Abstract
Heart morphogenesis requires the coordinated regulation of cell movements and cell–cell interactions between distinct populations of cardiac precursor cells. Little is known about the mechanisms that organize cardiac cells into this complex structure. In this study, we analyzed the role of Slit, an extracellular matrix protein and its transmembrane receptors Roundabout (Robo) and Roundabout2 (Robo2) during morphogenesis of the Drosophila heart tube, a process analogous to early heart formation in vertebrates. During heart assembly, two types of progenitor cells align into rows and coordinately migrate to the dorsal midline of the embryo, where they merge to assemble a linear heart tube. Here we show that cardiac-specific expression of Slit is required to maintain adhesion between cells within each row during dorsal migration. Moreover, differential Robo expression determines the relative distance each row is positioned from the dorsal midline. The innermost CBs express only Robo, whereas the flanking pericardial cells express both receptors. Removal of robo2 causes pericardial cells to shift toward the midline, whereas ectopic robo2 in CBs drives them laterally, resulting in an unfused heart tube. We propose a model in which Slit has a dual role during assembly of the linear heart tube, functioning to regulate both cell positioning and adhesive interactions between migrating cardiac precursor cells.
Keywords: cell adhesion, dorsal vessel, heart morphogenesis
Maintaining precise positioning between different populations of migrating cells is a fundamental aspect of organogenesis. Unlike single cells that respond individually to guidance cues, cells within a developing tissue often must travel in cohesive groups. These cells are not merely attached to each other but rather are organized into distinct patterns, which shape the developing tissue. Regulation of adhesive interactions between cells has been shown to be essential for tissue morphogenesis (1). Likewise, several genes encoding cell adhesion proteins have been implicated in heart tube assembly in both insects and vertebrates (2–5), yet we know very little about the underlying cellular mechanisms responsible for this process.
The steps leading to the formation of the primitive heart tube are highly conserved between insects and vertebrates (6). In each case, bilateral groups of heart precursor cells migrate toward the midline of the embryo, where they fuse to form a linear heart tube. During assembly of the Drosophila heart tube, cardioblasts (CBs) and pericardial cells (PCs), which are generated within bilateral fields in the lateral mesoderm, form continuous rows that converge toward the dorsal midline of the embryo to form a beating linear heart with a central lumen. In this study, we use genetic analysis to show that loss of slit, or both Roundabout (robo) and Roundabout2 (robo2), causes defects in cell adhesion, resulting in gaps in the rows of CBs and PCs. Moreover, the differential expression of Robo and Robo2 is important for maintaining the relative positioning of the two distinct populations of cells during dorsal migration. CBs, which express the single Robo receptor, are closer to the dorsal midline than the PCs, which express both Robo and Robo2. Removal of robo2 causes individual PCs to shift toward the midline, whereas ectopic expression of Robo2 in CBs drives the rows of cells laterally, resulting in an unfused heart tube. These data support the model whereby the combinatorial expression of Robo receptors controls the relative position of individual rows of migrating cells from the dorsal midline during heart tube assembly. This model was first proposed for the role of Slit and Robos in determining axonal positioning at the ventral midline of the Drosophila CNS (7, 8) and provides a paradigm for the organization of bilateral populations of cells on either side of a midline.
Results
Expression of Slit and Robo Proteins in the Embryonic Heart.
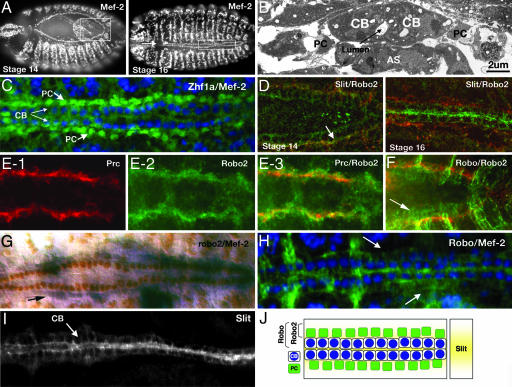
The Slit protein is highly conserved and functions as both an attractive and repulsive guidance signal for migrating cells in many different tissues (9–13). It was previously reported that slit was expressed in the CBs of the developing Drosophila heart (14); however, its function in cardiac development remains unknown. We first reexamined these findings in more detail. The Drosophila heart is composed of two major cell types that converge at the dorsal midline of the embryo to form the heart tube. The CBs are aligned in two highly ordered rows that merge to form the lumen of the heart (Fig. 1A–C). These cells are contractile and express muscle-specific proteins. Similar to their vertebrate counterparts (5), CBs are closely connected with each other and adopt the characteristics of polarized epithelia with apical and basal surfaces (15). The CBs are flanked by two rows of nonmuscle PCs, which are loosely associated with the CBs (ref. 3; Fig. 1 B and C). At stage 14, we detected Slit protein on the surface of the CBs as they are migrating dorsally toward the midline (Fig. 1D). We also detected Slit on the surface of the PCs, and this expression is stronger on the side that is adjacent to the CBs (Fig. 1D). As they migrate dorsally, the two rows of CBs become polarized before fusing at the dorsal midline of the embryo to form the heart tube (15). At this time (stage 16), we no longer detected Slit on the PCs. However, on the CBs, Slit localization becomes polarized, with most of the protein concentrated on the apical surface (Fig. 1 D and I). This surface corresponds to the regions of contact between opposing pairs of CBs where the lumen will form.
