Abstract
Toll-like receptors (TLRs) are key components of the immune system that detect microbial infection and trigger antimicrobial host defense responses. TLR5 is a sensor for monomeric flagellin, which is a component of bacterial flagella known to be a virulence factor. In this study we generated TLR5-deficient mice and investigated the role of TLR5 signaling in the detection of flagellin and antibacterial immune responses to Salmonella typhimurium and Pseudomonas aeruginosa. We found that TLR5 is essential for the recognition of bacterial flagellin both in vivo and ex vivo. TLR5 contribution to antibacterial host response to i.p. infection with S. typhimurium or intranasal administration of P. aeruginosa may be masked by TLR4 or other sensing mechanisms. By using radiation bone marrow chimera, we showed that upon i.p. injection of flagellin immune responses are mediated by lymphoid cells, whereas resident cells are required for the initiation of response upon intranasal flagellin administration. These results suggest that flagellin recognition in different organs is mediated by distinct TLR5-expressing cells and provide insights into the cooperation of the TLR5 and TLR4 signaling pathways used by the innate immune system in the recognition of bacterial pathogens.
Keywords: bacterial infection, flagellin
Recognition of microbial infection and initiation of immune response are controlled by multiple mechanisms. Toll-like receptors (TLRs) have recently emerged as key components of the innate immune system that recognize common molecular structures detected in certain groups of microorganisms and trigger the activation of adaptive immunity (1). Each TLR detects specific microbial components. For example, TLR4 recognizes LPS, TLR2 recognizes bacterial lipoproteins and lipoteichoic acid, and TLR3 recognizes viral double-stranded RNA. All TLRs share a common intracellular domain, the Toll-IL-1 receptor homology domain, and upon activation initiate signaling cascades that lead to common responses such as the induction of inflammatory cytokines and up-regulation of costimulatory molecules. Moreover, TLRs also have specific functions as exemplified by their different ability to induce type I IFN (1). Thus, TLRs activate multiple steps in the inflammatory reactions that help to eliminate the invading pathogens and coordinate systemic defenses.
Among TLRs, TLR5 is the receptor for flagellin, the major constituent of bacterial flagella and a virulence factor for Gram-negative and Gram-positive bacteria (2, 3). TLR5 engagement by bacterial flagellin activates the MyD88-dependent signaling pathway, which leads to the nuclear translocation of NF-κB and the activation of the MAPKs, ultimately inducing the maturation of antigen-presenting cells and the secretion of proinflammatory cytokines and chemokines (4–8). TLR5 is expressed by a variety of cells including monocytes, dendritic cells (DCs), epithelial cells, and mast cells (5, 9–13). Interestingly, TLR5 is expressed on the basolateral side of intestinal epithelial cells, which are chronically exposed to commensal bacteria (6, 14), but it is expressed on the apical side of lung epithelial cells, which are usually not exposed to microorganisms (15). The importance of TLR5 signaling in mucosal immunity has been highlighted by recent studies, where diverse human disorders have been attributed to the dysfunction of TLR5. It has been shown that a common stop codon polymorphism of TLR5 is associated with susceptibility to pneumonia caused by the flagellated bacterium Legionella pneumophila (16), whereas certain flagellated bacteria can escape from TLR5 recognition by amino acid change on flagellin and persist at the mucosal surfaces (17–19). Moreover, TLR5-dependent recognition of flagellin may play a role in the inappropriate mucosal immune response to commensal bacteria associated with inflammatory bowel disease, such as Crohn’s disease (20, 21). Nevertheless, a number of questions related to the importance of TLR5 signaling during infection with flagellated bacteria and especially in initiating and directing the immune responses remain unanswered.
In this study we demonstrate the requirement for TLR5 in the recognition of flagellin and TLR5–TLR4 cooperation in the antibacterial immune response. We generated TLR5-deficient (TLR5−/−) mice and characterized their responses upon flagellin stimulation in vitro and in vivo. Also, we investigated the importance of TLR5 and TLR4/TLR5 signaling upon systemic infection by Salmonella typhimurium or lung infection upon Pseudomonas aeruginosa administration. Finally, we addressed the role of TLR5 signaling on bone marrow (BM) and non-BM-derived cells in response to flagellin using radiation BM chimera. Our findings suggest an important role for TLR5–TLR4 cooperation in the recognition of a wide range of human flagellated Gram-negative bacteria.
Results
Generation and Characterization of TLR5−/− Mice.
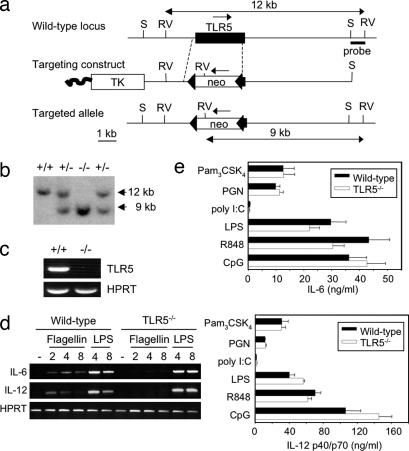
To characterize the biological function of TLR5 we generated TLR5−/− mice by homologous recombination in ES cells (Fig. 1a and b; see Methods). The expression of TLR5 messenger RNA in TLR5−/− and WT macrophages was assessed by RT-PCR, and we found that TLR5 transcripts were absent in TLR5−/− but not in WT cells (Fig. 1c). TLR5−/− mice had normal growth, size, fertility, and lifespan and showed no obvious behavioral abnormalities. Flow cytometric analysis of leukocytes from the spleen, lymph nodes, and thymus (stained for CD3, B220, CD4, and CD8) revealed that the composition of lymphocytes was similar in WT and TLR5−/− mice (data not shown).
