [Google Scholar]
- 2.Gorlich D., Kutay U. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 3.Adam S. A., Marr R. S., Gerace L. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weis K., Ryder U., Lamond A. I. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 6.Gorlich D., Pante N., Kutay U., Aebi U., Bischoff F. R. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 7.Belov G. A., Lidsky P. V., Mitkitas O. V., Egger D., Lukyanov K. A., Bienz K., Agol V. J. Virol. 2004;78:10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmenberg A. C. In: Positive Strand RNA Viruses, Brinton M. A., Rueckert R. R., editors. New York: Liss; 1987. pp. 25–34. [Google Scholar]
- 9.Molla A., Paul A. V., Wimmer E. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 10.Gustin K. E., Sarnow P. J. Virol. 2002;76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustin K. E., Sarnow P. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvorak C. M. T., Hall D. J., Hill M., Riddle M., Pranter A., Dillman J., Deibel M., Palmenberg A. C. Virology. 2001;290:261–271. doi: 10.1006/viro.2001.1193. [DOI] [PubMed] [Google Scholar]
- 13.Zoll J., Galama J. M. D., van Kuppeveld F. J. M., Melchers W. J. G. J. Virol. 1996;70:4948–4958. doi: 10.1128/jvi.70.8.4948-4952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoll J., Melchers W. J., Galama J. M., van Kuppeveld F. J. J. Virol. 2002;76:9664–9672. doi: 10.1128/JVI.76.19.9664-9672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delhaye S., van Pesch V., Michiels T. J. Virol. 2004;78:4357–4362. doi: 10.1128/JVI.78.8.4357-4362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lidsky P. L., Hato S., Bardina M. V., Aminev A. G., Palmenberg A. C., Sheval E. V., Polyakov V. Y., van Kuppeveld F. J., Agol V. J. Virol. 2006;80:2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duke G. M., Hoffman M. A., Palmenberg A. C. J. Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen G. C., Ransohoff R. M. Adv. Virus Res. 1993;42:57–101. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 19.Seewald M. J., Kraemer A., Farkasovsky M., Korner C., Wittinghofer A., Vetter I. R. Mol. Cell. Biol. 2003;23:8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macara I. G. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lounsbury K. M., Richards O. C., Carey K. L., Macara I. G. J. Biol. Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- 22.Kalab P., Weis K., Heald R. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 23.Caudron M., Bunt G., Bastiaens P., Karsenti E. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 24.Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 25.Nachury M., Maresca T., Salmon W., Waterman-Storer C., Heald R., Weis K. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 26.Gruss O., Carazo-Salas R., Schatz C., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 27.Wilde A., Lizarraga S. B., Zhang L., Wiese C., Gliksman N. R., Walczak C. E., Zheng Y. Nat. Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- 28.Dasso M. Curr. Biol. 2002;12:502–508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- 29.Aminev A. G., Amineva S. P., Palmenberg A. C. Virus Res. 2003;95:45–57. doi: 10.1016/s0168-1702(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 30.Rose J. K., Buonocore L., Whitt M. A. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 31.Hahn H., Palmenberg A. C. J. Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilde A., Zheng Y. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 33.Rueckert R. R., Pallansch M. A. Methods Enzymol. 1981;78:315–325. [PubMed] [Google Scholar]
- 34.Murray A. W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 35.O’Brien L. L., Albee A. J., Liu L., Tao W., Dobrzyn P., Lizarraga S. B., Wiese C. Mol. Biol. Cell. 2005;16:2836–2847. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]