Fig. 1.
Expression of Slit and Robos during heart formation. (A) Mef-2 labels the nuclei of CBs and somatic muscle cells in stage 14 (Left) and stage 16 (Right) embryos. Boxed regions highlight regions of the heart shown in D. (B) Electron micrograph of a stage 16 embryo showing the mature heart in cross-section. Two opposed CBs at the dorsal midline enclose a central lumen and are laterally flanked by PCs. The cells of the amnioserosa (AS) are visible under the heart tube. (C) Stage-16 embryo stained for Mef-2 (blue) and Zfh1a (green), which labels the PC nuclei. (D) At stage 14 (Left), Robo2 (red) is localized to the surface of PCs, whereas Slit (green) labels the surfaces of both CBs and PCs. Areas of Slit/Robo2 colocalization on PCs are shown in yellow (arrow). At stage 16 (Right), Robo2 is still localized to the PCs (red), whereas Slit expression becomes restricted to the surface of the CBs (green). (E) PC marker Pericardin (red, E-1) and Robo2 (green, E-2) colocalize to the PC surfaces (yellow, E-3) in stage-14 embryos. (F) Robo (green)/Robo2 (red) double-labeling showing colocalization (yellow) of both proteins on the surface of PCs at stage 14. Robo protein (arrow) is also seen on the surface of the CBs. (G) robo2 mRNA is restricted to the PCs (arrow); Mef-2 antibody (brown) is labeling the CB nuclei. (H) Mef-2 (blue) and Robo (green) expression. At stage 16, Robo expression is detected on surfaces of the CBs and more weakly on the PCs (arrows). (I) Slit localizes to CBs in a stage-16 embryo. The protein is concentrated at the regions of contact where the lumen will form. (J) Schematic showing a summary of the expression patterns of Slit, Robo, and Robo2. Slit (yellow), secreted by the CBs, is detected on the surface of both CBs and PCs. The Robo receptor is also expressed on the surfaces of both cell types. However, expression of the Robo2 receptor is restricted to the surfaces of the PCs.
There are three known receptors for Slit in Drosophila: Robo, Robo2, and Robo3 (11, 16, 17). We detected the expression of two of the three receptors, Robo and Robo2, in specific heart cell types during heart tube assembly. At stage 14, similar to the expression pattern for Slit, Robo protein is localized to the surface of both the CBs and PCs (Fig. 1F). Later, at stage 16, although Robo can still be detected on PC surfaces, the protein is more easily detected on CBs and concentrated on apical surfaces (Fig. 1H). At both stages, Robo2 expression is limited to the surface of the PCs, as confirmed by mRNA expression (Fig. 1G) as well as colocalization with Slit (Fig. 1D) and Pericardin, which localizes to the periphery of PCs (ref. 3; Fig. 1E). Thus, during the time when the rows of cells are migrating during heart tube assembly, both Slit and Robo are detected on the surfaces of CBs and PCs, whereas Robo2 is restricted to the PCs (summarized in Fig. 1J).
Slit Is Required in the CBs for Heart Cell Adhesion and Positioning.
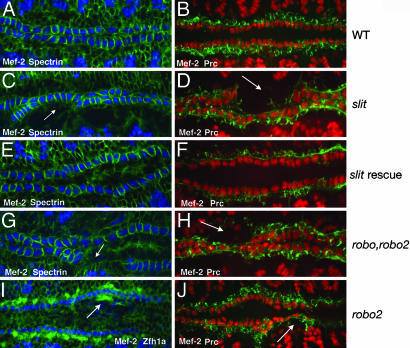
The temporal and spatial localization data of Slit, Robo, and Robo2 suggest these molecules play a role in the assembly of the heart tube after the heart precursors have been specified. To test this hypothesis, we first examined the heart in embryos mutant for slit. During heart tube assembly, CBs form two continuous rows of cells that coordinately migrate toward the dorsal midline to form the heart tube. This can be observed by staining for Mef-2, a marker for CB cell nuclei (18), and α-Spectrin (19), which localizes to the surface of these cells (Fig. 2A). α-Spectrin, which preferentially localizes to the basal-lateral surface of the CBs, also reveals the polarized nature of these cells, which adopt a columnar shape characteristic of epithelial cells (20). The CBs are flanked by two rows of more loosely connected PCs, as visualized by also staining for Pericardin (Fig. 2B), which labels PC surfaces (3). In slit mutant embryos, the appropriate numbers of CBs were specified, although we observed a loss of cell adhesion within each row of cells (Fig. 2 C and D). In 97% of slit/slit embryos (n = 102), the CBs no longer formed two continuous rows of cells, with frequent gaps and inappropriate cell clustering. We quantified this phenotype by counting the number of CBs that reached the dorsal midline in stage-16 embryos. In wild-type embryos, there are normally 104 Mef-2-positive CB nuclei that form two rows at the dorsal midline (ref. 21; Fig. 1A). However, in slit embryos, we counted an average of 87.9 ± 3.6 (n = 21) Mef-2-positive CBs at the dorsal midline. This phenotype was not due to a general loss of these cells, because when we counted Mef-2-positive cells at stage 13, before dorsal migration, both wild-type and slit mutant embryos had an identical number of CBs. In addition, the shape of these cells was abnormal, as seen with α-Spectrin staining (Fig. 2C). We also saw defects in the alignment of the flanking PCs as observed by Pericardin staining (Fig. 2D). As with the CBs, gaps were often observed in the rows of cells. These gaps frequently corresponded with the gaps in the rows of CBs.