Fig. 1.
Targeted disruption of the mouse TLR5 gene. (a) The structure of the TLR5 gene, the targeting construct, and the predicted mutated allele. Filled box, exon; filled triangles, loxP sites; open boxes, selection marker genes. The location of the 3′ external probe is shown. Restriction enzymes: S, SalI, RV, EcoRV. (b) Southern blot analysis of mouse genomic DNA digested with EcoRV gave a 12-kb band for WT (+/+), a 9-kb band for homozygous (−/−), and both bands for heterozygous (+/−) mice. (c) Expression of TLR5 and hypoxanthine phosphoribosyltransferase by BM-derived macrophages from WT (+/+) or TLR5−/− mice as determined by RT-PCR. (d) RNA was extracted from WT or TLR5−/− BMDCs stimulated with 100 ng/ml flagellin or 100 ng/ml LPS for 2, 4, or 8 h or left untreated (−), and the expression of IL-6 and IL-12 was determined by RT-PCR. (e) BMDCs from WT or TLR5−/− mice were stimulated with 10 ng/ml Pam3CSK4, 1 μg/ml peptidoglycan, 10 μg/ml polyI:C, 1 ng/ml LPS, 100 nM R848, or 1 μM CpG for 16 h, and concentrations of IL-6 and IL-12 in the culture supernatants were measured by ELISA.
Next we analyzed the responses of TLR5−/− cells to flagellin. BM-derived dendritic cells (BMDCs) or macrophages from WT and TLR5−/− mice were stimulated with 1 or 3 μg/ml flagellin. After 16 h the culture supernatants were collected, and the protein levels of IL-6, IL-12, and TNF-α were assessed by ELISA. Although no significant amounts of these cytokines were detectable (data not shown), the mRNA expression levels of IL-6 and IL-12 were significantly up-regulated in WT DCs stimulated with flagellin, but not in TLR5−/− cells, whereas they showed similar responses upon LPS stimulation (Fig. 1d). Similar results were obtained with WT and TLR5−/− macrophages (data not shown). To demonstrate the specificity of the ligand, we examined the responses of TLR5−/− BMDCs to various microbial molecules that are known to activate other TLR counterparts. WT and TLR5−/− DCs stimulated with Pam3CSK4 (TLR2 ligand), peptidoglycan (Nod ligand), poly I:C (TLR3 ligand), LPS (TLR4 ligand), R848 (TLR7 ligand), and unmethylated CpG DNA (TLR9) produced similar quantities of IL-6 and IL-12 (Fig. 1e). These results reveal that the unresponsiveness of the TLR5−/− cells is specific to the recognition of flagellin.
TLR5 Is Required for Responsiveness to Flagellin.
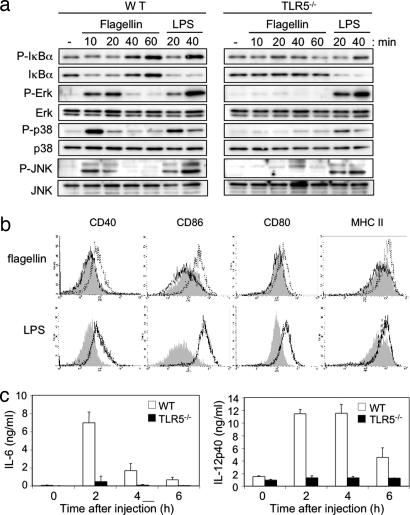
To dissect the signaling pathways that are affected in the absence of a functional TLR5 we analyzed the activation of the signaling cascades in TLR5−/− DCs upon stimulation with flagellin. We found that flagellin-induced degradation of IκΒα and phosphorylation of IκΒα, ERK, p38, and JNK were completely abolished in TLR5−/− DCs but not in WT cells (Fig. 2a). However, their responses to LPS were identical (Fig. 2a). Thus, flagellin-mediated signal transduction depends on TLR5.
Fig. 2.
Unresponsiveness of TLR5−/− cells and mice upon flagellin stimulation. (a) BMDCs from WT or TLR5−/− mice were stimulated with 100 ng/ml flagellin or 1 μg/ml LPS. At the indicated time points, cells were lysed; phosphorylation of IκBα, ERK, p38, and JNK and degradation of IκBα were analyzed by Western blot. (b) WT and TLR5−/− mice were injected i.p. with 1 μg of flagellin and 100 μg of LPS or PBS. After 6 h splenocytes were collected, stained, and analyzed by flow cytometry. Histograms show expression levels of CD40, CD86, CD80, and MHC class II on CD11chigh gated cells. Gray area, untreated cells; dotted line, WT cells; continuous line, TLR5−/− cells. (c) WT and TLR5−/− mice were injected i.p. with 1 μg of flagellin. Sera were collected at the indicated time points, and serum levels of IL-6 and IL-12p40 were determined by ELISA. The data are means ± SE of three mice per group.
