Abstract
Type III secretion systems are essential virulence determinants for many Gram-negative bacterial pathogens. The type III secretion system consists of cytoplasmic, transmembrane, and extracellular domains. The extracellular domain is a hollow needle protruding above the bacterial surface and is held within a basal body that traverses both bacterial membranes. Effector proteins are translocated, via this external needle, directly into host cells, where they subvert normal cell functions to aid infection. Physical contact with host cells initiates secretion and leads to formation of a pore, thought to be contiguous with the needle channel, in the host-cell membrane. Here, we report the crystal structure of the Shigella flexneri needle subunit MxiH and a complete model for the needle assembly built into our three-dimensional EM reconstruction. The model, combined with mutagenesis data, reveals that signaling of host-cell contact is relayed through the needle via intersubunit contacts and suggests a mode of binding for a tip complex.
Keywords: needle complex, protein secretion, Shigella
Bacillary dysentery is caused by Shigella species, which are responsible for over a million deaths worldwide annually (1). The genes required for invasion are clustered on a 31-kb fragment of a large virulence plasmid (2–4). Within this region, the mxi/spa operons encode a type III secretion system (T3SS) that is required to deliver virulence effectors into host cells. The secretion apparatus spans the two bacterial membranes and possesses an extracellular needle that, upon activation, inserts a pore into the host-cell membrane (Fig. 5, which is published as supporting information on the PNAS web site). Many of the 25 proteins from which the T3SS is constructed are similar in either sequence or function to proteins of the bacterial flagellum (5), and export of the axial flagellar components is highly analogous to export through a virulence T3SS (6). However, T3SSs (and not flagellar systems) undergo host-contact-mediated activation and translocation of effector proteins into host cells, apparently through direct contact of the external distal tip of the apparatus with the host cell (7–9). The role of the needle has been probed by using mutagenic studies of the needle-subunit proteins from Yersinia and Shigella (10, 11). In these systems, point mutations were identified that led to constitutive secretion of effectors and experimentally uninducible needles. These data suggest that the needle is important for transmission of the host-cell detection signal and activity of the T3SS of bacterial pathogens of animals. The T3SS needle is, therefore, a multifunctional structure that participates in the secretion and translocation of virulence proteins (12–15) and in the cell-contact regulation of type III secretion. However, the mechanism by which host-cell sensing and transmission of the activation signal to the secretion apparatus at the base of the T3SS occurs remains unknown.
We have previously demonstrated, by x-ray fiber diffraction and EM, that the Shigella flexneri needle shares an identical helical architecture (≈5.6 subunits per turn, 24-Å helical pitch) and inner channel diameter (≈2 nm) with the flagellar rod, hook, and filament (16). This architectural similarity is maintained despite the subunit of the T3SS needle being four to five times smaller and displaying no primary sequence homology with flagellar axial proteins. This finding suggests that the size of the subunit protein determines the outer diameter of the filament, whereas the inner diameter remains constant. The needle may, thus, represent the minimum core required to build a filament of this size with this architecture.
Atomic-level structural information for components of the secretory apparatus of the T3SS remain scarce despite there being a significant number of structures for effectors and their cytoplasmic chaperones (17). To increase our understanding of the T3SS, we have determined the structure of the S. flexneri T3SS needle subunit MxiH and docked this model into our three-dimensional EM reconstruction of the intact needle.