Fig. 2.
Slit and Robos are required for heart cell adhesion. Stage-16 embryos labeled with anti-Mef2 (blue) and anti-α-Spectrin (green) to visualize CB nuclei and cell membranes (A, C, E, and G), anti-Mef2 (red), and Pericardin (green) to visualize CB nuclei and PC surfaces (B, D, F, H, and J) or Mef2 (blue) and Zfh1a (green) to visualize CB and PC nuclei (I), respectively. (A) Wild-type embryo showing the two rows of CBs at the midline. The cells within each row are in close contact, as revealed by α-Spectrin. (B) The PCs form two rows of cells that flank the inner two rows of CBs. (C and D) In slit mutants, the CBs no longer form two continuous rows and now inappropriately intermingle with the PCs. Arrows emphasize areas where the CBs have lost contact with each other, resulting in a gap in the row of cells. (E and F) The cell adhesion defect is rescued by driving expression of UAS-slit in CBs with Mef-2-GAL4. (G and H) The robo,robo2 double-mutant phenotype is similar to slit. Arrows indicate gaps in the row of CBs. (I) In stage-14 robo2 mutants, PCs are sometimes mispositioned, extending too far toward the dorsal midline (arrow). We observed this phenotype in 29% of robo2 mutant embryos (n = 96). (J) Stage-15 robo2 mutant embryo showing mispositioned PCs (arrow).
slit mRNA is expressed by the CBs, but not the PCs, as they dorsally migrate to form the heart tube (14). However, we detected the Slit protein on both cell types (Fig. 1D). Consistent with Slit localization, we observed defects in the positioning of both cell types in slit mutants (Fig. 2 C and D). Together, these results suggest that Slit is secreted by the CBs and diffuses to the surface of neighboring PCs, where it is required for the correct migration of both cell types. To test this hypothesis, we performed a rescue experiment in which we used the GAL4-UAS system (22) to drive slit expression in CBs in slit mutant embryos. We used the Mef2-GAL4 driver, which drives expression in all CBs and somatic muscle cells before the time when they begin their initial migration toward the dorsal midline of the embryo (10). We first confirmed that Mef2-GAL4 is not active in PCs (data not shown). We also confirmed that UAS-slit was expressed in rescued embryos by examination of Slit localization. In slit-rescued embryos, we detected Slit staining on the surface of both CBs and PCs (data not shown). Unlike embryos homozygous mutant for slit (Fig. 2 C and D), we observed significant rescue of the mutant phenotype in 95% of slit/slit;Mef2-GAL4/UAS-slit embryos (n = 151). The CBs were now adhered with each other, and we observed no visible gaps between adjacent CBs and PCs (Fig. 2 E and F). Similar to wild-type embryos, we counted 103.8 ± 0.5 (n = 18) Mef-2-positive CBs at the dorsal midline. Furthermore, the columnar shape of these cells also resembled that of wild-type CBs (Fig. 2E). The rescue experiments are thus consistent with the idea that expression of Slit, a diffusible protein, by CBs creates a region of Slit expression extending to the boundary of the adjacent PCs, where it is required for the proper positioning of both cell types.
robo,robo2 Double Mutants Have a slit Phenotype.
We next examined whether heart cell alignment also requires Robo receptors. Robo2 protein is restricted to the surface of the PCs (Fig. 1E), whereas Robo is expressed on both the CBs and PCs (Fig. 1F). Consistent with the expression data, we also observed defects in CB and PC positioning in embryos mutant for both robo and robo2 (Fig. 2 G and H). Furthermore, we observed a reduction in the number of CB nuclei that reached the dorsal midline (86.2 ± 5.5, n = 20). These phenotypes are similar to those we observed in a slit mutant, supporting the idea that the specific interaction between Slit and these Robo receptors is required for the correct alignment of heart precursors during the assembly of the heart tube.
robo and robo2 Have Different Roles in Heart Cell Positioning.