We next tested the ability of TLR5−/− DCs to mature in vivo in response to flagellin. WT and TLR5−/− mice were injected i.p. with flagellin, LPS, or PBS, and 6 h later the expression levels of CD40, CD80, CD86, and MHC class II on CD11c+ splenic cells were analyzed. A significant increase in the expression of these activation markers was observed in WT CD11c+ cells upon flagellin and LPS stimulation. In contrast, TLR5−/− cells did not show any response to flagellin despite exhibiting normal responses to LPS (Fig. 2b). To investigate whether signaling by TLR5 is required for the in vivo inflammatory response to flagellin, mice were injected i.p. with flagellin, and the levels of IL-6 and IL-12 in their sera were assessed 2, 4, and 6 h later. WT mice showed a marked elevation of serum concentrations of IL-6 at 2 h and IL-12 at 2 and 4 h after flagellin challenge (Fig. 2c). In contrast, TLR5−/− mice were unresponsive to flagellin (Fig. 2c). These results demonstrate that TLR5 is essential for the recognition of bacterial flagellin both ex vivo and in vivo.
Survival of TLR5−/− Mice to Challenge with S. typhimurium.
To elucidate the role of TLR5 in host defense, we infected TLR5−/− mice with the Gram-negative bacterium S. typhimurium, a common human pathogen isolated from many cases of acute food-borne gastroenteritis. We used two virulent strains of S. typhimurium, a flagellated strain (SB300) and an isogenic mutant, nonflagellated derivative (SB762). TLR5−/− and WT mice were infected i.p. with S. typhimurium and monitored twice daily for morbidity and survival, and the time of death was recorded. Both groups of mice succumbed to infection with the WT and nonflagellated S. typhimurium strain in a similar time frame (Fig. 3a and b). Furthermore, we infected WT and TLR5−/− mice i.p. with flagellated S. typhimurium, and the bacterial load in spleens and livers was examined 24 h after the infection. In both organs the number of the bacterial cfu of infected TLR5−/− mice was similar to WT controls (data not shown). Previous studies with C3H/HeJ mice carrying a natural dominant-negative mutation in the cytoplasmic domain of TLR4 and TLR4-deficient mice have uncovered the critical role of TLR4 to host resistance to Salmonella (22, 23). To tackle the possibility that TLR4 signaling compensates for the loss of functional TLR5 in TLR5−/− mice during S. typhimurium infection we performed the above infection studies using double TLR4/TLR5-deficient (TLR4/5−/−) mice. As expected, TLR4−/− mice showed an increased susceptibility to S. typhimurium infection compared with WT or TLR5−/− mice (Fig. 3c). Interestingly, we also observed that double TLR4/5−/− mice were somewhat more susceptible than either single TLR4−/− or TLR5−/− mice (Fig. 3c). Next, we expanded our studies using the nonflagellated S. typhimurium, and with this strain we observed no difference in survival between double TLR4/5−/− and TLR4−/− mice (Fig. 3d). These findings suggest that the potential antibacterial effect of TLR5 in this infection model is compensated for by the presence of a functional TLR4 or other flagellin-sensing mechanisms.
Fig. 3.
Role of TLR5 in host defense upon S. typhimurium infection. WT, TLR5−/−, TLR4−/−, or double TLR4/5−/− mice were injected i.p. with 1 × 104 cfu per 20 g of body weight of flagellated SB300 (a and c) or nonflagellated SB762 (b and d) S. typhimurium. The group of mice were n = 22 (a), n = 19 (b), n = 8 (c), and n = 6 (d). Data in a and c are representative of three independent experiments.
Role of TLR5 in Inflammatory Lung Response Against Flagellin.
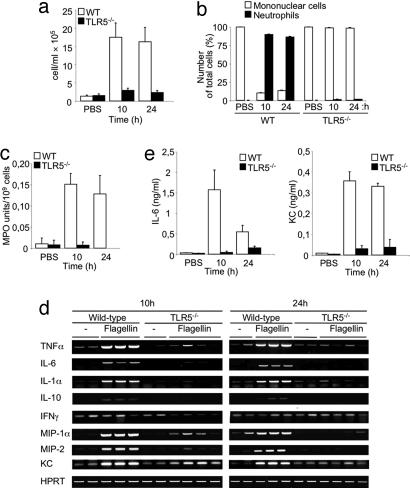
Recombinant flagellin stimulates a transient innate immune response in the lung of several strains of mice characterized by the infiltration of neutrophils and rapid production of cytokines and chemokines (24, 25). To determine the role of TLR5 in innate immunity in the lung we challenged WT and TLR5−/− mice intranasally (i.n.) with flagellin. Ten and 24 h later the animals were killed, and the bronchoalveolar lavage fluids (BALFs) were collected and analyzed. The total cell count was significantly elevated in WT BALFs at both 10 and 24 h after challenge, whereas no cell infiltration was observed in TLR5−/− BALFs (Fig. 4a). The total neutrophil counts performed in the same BALFs showed a significant neutrophil accumulation in the lungs of flagellin-treated control mice at both 10 and 24 h after treatment (Fig. 4b). In contrast, neutrophils were absent in TLR5−/− BALFs (Fig. 4b). These results were confirmed by measuring myeloperoxidase (MPO) activity in cell pellets from WT and TLR5−/− BALFs (Fig. 4c). Next we examined the expression levels of several cytokines and chemokines that are important for neutrophil migration in vivo. We observed significant increases in TNF-α, IL-6, IL-1α, IL-10, and IFN-γ cytokines and macrophage inflammatory protein (MIP) 1α, MIP-2, and keratinocyte chemoattractant (KC) chemokines at the mRNA level in WT lungs, at both 10 and 24 h upon flagellin instillation, whereas TLR5−/− lungs showed no or very slight expression of the same molecules (Fig. 4d). Moreover, flagellin administration induced a marked increase of IL-6 and KC at the protein level in the WT BALFs, whereas there was a very little production in TLR5−/− BALFs (Fig. 4e). These results suggest that the flagellin-induced pulmonary inflammatory response is TLR5-dependent.