Results and Discussion
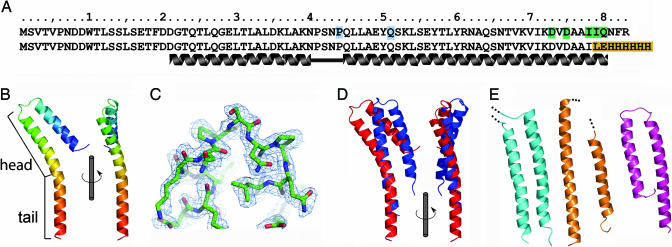
The construct of the S. flexneri T3SS needle subunit MxiH used in this study has been described (11, 18), and the sequence is shown in Fig. 1A (row 2) compared with the wild-type sequence (row 1). The crystal structure of MxiH has been determined to 1.9-Å resolution (Fig. 1, Table 1, and Tables 2 and 3, which are published as supporting information on the PNAS web site). The structure of MxiH (Fig. 1B) consists of two extended helices connected by a short Pro-Ser-Asn-Pro (PSNP) turn (Fig. 1C). Two different conformations of MxiH (hereafter called molecules A and B) were seen as two crystallographically distinct copies in our crystal structure. In both molecules A (residues 20–81) and B (residues 15–75) the N terminus is disordered despite being strongly predicted to form a helix at the primary sequence level (11). The “head” (Fig. 1B, residues 26–57) superposes well between the two crystallographically distinct copies of MxiH (Fig. 6, which is published as supporting information on the PNAS web site) (rms difference of 0.2 Å over Cα atoms), and the orientation of the head with respect to the “tail” is determined by differences in the degree of bending of the long C-terminal helix (Fig. 1D). This finding implies the presence of a molecular hinge between the head and tail of MxiH that may be important for signaling within the intact needle, as discussed below.
Fig. 1.
Crystal structure of the MxiH monomer and comparison with related proteins. (A) Sequence of full-length MxiH and the truncated, His-tagged construct used in this study (different residues are highlighted in orange). Residues for which mutagenesis data are discussed are colored according to the following proposed role: signaling (green) and tip interaction (blue). (B) Ribbon diagram of MxiH molecule A colored N (blue) to C (red) termini. Views rotated by 90° about the long axis of the molecule are shown. The head (residues 26–57) and tail are labeled. (C) SHARP/SOLOMON solvent-flattened electron density for the PSNP loop is shown at 1.0 σ. The final model for this region of molecule A is shown as a stick representation colored by atom type (C, green; O, red; N, blue). (D) Change in the relative domain arrangement of MxiH molecules A (red) and B (blue). Molecules are aligned over the tail region. (E) Ribbon diagrams of the D0 domain of flagellin (19) (cyan), the ordered (chaperone-bound) region of EspA (20) (orange), and YscE (21) (magenta). Figures were prepared by using PyMOL (31).
Table 1.
MxiH refinement statistics
| Space group (Z) | C2 (8) |
|---|---|
| Cell dimensions | a = 183.2 Å, b = 28.1 Å, c = 27.7 Å, α = γ = 90.0° β = 96.5° |
| R, 28–1.9 Å (2.1–1.9 Å) | 0.19 (0.21) |
| Rwork,28–1.9 Å (2.1–1.9 Å) | 0.19 (0.21) |
| Rfree, 28–1.9 Å (2.1–1.9 Å) | 0.23 (0.23) |
| rmsd bonds, Å (n) | 0.008 (988) |
| rmsd angles, ° (n) | 1.00 (1,332) |
| No. of atoms in model | 1,042 |
| No. of atoms in protein | 977 |
| No. of waters | 49 |
| Ligands | 1 glycerol, 1 isopropanol, 2 Na+ ions |
| 〈B〉 protein (main; side chains) | |
| Molecule A | 27 (37; 32) |
| Molecule B | 42 (50; 46) |
| 〈B〉 waters | 68 |
| Ramachandran plot | |
| Residues in most favored regions, % | 93.7 |
| Residues in additionally allowed regions, % | 6.3 |
| Residues in generously allowed regions, % | 0 |
| Residues in disallowed regions, % | 0 |
The crystal structure of MxiH (which is at least 24% identical in primary sequence to most animal-pathogen T3SS needle subunits) provides the first detailed picture of a T3SS needle subunit and reveals similarities to two other proteins that form polymeric assemblies with identical helical architecture to the T3SS needle. The D0 portion of flagellin (19) and the enteropathogenic Escherichia coli T3SS needle-extension component EspA (20) (Fig. 1E) both possess two long antiparallel helices as seen for MxiH, suggesting that this structural feature is required to build such a helical assembly. MxiH also has structural similarity to the Yersinia T3SS protein YscE (21) (Fig. 1E), which has recently been proposed to be the chaperone for YscF, the Yersinia needle subunit (22, 23). The strong structural similarity between a needle subunit, MxiH, and a needle-subunit chaperone, YscE, suggests that the subunit–chaperone interactions might mimic the subunit–subunit interactions in the assembled needle to block further polymerization. Details of these similarities are discussed further in Supporting Methods, which is published as supporting information on the PNAS web site.