The differential expression of Robo receptors in the two cell types in the heart led us to question the individual roles of each receptor in this process. The CBs express the single Robo receptor, whereas the PCs express both Robo and Robo2. During heart cell migration, the rows of CBs are closer to the dorsal midline of the embryo than the PC rows. In embryos mutant for both receptors, the relative positioning of each cell type is no longer maintained, and we observed severe defects in CB and PC alignment (Fig. 2 G and H). In contrast, embryos mutant for either robo or robo2, we observed much milder defects. In robo mutants, we could occasionally detect mispositioned CBs (data not shown). In mutants for robo2, we sometimes observed that one or more PCs migrated too far dorsally and could be found closer to the dorsal midline in front of the rows of CBs, as visualized by staining for Zfh1a, which labels PC nuclei (23) and Pericardin (Fig. 2 I and J). These defects were not observed in wild-type embryos or embryos heterozygous for robo2. However, whereas we occasionally observed some alignment defects in the rows of CBs in robo2 mutants, we failed to observe gaps between these cells. Together with the expression data, these results are consistent with the hypothesis that the relative distance a CB or a PC maintains from the midline during dorsal migration could at least partially depend upon which combination of Robo receptors are expressed.
Misexpression of Robo2, but Not Robo, in CBs Disrupts Heart Cell Positioning.
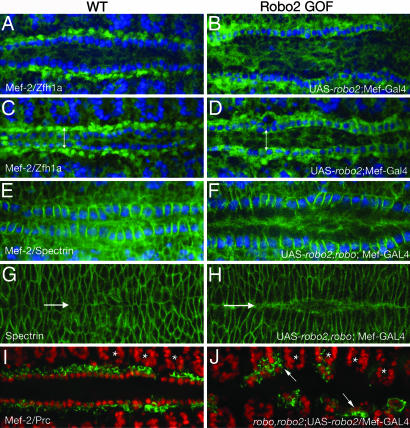
To test this hypothesis and determine the relative role that each Robo receptor plays during heart cell migration, we performed overexpression analysis. Both the CBs and PCs normally express robo, and driving expression of a UAS-robo transgene in CBs with Mef2-GAL4 does not result in any observable defects in the heart tube (data not shown). However, misexpression of UAS-robo2 in the CBs, which normally express only robo, resulted in a lateral shift of the row of CBs away from the dorsal midline to a position normally occupied by the PCs (Fig. 3A–D). To quantify this change in positioning, we compared the distance between opposing pairs of CB or PC nuclei in stage-16, wild-type, and robo2 overexpressing embryos. At this stage, both cell types have nearly completed their dorsal migration, but the inner rows of CBs have not yet fused to form the heart tube. In wild-type embryos, the average distance between opposing CB nuclei (as visualized by Mef-2 staining) was 9.21 ± 1.10 μm, and the opposing PC nuclei (as visualized by Zfh1a staining) are normally 21.39 ± 1.97 μm apart (Fig. 3C). When we ectopically expressed robo2 in the CBs, the distance between these cells increased to 22.28 ± 1.09 μm, a value comparable to the distance normally maintained by the PCs at this stage (Fig. 3D). This space between the CB rows is maintained through stage 17, when the opposing pairs of cells should have fused at the midline to form the heart tube. Interestingly, this phenotype is reminiscent of that seen in cardia bifida, a vertebrate developmental disorder due to a failure of convergent migration by the left and right cardiac primordial (24).
Fig. 3.
Robo expression directs heart cell positioning. Wild-type (A, C, E, G, and I) and Robo2 gain-of-function (GOF; B, D, F, H, and J). Stage-15 (A and B) and -16 (C and D) embryos labeled with anti-Mef-2 (blue, CB nuclei) and anti-Zfh1a (green, PC nuclei). At stage 16 (C), the average distance between opposing CB nuclei was 9.21 ± 1.10 μm, and between PC nuclei, 21.39 ± 1.97 μm (represented by double-headed arrow). (B and D) Ectopic expression of robo2 in CBs with the Mef2-Gal4 driver results in an increased distance between the CBs. At stage 16 (D), the average distance between opposed CBs was 22.28 ± 1.09 μm (double-headed arrow), a value comparable to the distance normally maintained by the PCs at this stage (C). We observed this defect in 84% of the embryos scored (n = 72). (E) Stage-15–16 embryo stained with Mef-2 (blue) and spectrin (green). Ectopic expression of robo2 in CBs in a robo mutant background also results in a similar increase in the distance between the rows of CBs (F). Dorsal closure in these embryos is observed by Spectrin staining in the leading edge dorsal ectodermal cells (G and H). Arrows mark the dorsal midline. In robo2 overexpressing embryos, dorsal closure still occurs, although slightly delayed compared with wild type. E and G and F and H are the same embryos imaged in two different focal planes. (I and J) Mef-2 (red) and Pericardin (green) staining in stage-15 wild-type embryos (I) and robo2 GOF embryos in a robo,robo2 double-mutant background (J). Simultaneous removal of both receptors and ectopic expression of Robo2 reveals both cell positioning and cell adhesion defects. CBs and PCs are no longer adhered to each other and are driven away from the dorsal midline. Often, these cells are found too far laterally in the somatic muscle territory (arrow). The relative positioning of the somatic muscle nuclei (asterisks) appears normal compared with wild type.