Fig. 4.
Unresponsiveness of TLR5−/− mice to i.n. administration of flagellin. WT and TLR5−/− mice were treated i.n. with 1 μg of flagellin or PBS, and 10 or 24 h later BALFs and lungs were collected. (a–c) Total cell count (a), cellular composition (b), and determination of MPO activity (c) in WT and TLR5−/− BALFs. (d) RNA was extracted from treated lungs, and the expression of TNF-α, IL-6, IL-1α, IL-10, IFN-γ, MIP-1α, MIP-2, and KC was determined by RT-PCR. Each column represents an individual mouse. (e) IL-6 and KC levels in BALFs were quantified by ELISA. Each bar represents the mean ± SEM (n = 4–6).
Survival of TLR5−/− Mice upon P. aeruginosa Lung Infection.
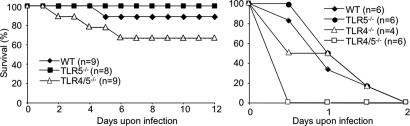
Previous studies have shown that the resistance of mice to P. aeruginosa acute lung infection is MyD88-dependent and does not seem to require TLR2 or TLR4, because mice lacking both TLR2 and TLR4 did not show increased susceptibility (26). To explore the role of TLR5 in host defense against P. aeruginosa lung inflammation, WT, TLR5−/−, and double TLR4/5−/− mice were inoculated i.n. with 1 × 106 cfu of P. aeruginosa strain PAK and monitored twice daily for morbidity and survival, and the time of death was recorded. Both WT and TLR5−/− mice showed similar rates of survival, whereas TLR4/5−/− mice showed higher susceptibility (Fig. 5Left). Next we infected i.n. WT, TLR4−/−, TLR5−/−, and double TLR4/5−/− mice with a higher dose of P. aeruginosa (5 × 106 cfu). Interestingly, WT, TLR4−/−, and TLR5−/− mice showed quite similar rates of survival, and all of them died by day 2, whereas TLR4/5−/− mice showed increased susceptibility and succumbed by 12 h after the infection (Fig. 5 Right). These data suggest that cooperation between TLR4 and TLR5 is important for effective innate response to P. aeruginosa lung infection.
Fig. 5.
Survival curves of WT, TLR5−/−, TLR4−/−, and TLR4/5−/− mice upon i.n. P. aeruginosa infection. Survival of 7-week-old mice infected i.n. with 1 × 106 cfu (Left) and 6 × 106 cfu (Right) of P. aeruginosa was monitored every 12 h for 12 days.
Role of TLR5 Expression in BM and Non-BM Cells upon in Vivo Flagellin Challenge.
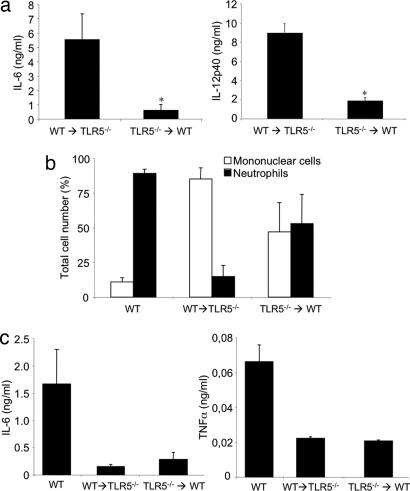
In an attempt to clarify the relative contribution of BM and non-BM-derived TLR5-dependent signaling in the production of proinflammatory cytokines upon flagellin stimulation in vivo we constructed radiation BM chimera between TLR5−/− (CD45.2) and WT congenic CD45.1 mice. Two groups of mice were compared: WT→TLR5−/− (WT BM into a TLR5−/− mouse) and TLR5−/−→WT (TLR5−/− BM into a WT mouse). Six weeks after irradiation and BM transfer, reconstitution of circulating leukocytes was determined by staining leukocytes for CD45.1 and CD45.2. We found that macrophages, DCs, B cells, and T cells were reconstituted at 98%, 88%, 99%, and 85%, respectively, with BM-derived cells in all mixed chimera. At week 7 after reconstitution chimeras were injected i.p. with flagellin, and production of IL-6 and IL-12 in their sera was measured 2 h later. WT→TLR5−/− chimeras produced significant amounts of IL-6 (5.5 ± 1.9 ng/ml) and IL-12 (8.9 ± 1.9 ng/ml) in their sera, whereas TLR5−/−→WT chimeras showed a very slight response (0.6 ± 0.5 ng/ml for IL-6 and 0.9 ± 0.2 ng/ml for IL-12) (Fig. 6a). These data demonstrate that the major producers of serum IL-6 and IL-12 upon i.p. injection of flagellin are BM-derived cells.