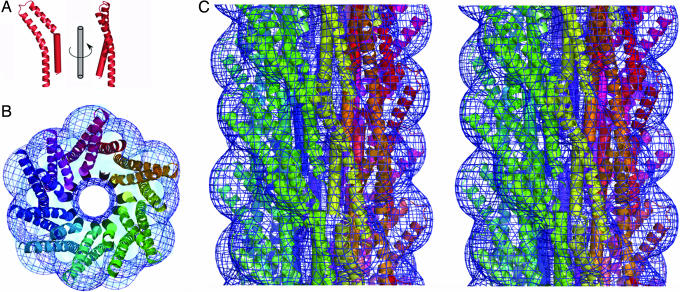
A model of the Shigella T3SS needle was generated by docking the crystal structure for MxiH into the 16-Å density map from our earlier three-dimensional EM reconstruction of the needle (16) (see also Methods and Supporting Methods). Initial docking was performed manually and optimized by using rigid-body refinement with strict helical constraints to maintain the overall symmetry of the needle. The optimal orientation of the crystal structure in the reconstruction was with the major axis of the monomer in line with the major axis of the needle (as oriented in Fig. 2A) and the tail buried in the core of the assembly with the head group decorating the surface [the correlation coefficient (CC) between this model and the experimental EM density is 71%, with molecule A]. A possible alternative orientation, with the head group facing the channel, is not compatible with the experimental density (CC = 35%). This optimal orientation agrees with that described for both the hook and flagellin subunits in the bacterial flagellum (19, 24). The bend of the A molecule of MxiH fits the shape of the EM density well (as demonstrated by the CC of 71% for the optimal position), as the orientation of the head matches the surface features of the needle (Fig. 2 B and C), whereas the B molecule is not consistent with the observed needle density (CC = 47% for optimized positioning of molecule B). Although the conformation of the B molecule may be induced by crystal packing, it is also possible that it reflects the structure of the needle subunit when the needle is in a different activation state from that for which we have an EM reconstruction (16, 25). The C-terminal helix forms the outer shell of the needle core, whereas the PSNP loop directs the N-terminal helix to line the inner wall of the needle channel. This arrangement is unlike the flagellum, where the C-terminal flagellin helix lines the central channel (19). The model for the needle contained a gap of the appropriate size and position to accommodate the N terminus of MxiH, which was modeled as a helical extension to our experimental structure (Fig. 2A). This extended model for full-length MxiH is improved in its fit to the observed EM density (CC = 84%; Fig. 2 B and C and Movie 1, which is published as supporting information on the PNAS web site) and constitutes the first atomic model for an intact T3SS needle.
Fig. 2.
Docking of the atomic model of MxiH into the EM density of the Shigella T3SS needle. (A) Molecule A of MxiH (ribbon) with the modeled N-terminal helix (cylinder) is shown as two views rotated by 90° about the long axis of the molecule. (B) End-on view of a 40-Å-thick section of the assembled needle. Each MxiH monomer is shown as in A and colored differently, starting from red and circling the needle to purple. EM density is shown as a blue mesh. (C) Stereo diagram of the side view of the assembled needle, colored as for B. Note that B and C are not shown at the same scale, and the needle assembly has an exterior diameter of ≈70 Å (16).