In robo2-overexpressing embryos, although the rows of PCs became displaced laterally, we observed no visible gaps indicating that adhesion between these cells is maintained (Fig. 3 B and D). We observed similar defects when we overexpressed robo2 in embryos mutant for robo (Fig. 3 E and F). The rows of CBs were laterally displaced, but cell adhesion between these cells was maintained. However, when we performed the same experiment in embryos missing both robo and robo2, we now observed both phenotypes (Fig. 3 I and J). Similar to robo,robo2 double mutants (Fig. 2H), we observed gaps in the mispositioned rows of CBs indicating a loss of adhesion between adjacent cells. However, unlike the loss-of-function robo,robo2 phenotype, the additional ectopic expression of Robo2 in CBs resulted in a mispositioning of these cells away from the dorsal midline. This aspect of the phenotype was strikingly severe as compared with the ectopic expression of Robo2 in a wild-type background (Fig. 3 B and D), presumably because of the additional loss of adhesion between these cells. These cells were displaced even more laterally, and we often observed CBs and PCs inappropriately in somatic muscle territory (Fig. 3J).
In a recent study, Chartier et al. (3) provided evidence that the dorsal ectoderm coordinately migrates with the heart cells during dorsal closure. Thus, we looked at this process in embryos overexpressing robo2, to ensure that the cardia bifidia phenotype we observed was not potentially due to dorsal closure defects. To do this, we examined the leading-edge dorsal ectodermal cells with α-Spectrin in stage- 15–16 embryos, when these cells should have come in contact with one another. We observed that in embryos overexpressing robo2, although the rows of CBs have not come together, the leading edges of the epidermal cells have made contact with each other (Fig. 3 G and H). However, the final steps of dorsal closure may be slightly delayed as compared with wild-type embryos, because we sometimes observed a slight gap between the dorsal ectoderm in late stages.
Discussion
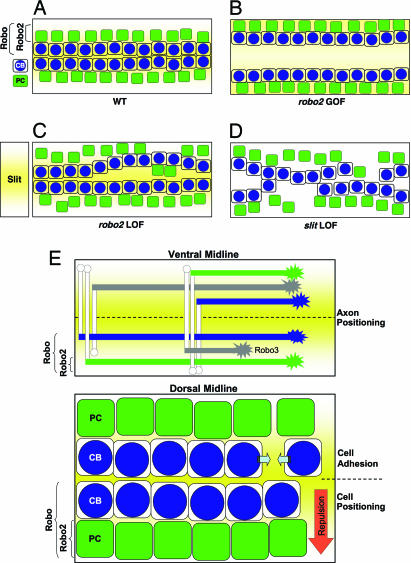
Morphogenesis of the heart is a complex process requiring the coordinated regulation of cell positioning and adhesive interactions between distinct populations of migrating precursor cells. In this study, our results are consistent with the model that Slit and Robos are required for both of these functions (Fig. 4E). During assembly of the Drosophila heart, two types of progenitor cells align into rows and coordinately migrate to the dorsal midline of the embryo, where they merge to assemble a linear heart tube. We found that the differential expression of Robo and Robo2 is important for maintaining the relative positioning of the two distinct populations of cells during this dorsal migration (Fig. 4A). The inner rows of CBs express the single Robo receptor, whereas the PCs, which are positioned more laterally, express both receptors. Removal of robo2 may cause individual PCs to shift toward the midline (Fig. 4C), whereas ectopic expression of Robo2 in CBs drives the rows of cells laterally, resulting in an unfused heart tube (Fig. 4B).
Fig. 4.