Fig. 6.
Role of TLR5 in BM and non-BM cells in response to flagellin stimulation in vivo. (a) BM WT→TLR5−/− and BM TLR5−/−→WT chimera were injected i.p. with 1 μg of flagellin. Two hours later sera were collected, and production of IL-6 (Left) and IL-12p40 (Right) was assessed by ELISA. (b and c) WT mice and BM chimeras were instilled i.n. with 1 μg of flagellin, and 10 h later BALFs were collected and used for the determination of cellular composition (b) and IL-6 and TNF-α production (c). The data are means ± SE. ∗, P < 0.05. n = 5 WT→TLR5−/−, and n = 4 (a) and 3 (b and c) BM TLR5−/−→WT.
Next we examined the contribution of BM- and non-BM–TLR5-expressing cells in the model of flagellin-induced lung inflammation. At week 12 after reconstitution BM chimera were instilled i.n. with flagellin, and pulmonary neutrophilic infiltration and cytokine production in BALFs were evaluated. WT→TLR5−/− showed no significant neutrophil accumulation in airspaces or IL-6 production, whereas TLR5−/−→WT showed a considerable neutrophilic infiltration that was accompanied by a slight production of IL-6 and TNF-α compared with WT treated mice (Fig. 6 b and c). These data suggest that recognition of flagellin by non-BM–TLR5-expressing cells is required for the initiation of a lung inflammatory response.
Discussion
Although it is well established that TLR5 recognizes bacterial flagellin, recent studies have identified Ipaf as an essential sensor for cytoplasmic flagellin (27, 28). Thus, mammalian cells sense extracellular flagellin through TLR5 and intracellular flagellin through Ipaf. TLR5 activates NF-κB and MAPKs, leading to the secretion of many cytokines, including IL-6, IL-12, and TNF-α, whereas Ipaf permits the activation of caspase-1 and secretion of mature IL-1β. In the current study we describe the generation and characterization of TLR5-deficient mice and show that TLR5 is crucial for the in vivo recognition of flagellin but also may participate in the detection of systemic infection by S. typhimurium and lung infection by P. aeruginosa, although these phenomena can be masked by the presence of a functional TLR4 gene or other flagellin-sensing mechanisms.
In our studies, although in vitro stimulation of WT BMDCs by flagellin led to a strong early activation of IκBα and MAPKs and expression of IL-6 and IL-12 cytokines at the mRNA level, the protein levels of these cytokines were undetectable in the culture supernatants (Figs. 1d and 2a). In contrast, we observed that in vivo stimulation of control mice with flagellin led to strong secretion of these cytokines in sera and maturation of splenic DCs, responses that are undetectable in TLR5 mice (Fig. 2 b and c). These results can be explained by the absence of cellular interactions and factors in our culturing conditions that block excessive flagellin-induced activation of TLR5 in vitro. Indeed, TLR5 signaling induces rapid activation of phosphoinositide 3-kinase that serves to limit MAPK signaling, thus limiting the potential negative consequences of excessive cytokine production (29).
To elucidate the role of TLR5 in the induction of antibacterial host defense, we tested the susceptibility of TLR5−/− mice upon i.p. infection with the bacterium S. typhimurium. We found no difference in survival between WT and TLR5−/− mice upon infection with flagellated or nonflagellated S. typhimurium. This finding is in accord with a recent report showing that the extreme susceptibility to infection of MOLF/Ei mice, a natural TLR5-deficient mouse, to S. typhimurium was not due to the absence of TLR5 (30). However, LPS is an important virulence factor of Salmonella, and the absence of a functional TLR4 leads to increased susceptibility upon S. typhimurium infection (22, 23). Because a general concept for TLR function is that multiple TLRs act in partnership in determining pathogen control, we speculated that the role of TLR5 in host resistance to S. typhimurium could be masked by the presence of a functional TLR4 gene. To test our hypothesis, we compared the survival rates of double TLR4/5−/− versus single TLR4−/− and TLR5−/− mice to S. typhimurium infection. Infected double TLR4/5−/− mice displayed enhanced susceptibility in direct contrast to TLR4−/− mice, which were slightly more susceptible than TLR5−/− or control animals (Fig. 3c). The rather modest effect of the absence of TLR5 in S. typhimurium infections may be due to the existence of redundant mechanisms to sense the bacteria and flagellin itself by the innate immune system. The fact that the nonflagellated mutant of Salmonella somewhat masked the increased susceptibility of the TLR4/5−/− mice (Fig. 3d) supports this hypothesis. Thus, although TLR5 may have a protective role in host resistance to S. typhimurium, it is clear that its action can be substituted by the action of TLR4 and probably other flagellin-sensing mechanisms.