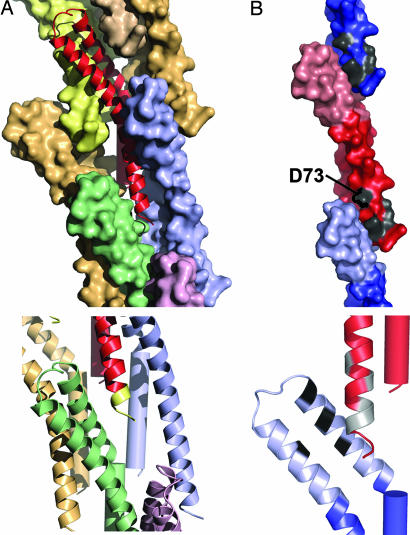
The model reveals that the C terminus of MxiH is involved in extensive intersubunit contacts (Fig. 3A). This packing arrangement, in combination with the nonpolymerizing phenotype of C-terminal truncations of MxiH (11), suggests that this interaction plays a key role in stabilizing the T3SS needle. The observation that the N terminus remains disordered within the context of an almost full-length MxiH monomer suggests that either the five C-terminal residues are essential for N-terminal helix formation or that this region becomes folded only within the intact needle (as it is thought to occur for flagellin) (26–28). In the latter case, the unfolded N terminus might facilitate ease of movement through the central channel, and folding of the N-terminal helix may be important for correct packing of monomers in the assembled needle. It is worth noting that the size of the channel in the center of the assembly is likely to allow secretion of only single α-helices and random coil, rather than fully folded structures (Fig. 7, which is published as supporting information on the PNAS web site). However, because the N-terminal helix was not visible in the crystal structure, we cannot speculate as to the precise chemical nature of the channel lining. A large proportion (57% covering 2,654 Å2) (see Supporting Methods) of the subunit surface area is buried in the context of the needle assembly, consistent with the observation that many single mutations (W10A, Y57A, K69A, K72A, and R83A) are capable of severely altering needle polymerization (11).
Fig. 3.
Interactions between subunits in the assembled needle. (A) A monomer of MxiH (ribbon diagram, red) is surrounded by seven identical subunits (shown as colored surface representations) within the needle assembly (Upper). The C terminus of MxiH, magnified (Lower), with the five C-terminal residues colored yellow, makes direct contact with three surrounding monomers (shown as ribbon diagrams in blue, green, and purple). (B) The tail region of MxiH (blue for the monomer above and red for the central monomer) contacts the head region of the monomer below (light red for the central monomer and light blue for the monomer below). Mutations that cause severe defects in hemolysis/invasion despite normal needle assembly (11) are shown in gray (D73A, D75A, I78A, I79A, and Q80A) (Upper). The magnification of the interface (as a ribbon diagram) (Lower) is rotated to show the patch of residues (black, L30, L34, A38, and Y50) on the head (light blue) that contact the residues listed above (colored gray on the red ribbon diagram).
The mechanism by which host-cell sensing, occurring at the tip of the needle, is transmitted to the secretion apparatus at the base is unknown (7–9). We have previously proposed that signaling occurred via changes in the helical architecture of the needle in ways analogous to those known to control flagellar filament switching. EM studies failed to detect alterations in the helical architecture of functionally altered mutant needles (25). However, mapping onto our needle model of a mutant (D73A) that leads to uncontrolled secretion and also renders the needle insensitive to all further forms of activation (11) provides some insight. The location of D73 within the needle assembly strongly suggests that this mutation acts by modulation of the contacts between the tail of one monomer and the head of the subunit packing below (Fig. 3B). This mutation and also four others that lead to severe defects in hemolysis and invasion despite normal needle assembly (11) (D75A, I78A, I79A, and Q80A) cluster within the same region of the needle (Fig. 3B), implying that this region is key to signaling the activation state of the assembly. These mutational data, combined with analysis of the dynamics of the monomer (showing that the major motion is a movement of the head with respect to the tail) (Movie 2, which is published as supporting information on the PNAS web site, and Supporting Methods) and the dual conformations of the head of MxiH seen in the crystal structure now lead us to propose that transmission of the signal from the tip to the base will be defined by reorientation of the subunit head relative to its tail, inducing subtle changes in the packing of the needle without altering the helical parameters defining the structure.