Slit is required for cell adhesion and cardiac cell positioning. The illustration shows the dual role for Slit and Robo receptors during heart tube formation. (A) Wild-type embryo showing the two inner rows of CBs (blue) flanked by two rows of PCs (green). Slit (yellow) is secreted by the CBs creating a region of Slit expression extending to the boundary of the adjacent PCs. PCs, which express both Robo and Robo2, are positioned farther away from the dorsal midline than the CBs that express only Robo. (B) Ectopic expression of Robo2 in CBs drives the two rows of cells away from the dorsal midline to a distance normally maintained by the PCs at this stage. (C) In a robo2 mutant, individual PCs sometimes migrate too far dorsally. (D) Loss of slit (or robo and robo2) results in defects in cell adhesion within rows of CBs as well as mispositioning of both cell types. (E) Model showing the role of the Robo code in both CNS and heart patterning. In the CNS (Upper), Robo, Robo2, and Robo3 are differentially expressed by migrating axons. This combinatorial code of Robo receptors determines lateral axonal positioning by responding to Slit secreted from the ventral midline. Similarly, differential expression of Robo receptors in the heart (Lower) accounts for the relative cell position of CBs and PCs from a presumptive dorsal gradient of Slit. Robo- and Robo2-expressing PCs maintain their position further away from the dorsal midline than Robo-expressing CBs. Slit and Robos are also implicated in cell adhesion between adjacent cells (facing arrows).
Furthermore, we found that loss of slit, or both robo and robo2, causes defects in cell adhesion, resulting in gaps in the rows of CBs and PCs (Fig. 4D). We observed that often the gaps in the rows of CBs corresponded with the gaps in the PC rows, suggesting that these two cell types must also be adhered to each other. The phenotypes we observed may also be due to a loss of adhesion between these cardiac cells and the overlying dorsal ectoderm. Indeed, in a recent study, Chartier et al. (3) provided evidence that the dorsal ectoderm coordinately migrates with the heart cells during dorsal closure. Although we do not observe any significant defects in dorsal closure in slit mutants (data not shown), it is possible that Slit may also be playing a role in adhesion between the overlying dorsal epithelium and the cardiac cells. How is cell adhesion between adjoining groups of cells regulated? It is likely that Slit or Robo receptors have parallel or cooperative roles with cell adhesion systems during heart formation. In vitro, activation of Robo by Slit interferes with N-cadherin-mediated adhesion (25). Future studies will reveal whether Slit and Robos cooperate with homophilic cell adhesion molecules in the developing heart.
Before this submission, two papers have been recently published that also implicate Slit in Drosophila heart patterning (26, 27). Both of these studies support our findings that Slit plays an important role in regulating cell adhesion between migrating groups of CBs during heart tube assembly. However, these papers differ somewhat, both from each other and from our current study, in their assessment of the role of the two receptors for Slit, Robo and Robo2, during this process. One issue on which these studies disagree is in the expression patterns of Robo and Robo2 in the cells of the heart. In this study, we report that Robo and Robo2 are differentially expressed in the heart. Specifically, our analysis of the expression of both receptors reveals that at the dorsal midline, the inner rows of CBs express Robo, whereas the flanking rows of PCs express both receptors. Interestingly, this expression pattern is similar to what is observed for Robo and Robo2 in the ventral midline of the CNS (16, 17), and we believe this similarity also reflects a comparable function for these receptors at both the ventral and dorsal midlines (Fig. 4E). These findings, which we have confirmed at both the protein and mRNA levels, were not observed in the two other studies. For example, the authors of one study failed to report the coexpression of Robo with Robo2 that we observed in PCs (26). The weaker expression of Robo in PCs as compared with its expression in CBs may account for the fact that this expression pattern was not reported. In the more recently published study, the authors report that robo2 (and not Robo, as we report) is coexpressed with Slit in CBs (27). Surprisingly, the expression of robo was not examined in this study. That these results were based solely on in situ hybridization and were not supported by analysis of protein expression may account for the significant differences between our findings. Another difference between these studies lies in our gain-of-function experiments with Robo2. Specifically, our analysis revealed an important role for Robo2 in specifying the distance a migrating PC maintains from the midline. This phenotype is similar to what is observed at the ventral midline for CNS axons ectopically expressing Robo2 and provides strong support for our positioning model presented in this study.
Together, these differences in our study have led us to propose an alternative model for Slit and Robo receptors in heart cell positioning at the dorsal midline, whereby the combinatorial expression of Robo receptors controls the relative position of individual rows of migrating cells from the dorsal midline during heart tube assembly. Why do cells that express both Robo and Robo2 receptors stay farther away from the dorsal midline than cells that express only Robo? Our results are similar to what is observed at the ventral midline of the Drosophila CNS, where Slit, secreted from the midline glial cells, functions as a repellent to specify the lateral positioning of axons according to the specific combination of Robo receptors that these axons express (refs. 7 and 8; Fig. 4E). During development of the CNS, medial axons expressing the Robo receptor are positioned closer to the ventral midline than lateral axons expressing both Robo and Robo2. From our loss- and gain-of-function genetic experiments presented here, we propose a similar model for heart cell positioning at the dorsal midline during heart tube formation (Fig. 4E). Rows of CBs that express the single Robo receptor migrate closer to the dorsal midline than PCs that express both Robo and Robo2. These development events, although seemingly diverse, share a key similarity. In both cases, bilateral populations of migrating cells are organizing themselves relative to a midline. However, there is also a notable difference between these two circumstances. In the CNS, Slit secreted from the ventral midline glial cells prevents migrating Robo-expressing axonal growth cones from crossing into ligand-expressing territory. At the dorsal midline, Slit is secreted by the innermost CB cells, which are also cells that respond to Slit. This represents a novel intrinsic function for Slit-Robo signaling. Cells expressing Slit are organizing themselves and neighboring cells by virtue of which Robo receptor they express. Further study will reveal the precise nature of Slit’s role in this process and will have important implications for understanding mechanisms of organ self assembly.