It has been shown previously (24) and confirmed in this study that i.n. administration of purified flagellin in mice induces a transient innate immune response in the lung characterized by production of inflammatory mediators that affect neutrophil recruitment. Indeed, we showed here that all these responses were absent in the lungs of TLR5−/− mice that have been i.n. challenged with flagellin, demonstrating that the inflammatory lung response against flagellin is mediated by TLR5 (Fig. 4) and suggesting that TLR5 might play a critical role in the clearance of pulmonary infection that has been induced by flagellated bacteria. Although MyD88, the major adaptor molecule required for signaling events by most TLR/IL-1R family members, appears to play a major role in host resistance to P. aeruginosa, it has been difficult to attribute this requirement to the function of a single TLR. Thus, mice deficient in TLR2, TLR4, TLR2/TLR4, or IL-18, although in some cases displaying specific defects in antibacterial responses, exhibit no or only minor increases in susceptibility to i.n. challenge (26, 31). In contrast, absence or reduction in endogenous IL-1 activity improves host defense against Pseudomonas pneumonia (32). Our results showed that the lack of TLR5 alone cannot explain the increased susceptibility of MyD88−/− mice to P. aeruginosa lung infections. We also observed that TLR4/5−/− mice were more susceptible to P. aeruginosa lung infection than WT or TLR5−/− mice (Fig. 5). These results suggest that an efficient lung immune response against P. aeruginosa requires the simultaneous activation of TLR4, TLR5, and presumably other innate immune receptors and that the activation of one can compensate for the absence of activation of any of the others. Based on the data that we obtained from two infectious models, i.p. Salmonella and i.n. Pseudomonas, in the current study we could speculate that, depending on the bacterium that causes the infection, the route of infection, and the primary organ that is affected, the protective action of TLR5 can be substituted in a certain degree by another innate immune receptor (in our examples, TLR4).
We used radiation BM chimera between WT and TLR5−/− mice and found that, upon i.p. flagellin challenge, the early cytokine production in vivo requires TLR5 expression by BM-derived cells. The complex mechanisms of inflammatory lung response presumably involve different cell types, including resident and hematopoietic cells. Indeed, we observed that the initiation of the inflammatory response in the lung, characterized by neutrophil infiltration, is mediated by TLR5 signaling by resident cells (Fig. 6b). We hypothesized that epithelial cells mediate this response, because they are the first non-BM-derived cells to directly contact flagellin. This finding is in agreement with the apical localization of TLR5 on the airway epithelium that can respond specifically to bacterial flagellin (33). This is not the case for other TLRs, because upon inhaled LPS the genotype of the donor BM cells seems to influence only in part the neutrophil recruitment, and both resident and myeloid cells are required for a full recruitment of neutrophils via MyD88 signaling (34). However, a recent study (where the requirement of MyD88 expression in stromal and hematopoietic cells was addressed in BM chimeras that have been challenged by P. aeruginosa) has shown that the early recruitment of polymorphonuclear cells required MyD88 expression by non-BM-derived cells and that early cytokine production was BM-dependent, whereas chemokine production was a non-BM-dependent phenomenon (35).
The host relies on two sensors for the detection of invading flagellated bacteria: TLR5 on the cell membrane and Ipaf in the cytoplasm. The relative importance of each one of these two molecules in the antibacterial host response likely depends on the type of the pathogen that is infecting the host, where extracellular bacteria might activate TLR5, whereas intracellular bacteria could trigger both TLR5 and Ipaf. Therefore, it would be interesting to address the host immune response to flagellated bacteria in the context of a system that both TLR5 and Ipaf signaling pathways are abrogated.
Methods
Mice.
For the generation of the TLR5−/− mice, a genomic DNA fragment containing the TLR5 gene was screened from a 129/Sv mouse genomic library. The targeting vector was constructed by replacing a 2.6-kb fragment of the TLR5 exon, including the start codon of the TLR5 gene, with a neomycin-resistance gene cassette flanked by two loxP sites. A herpes simplex virus thymidine kinase gene, driven by an MC1 promoter, allowed for negative selection. The targeting vector was transfected into embryonic W9.5 ES cells. Targeted ES cells were identified by Southern blotting and subsequently injected into C57BL/6 blastocysts. Male chimeric mice were then mated to C57BL/6 female mice. Heterozygous TLR5 mice were intercrossed to generate TLR5-deficient mice. TLR5−/− mice on the C57BL/6 background (10 generations) and WT C57BL/6 mice were used for all of the experiments. TLR4−/− mice (36) (C57BL/6) were a gift from S. Akira (Osaka University, Osaka, Japan). WT C57BL/6 congenic CD45.1 and CD45.2 mice were purchased from Charles River (L’Arbresle, France). Double TLR4/5−/− mice were obtained by intercrossing TLR5−/− with TLR4−/− mice (36).
Reagents.
Endotoxin-free flagellin fliB from S. typhimurium was kindly provided by VaxInnate (New Haven, CT). LPS from E. coli was from Sigma (St. Louis, MO); peptidoglycan from Staphylococcus aureus was from Fluka (Buchs, Switzerland); polyI:C, R848, and Pam3CSK4 were from Invivogen (San Diego, CA); and CpG DNA was synthesized by Sigma-Genosys. (St. Louis, MO).
Salmonella and Pseudomonas Challenge of Mice in Vivo.
Age- and sex-matched groups of mice were infected i.p. with S. typhimurium or i.n. with P. aeruginosa. The P. aeruginosa PAK strain was a gift from Alain Filloux (Institut de Biologie Structurale et Microbiologie–Centre National de la Recherche Scientifique, Marseille, France). For i.n. challenge, mice were anesthetized by i.p. injection of ketamine–xylazine, and a 40-μl suspension of 1 μg of flagellin or P. aeruginosa was applied i.n. Lungs were collected at 10 or 24 h after challenge and homogenized, and RNA was extracted by TRIzol. BALFs were collected by five instillations of 0.5 ml of normal saline.