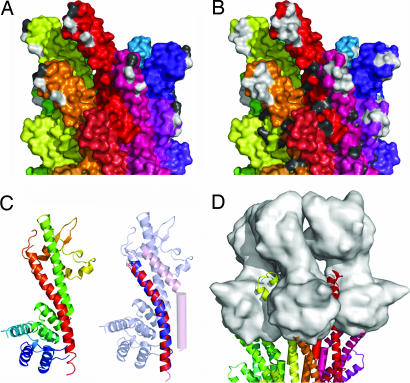
Mutations to alanine at P44 or Q51 lead to secretion of effectors before activation by the artificial inducer Congo red, although this stimulus is able to further increase secretion levels (11). As residues P44 and Q51 are exposed on the outer surface at the top of the needle (Fig. 4A), it is possible that they act to produce the functional phenotype by altering the interactions with protein(s) postulated to lie at the tip of the needle. A similar mechanism was proposed to explain the constitutive secretion phenotype of the mutation D46A in the same region of the homologous Yersinia needle subunit YscF (10) (an ≈9-kD protein with 28% sequence identity to MxiH; Fig. 4 A and B and Fig. 8, which is published as supporting information on the PNAS web site). To date, tip–protein complexes have been visualized for a limited subset of highly related T3SSs, typified by the Yersinia system, where the protein LcrV is critical to assembly of the tip complex (29). Despite lacking any sequence homology, the crystal structure of LcrV (30) possesses a long, bent α-helix at the C terminus that is structurally homologous (rms difference of 0.76 Å over 31 Cα atoms) (Supporting Methods) to the C-terminal helix of MxiH that is critical for stabilizing MxiH subunits in the Shigella needle (Fig. 4C). By using MxiH as a model for the Yersinia needle subunit, YscF, a Yersinia tip complex was built by superimposing the LcrV C-terminal helix onto the structurally equivalent helix of the needle subunits at the tip of the needle. The needle monomers used for placing LcrV monomers were then removed, producing a model with no significant atomic clashes between tip-complex subunits. Addition of more than five LcrV monomers is not possible because of the size of the N- and C-terminal domains of the already placed LcrV monomers. Although the biological data to date do not allow us to know how many copies of LcrV are present at the tip of the Yersinia needle, it is of note that this simple tip-complex model is visually reminiscent of the EM images of the Yersinia tip complex (29) (Fig. 4D). The model packs both the putatively tip-interacting residues (P44 and Q51 of Shigella and D46 of Yersinia) and a patch of residues strongly conserved among all T3SS needle components (Fig. 9, which is published as supporting information on the PNAS web site) between two subunits of the tip complex. Mutations at the surface of the head of the needle subunit are likely to lead to altered secretion by directly altering binding, or conformation, of the tip complex. This arrangement provides a mechanism for initiation of a signal, via the tip complex, directly to the head of the needle subunits at the tip of the needle that can then be propagated down the needle.
Fig. 4.
Characterization of the putative tip-interaction interface of a T3SS needle. (A) Surface representation of the side view of the T3SS needle with each monomer colored differently, starting from red and circling the needle to purple. Residues likely to effect interactions with the tip complex are highlighted as follows: P44 and Q51 in Shigella (white) and the equivalent of D46 in Yersinia (gray). (B) View and coloring as for A, with residues conserved between Shigella MxiH and Yersinia YscF highlighted. Residues conserved in the head domain: L37, P41, N43, P44, L46, L47, A48, and Q51 (white); in the tail domain, N62, S65, V68, K72, D73, I78, Q80, and F82 (gray) are shown for the top circle of the needle. (C Left) Ribbon diagram of LcrV (30) colored N (blue) to C (red) termini. (C Right) Overlay of the C-terminal helices of MxiH (red, residues 45–75) and LcrV (blue, residues 287–317), with all but the overlaid region made transparent to aid visualization. (D) Model of an LcrV tip complex (surface representation, gray) onto the tip of a T3SS needle.