A major weakness of our positioning model is in our analysis of the weak robo or robo2 loss-of-function phenotypes. For example, our model would predict that removal of robo2 from PCs would cause these cells to shift to a position closer to the dorsal midline. Although we do occasionally detect mispositioned PCs in robo2 mutants, the phenotypes we observe are not very penetrant or striking. Likewise, robo also has a very mild cardiac phenotype. Although these observations may reflect a flaw in our model, the lack of strong phenotypes for robo or robo2 single mutants could also be explained by the additional roles we believe these molecules play in cell–cell adhesion. By this reasoning, the loss of a single receptor may not be enough to disrupt the adhesion between adjacent cells. We observed the same findings in our gain-of-function experiments with Robo2. Overexpression of Robo2 in CBs in a robo mutant background results in cardiac cell mispositioning, but the adhesion between the rows of cell is maintained. However, ectopic Robo2 in a robo,robo2 double mutant background revealed strong defects in both processes.
Materials and Methods
Drosophila Strains.
The slit mutant used in this study is the null allele slit2 (10). The robo and robo2 alleles used in this study were described previously (26). The UAS-slit and UAS-robo and -robo2 transgenes were described previously (9, 10). The UAS-GAL4 system was used to drive expression of robo, robo2, or slit transgenes with Mef2-GAL4 (22). slit2/CyoWgBgal;UAS-slit was crossed with slit2/CyoWgBgal; Mef2-GAL4 to get slit2/slit2; Mef2-Gal4/UAS-slit embryos. roboZ570/CyoWgBgal; UAS-robo2 was crossed with roboZ570/CyoWgBgal;Mef2-Gal4 to get roboZ570/roboZ570;Mef2-Gal4/UAS-robo2 embryos. robo2X123,roboZ570/CyoWgBgal;UAS-robo2 was crossed with robo2X123,roboZ570/CyoWgBgal;Mef2-Gal4 to get robo2X123,roboZ570;Mef2-Gal4/UAS-robo2 embryos.
Immunofluorescence and RNA Localization.
Embryos were fixed and stained according to standard procedures, as described (9). The following antibodies and dilutions were used: mouse and rat anti-Slit (10), mouse anti-Robo (1:10) and anti-Robo2 (1:1,000; ref. 16), rabbit anti-Mef-2 (1:1,000), mouse anti Zfh1a (1:200), mouse anti-α-Spectrin (1:10), mouse anti-Pericardin/EC11 (1:10; obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Iowa City, IA), FITC anti-mouse (1:500), Alexa 488 anti-rabbit (1:500), Cy3 anti-mouse (1:500) and rabbit (1:500; Molecular Probes, Carlsbad, CA), and anti-b-Gal (1:10,000; Cappel, Solon, OH). Confocal Z sections were collected on an Olympus IX81 with a CARV Nipkow disk confocal unit (Atto Biosciences, Rockville, MD) and SensiCam QE camera (Cooke, Eugene, OR) and analyzed by using IPLab image analysis software (Scanalytics, Billerica, MA). In situ hybridization and antibody staining for robo2 and Mef-2 were performed as described (18).
Measurements.
IPLab image analysis software was used on confocal images to measure the distances between the center of two opposing CB or PC nuclei. For each distance reported, measurements were made between at least 100 pairs of nuclei in 10 different embryos, and the average number is reported. We limited our measurements to the cells in abdominal segments A4–A5 to ensure that the changes in distances we measured were not due to changes resulting from the normal curvature of the embryo. The embryos were carefully staged before measurement by examining gastrulation events, including the formation of head folds and development of the gut.
Electron Microscopy.
Embryos were dechorionated and rinsed briefly with 0.1% TritonX-100 before being fixed in 12.5% glutaraldehyde. After fixation, embryos were staged and dechorionated with a tungsten needle and then embedded in Epon–Spurr resin (Electron Microscopy Services, Hatfield, PA). Ninety-nanometer sections were cut by using Richert Ultracut E Microtome and picked up on a copper grid. Sections were stained with uranyl acetate and lead citrate and examined and photographed by using a JEOL 1200EX electron microscope at 80 kv.