BMDCs and Macrophages.
BMDCs and BM macrophages were generated as previously described (37). Immature BMDCs and BM macrophages were collected at days 6 and 5, respectively, and plated at 1 × 106 cells per milliliter in the presence or absence of stimuli.
RT-PCR.
Total RNA from BMDCs or mouse lungs was isolated with TRIzol reagent (Invitrogen), and contaminant DNA was removed by DNase I (Ambion, Huntingdon, U.K.) according to the manufacturer’s instructions. Total RNA (5 μg) was reverse transcribed by using SuperScript II reverse transcriptase (Invitrogen). cDNAs were amplified with specific primers for cytokines, chemokines, or hypoxanthine phosphoribosyltransferase.
Measurement of Cytokine, Chemokine, and MPO Production.
Concentrations of IL-6 (eBioscience, San Diego, CA), IL-12p40 (BD Bioscience, San Jose, CA), and KC (Biosource, Nivelles, Belgium) in culture supernatants, sera, or cell-free BALF were measured by ELISA. The MPO activity was determined in cell pellets derived from BALFs resuspended in 200 μl of substrate solution containing 0.5% hexadecyltrimethylammonium bromide, 0.17 mg/ml O-dianisidine dihydrochloride, and 0.05% H2O2. The enzymatic activity was determined by measuring the change in absorbance at 460 nm every minute.
Western Blotting.
Cytosolic protein extracts (20 μg) from BMDCs were resolved on 10% SDS/PAGE gels and transferred to Immunobilon P membrane (Millipore, Billerica, MA). Blotting was performed with the indicated antibodies (Cell Signaling, Danvers, MA). Bands were visualized with secondary HRP-conjugated antibodies and the ECL System (Amersham Pharmacia, Piscataway, NJ).
BM Chimeras.
Female mice (8–9 weeks of age) were lethally irradiated with 600 rads using a 137Cs source and injected i.v. 3–4 h later with 2 × 106 BM cells. All mice were placed on Baytril for 3 weeks after irradiation. The degree of chimerism for different hematopoietic-derived cells type (B cells, T cells, DCs, and macrophages) was assessed by measuring CD45.1 and CD45.2 expression by blood leukocytes. Red blood cells were stained for CD45.1, CD45.2, B220, CD3, CD11b, or CD11c (Pharmingen, San Diego, CA) at the appropriate dilution and analyzed on a FACSCalibur cytometer (Becton Dickinson, Franklin Lakes, NJ).
Acknowledgments
We thank S. Akira for providing the TLR4−/− mice; A. Filloux for providing the P. aeruginosa strains; E. A. Hughes and J. Stein for initial help in this study; and J. P. Gorvel and S. P. Salcedo for help. V.F. received a Fondation Recherche Médicale postdoctoral fellowship, and S.M. received a Centre National de la Recherche Scientifique postdoctoral fellowship. R.A.F. is an Investigator of the Howard Hughes Medical Institute. L.A. is supported in part by an Action Thématique et Incitative sur le Programme (ATIP) grant from the Centre National de la Research Scientific and a Chair of Excellence grant from the French Ministry of Research.
Glossary
Abbreviations
- BALF
bronchoalveolar lavage fluid
- BM
bone marrow
- DC
dendritic cell
- BMDC
BM-derived DC
- i.n.
intranasal
- TLR
Toll-like receptor
- MPO
myeloperoxidase
- MIP
macrophage inflammatory protein
- KC
keratinocyte chemoattractant.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Akira S., Uematsu S., Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 3.Ramos H. C., Rumbo M., Sirard J. C. Trends Microbiol. 2004;12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Didierlaurent A., Ferrero I., Otten L. A., Dubois B., Reinhardt M., Carlsen H., Blomhoff R., Akira S., Kraehenbuhl J. P., Sirard J. C. J. Immunol. 2004;172:6922–6930. doi: 10.4049/jimmunol.172.11.6922. [DOI] [PubMed] [Google Scholar]
- 5.Eaves-Pyles T., Murthy K., Liaudet L., Virag L., Ross G., Soriano F. G., Szabo C., Salzman A. L. J. Immunol. 2001;166:1248–1260. doi: 10.4049/jimmunol.166.2.1248. [DOI] [PubMed] [Google Scholar]
- 6.Gewirtz A. T., Navas T. A., Lyons S., Godowski P. J., Madara J. L. J. Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 7.Tallant T., Deb A., Kar N., Lupica J., de Veer M. J., DiDonato J. A. BMC Microbiol. 2004;4:1–24. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y., Zeng H., Lyons S., Carlson A., Merlin D., Neish A. S., Gewirtz A. T. Am. J. Physiol. 2003;285:G282–G290. doi: 10.1152/ajpgi.00503.2002. [DOI] [PubMed] [Google Scholar]
- 9.Applequist S. E., Wallin R. P., Ljunggren H. G. Int. Immunol. 2002;14:1065–1074. doi: 10.1093/intimm/dxf069. [DOI] [PubMed] [Google Scholar]