Acknowledgments
We thank Rajesh Patel for assistance with EM; Bruce Paterson (National Cancer Institute–National Institutes of Health, Bethesda, MD), Zhi-Chun Lai (Pennsylvania State University, University Park, PA), and the University of Iowa Hybridoma Bank (Iowa City, IA) for antibodies; Thomas Kidd and Shaun Cordes for early observations; Colleen Guerin and Donald Winkelmann for helpful discussions; and William Wadsworth for the critical reading of the manuscript. We also thank Corey Goodman for his encouragement and support during early stages of this work. This work was supported by a grant from the Foundation of the University of Medicine and Dentistry of New Jersey, by a Scientist Development Grant from the American Heart Association Heritage Affiliate (to S.G.K.), and by National Institutes of Health Predoctoral Fellowship Grant F31/GM75393 (to E.S.M.).
Glossary
Abbreviations
- CB
cardioblast
- PC
pericardial cell
- Robo
Roundabout
- Robo2
Roundabout2.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gumbiner B. M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 2.Haag T. A., Haag N. P., Lekven A. C., Hartenstein V. Dev. Biol. 1999;208:56–69. doi: 10.1006/dbio.1998.9188. [DOI] [PubMed] [Google Scholar]
- 3.Chartier A., Zaffran S., Astier M., Sémériva M., Gratecos D. Development (Cambridge, U.K.) 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- 4.Wang J., Tao Y., Reim I., Gajweski K., Frasch M., Schulz R. A. Mol. Cell. Biol. 2005;25:4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trinh L. A., Stainier D. Y. R. Dev. Cell. 2004;6:371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 6.Zaffran S., Frash M. Circ. Res. 2002;91:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan S., Vivancos V., Nicolas E., Dickson B. J. Cell. 2000;7:1033–1045. doi: 10.1016/s0092-8674(00)00207-5. [DOI] [PubMed] [Google Scholar]
- 8.Simpson J. H., Bland K. S., Fetter R. D., Goodman C. S. Cell. 2000;7:1019–1032. doi: 10.1016/s0092-8674(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 9.Wong K., Park H. T., Wu J. Y., Rao Y. Curr. Opin. Genet. Dev. 2002;12:583–591. doi: 10.1016/s0959-437x(02)00343-x. [DOI] [PubMed] [Google Scholar]
- 10.Kramer S. G., Kidd T., Simpson J., Goodman C. S. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [DOI] [PubMed] [Google Scholar]
- 11.Kidd T., Bland K. S., Goodman C. S. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 12.Brose K., Bland K. S., Wang K. H., Arnott D., Henzel W., Goodman C. S., Tessier-Lavigne M., Kidd T. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 13.Tayler T. D., Robichaux M. B., Garrity P. A. Development (Cambridge, U.K.) 2004;131:5935–5945. doi: 10.1242/dev.01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothberg J. M., Jacobs J. R., Goodman C. S., Artavanis-Tsakonas S. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- 15.Fremion F., Astier M., Zaffran S., Guillen A., Homburger V., Semeriva M. J. Cell Biol. 1999;145:1063–1076. doi: 10.1083/jcb.145.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopalan S., Nicolas E., Vivancos V., Berger J., Dickson B. J. Neuron. 2000;28:767–777. doi: 10.1016/s0896-6273(00)00152-5. [DOI] [PubMed] [Google Scholar]
- 17.Simpson J. H., Kidd T., Bland K. S., Goodman C. S. Neuron. 2000;28:753–766. doi: 10.1016/s0896-6273(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 18.Lilly B., Zhao B., Ranganayakulu G., Paterson B. M., Schulz R. A., Olson E. N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 19.Lee J. K., Coyne R. S., Dubreuil R. R., Goldstein L. S., Branton D. J. Cell Biol. 1993;123:1797–1809. doi: 10.1083/jcb.123.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tepass U. BioEssays. 1997;8:673–682. doi: 10.1002/bies.950190807. [DOI] [PubMed] [Google Scholar]
- 21.Fossett N., Zhang Q., Gajewski K., Choi C. Y., Kim Y., Schulz R. A. Proc. Natl. Acad. Sci. USA. 2000;97:7348–7353. doi: 10.1073/pnas.97.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand A. H., Perrimon N. Development (Cambridge, U.K.) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Lai Z. C., Fortini M. E., Rubin G. M. Mech. Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 24.Compernolle V., Brusselmans K., Franco D., Moorman A., Dewerchin M., Collen D., Carmeliet P. Cardiovasc. Res. 2003;60:569–579. doi: 10.1016/j.cardiores.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Rhee J., Mahfooz N. S., Arregui C., Lillien J., Balsamo J., VanBerkum M. F. Nat. Cell Biol. 2002;4:798–805. doi: 10.1038/ncb858. [DOI] [PubMed] [Google Scholar]
- 26.Qian Li., Liu J., Bodmer R. Curr. Biol. 2005;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 27.MacMullin A., Jacobs J. R. Dev. Biol. 2006;293:154–164. doi: 10.1016/j.ydbio.2006.01.027. [DOI] [PubMed] [Google Scholar]