- 10.Gewirtz A. T., Simon P. O., Jr., Schmitt C. K., Taylor L. J., Hagedorn C. H., O’Brien A. D., Neish A. S., Madara J. L. J. Clin. Invest. 2001;107:99–109. doi: 10.1172/JCI10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulka M., Alexopoulou L., Flavell R. A., Metcalfe D. D. J. Allergy Clin. Immunol. 2004;114:174–182. doi: 10.1016/j.jaci.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Means T. K., Hayashi F., Smith K. D., Aderem A., Luster A. D. J. Immunol. 2003;170:5165–5175. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Boado Y. S., Cobb L. M., Deora R. Infect. Immun. 2005;73:7525–7534. doi: 10.1128/IAI.73.11.7525-7534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed K. A., Hobert M. E., Kolenda C. E., Sands K. A., Rathman M., O’Connor M., Lyons S., Gewirtz A. T., Sansonetti P. J., Madara J. L. J. Biol. Chem. 2002;277:13346–13353. doi: 10.1074/jbc.M200149200. [DOI] [PubMed] [Google Scholar]
- 15.Adamo R., Sokol S., Soong G., Gomez M. I., Prince A. Am. J. Respir. Cell Mol. Biol. 2004;30:627–634. doi: 10.1165/rcmb.2003-0260OC. [DOI] [PubMed] [Google Scholar]
- 16.Hawn T. R., Verbon A., Lettinga K. D., Zhao L. P., Li S. S., Laws R. J., Skerrett S. J., Beutler B., Schroeder L., Nachman A., et al. J. Exp. Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen-Nissen E., Smith K. D., Strobe K. L., Barrett S. L., Cookson B. T., Logan S. M., Aderem A. Proc. Natl. Acad. Sci. USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gewirtz A. T., Yu Y., Krishna U. S., Israel D. A., Lyons S. L., Peek R. M., Jr. J. Infect. Dis. 2004;189:1914–1920. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- 19.Watson R. O., Galan J. E. Cell. Microbiol. 2005;7:655–665. doi: 10.1111/j.1462-5822.2004.00498.x. [DOI] [PubMed] [Google Scholar]
- 20.Sitaraman S. V., Klapproth J. M., Moore D. A., III, Landers C., Targan S., Williams I. R., Gewirtz A. T. Am. J. Physiol. 2005;288:G403–G406. doi: 10.1152/ajpgi.00357.2004. [DOI] [PubMed] [Google Scholar]
- 21.Lodes M. J., Cong Y., Elson C. O., Mohamath R., Landers C. J., Targan S. R., Fort M., Hershberg R. M. J. Clin. Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien A. D., Rosenstreich D. L., Scher I., Campbell G. H., MacDermott R. P., Formal S. B. J. Immunol. 1980;124:20–24. [PubMed] [Google Scholar]
- 23.Vazquez-Torres A., Vallance B. A., Bergman M. A., Finlay B. B., Cookson B. T., Jones-Carson J., Fang F. C. J. Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- 24.Honko A. N., Mizel S. B. Infect. Immun. 2004;72:6676–6679. doi: 10.1128/IAI.72.11.6676-6679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liaudet L., Szabo C., Evgenov O. V., Murthy K. G., Pacher P., Virag L., Mabley J. G., Marton A., Soriano F. G., Kirov M. Y., et al. Shock. 2003;19:131–137. doi: 10.1097/00024382-200302000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Ramphal R., Balloy V., Huerre M., Si-Tahar M., Chignard M. J. Immunol. 2005;175:3927–3934. doi: 10.4049/jimmunol.175.6.3927. [DOI] [PubMed] [Google Scholar]
- 27.Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., Aderem A. Nat. Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 28.Franchi L., Amer A., Body-Malapel M., Kanneganti T. D., Ozoren N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. Nat. Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y., Nagai S., Wu H., Neish A. S., Koyasu S., Gewirtz A. T. J. Immunol. 2006;176:6194–6201. doi: 10.4049/jimmunol.176.10.6194. [DOI] [PubMed] [Google Scholar]
- 30.Angers I., Sancho-Shimizu V., Descoteaux A., Gewirtz A. T., Malo D. Mamm. Genome. 2006;17:385–397. doi: 10.1007/s00335-005-0132-x. [DOI] [PubMed] [Google Scholar]
- 31.Nakasone C., Kawakami K., Hoshino T., Kawase Y., Yokota K., Yoshino K., Takeda K., Akira S., Saito A. Infect. Immun. 2004;72:6176–6180. doi: 10.1128/IAI.72.10.6176-6180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz M. J., Rijneveld A. W., Florquin S., Edwards C. K., Dinarello C. A., van der Poll T. Am. J. Physiol. 2002;282:L285–L290. doi: 10.1152/ajplung.00461.2000. [DOI] [PubMed] [Google Scholar]
- 33.Prince A. Am. J. Respir. Cell Mol. Biol. 2006;34:548–551. doi: 10.1165/rcmb.2006-0022SF. [DOI] [PubMed] [Google Scholar]
- 34.Noulin N., Quesniaux V. F., Schnyder-Candrian S., Schnyder B., Maillet I., Robert T., Vargaftig B. B., Ryffel B., Couillin I. J. Immunol. 2005;175:6861–6869. doi: 10.4049/jimmunol.175.10.6861. [DOI] [PubMed] [Google Scholar]
- 35.Hajjar A. M., Harowicz H., Liggitt H. D., Fink P. J., Wilson C. B., Skerrett S. J. Am. J. Respir. Cell Mol. Biol. 2005;33:470–475. doi: 10.1165/rcmb.2005-0199OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 37.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]