Materials and Methods
Crystal Structure Determination.
Protein preparation and crystallization are as described in ref. 18 and in Supporting Methods. Initial phases were computed with the program SHARP with the multiple isomorphous replacement (MIRAS) method, by using data from a native crystal, two selenomethionine (SeMet)-labeled crystals (three wavelengths, one Se site), and a uranyl acetate derivative (one U site). For details of data collection statistics and heavy-atom finding, see ref. 18 and Supporting Methods. Solvent-flattening using programs CCP4-DM and SOLOMON gave a 3.2-Å map that was used for manual building, in Xfit, of an initial model, comprising residues 31–53 of molecule A and residues 21–38 and 41–69 of molecule B. After refinement in Buster-TNT, this initial model was used to aid mask-determination in a second round of solvent-flattening of the experimental phases which produced a 2.1-Å map that enabled automated model-building in ARP/wARP (101 residues were built, 61 of which were docked into sequence). Alternate cycles of refinement and manual building led to the final model for residues A20–81 and B15–75. Literature citations for crystallographic computing programs are provided in Supporting Methods.
Fitting of the Model in the EM Needle Reconstruction.
The EM needle density (in both hands) was put in a P1 cell of dimensions: a = b = 159.0 Å, c = 120.7 Å, α = β = γ = 90.0°, with the needle axis directed along c. The model to be fit was manually placed in the needle density, to give an initial model for the needle constructed by generating 27 more copies obeying the helical symmetry, the nth copy using the helical operator: Euler angles: α = 0.0°, β = 0.0°, γ = n·64.3°; translation vector tx = 0.0, ty = 0.0, tz = n·4.31 Å. A set of 1,563 structure-factor intensities was computed from the P1 needle density in the resolution range 158–16 Å by using the program CCP4-FFT; the associated statistical uncertainty (su) was computed as σ(Ihkl) = in CCP4-SFTOOLS; structure-factor amplitudes (and associated su) were computed from these intensities by using the program CCP4-TRUNCATE. Models were rigid-body refined in Buster-TNT by using hard NCS constraints; the calculated su were used in computing the likelihood in a standard fashion. Refinement was driven by maximum likelihood with nonbonding restraints to minimize clashes. The refined models were then scored by using CCs between calculated and observed maps, computed with the program MAPMAN. A variety of starting positions, including head group on the surface and in the core (each in both vertical orientations), were tested. The hand of the helical structure was determined empirically by attempting fits of the two crystallographic monomers to the density in both hands. For both the A and B monomers, the best fit was to the right (R)-handed (1-start) reconstruction [A: CC = 71% (R) vs. 61% left (L), B: CC = 47% (R) vs. 23% (L)].
Supplementary Material
Acknowledgments
We thank Stephen Graham for assistance with figure preparation. This work was supported by grants from the Wellcome Trust (to P.R. and S.M.L), the Medical Research Council of the United Kingdom (to S.J. and S.M.L.), the U.S. Public Health Service (to W.D.P. and W.L.P.), the University of Kansas Research Development Fund, and the Guy G. F. Newton Research Trust (to A.B.). J.E.D. is funded by an Australian National Health and Medical Research Council Postdoctoral Fellowship, F.S.C. by a William R. Miller Junior Research Fellowship, and R.K. by a Barbara Johnson Bishop Scholarship.
Glossary
Abbreviations
- CC
correlation coefficient
- T3SS
type III secretion system.
Note Added in Proof.
Since submission of the manuscript, a partial NMR structure for the Burkholderia pseudomallei needle subunit has been published (32). The ordered portion adopts the same structure as the head of the MxiH subunit reported herein. Recent data have also formally demonstrated that IpaD sits at the tip of the Shigella needle in an analogous fashion to LcrV at the tip of the Yersinia needle (ref. 33 and A. K. J. Veenendaal, J. L. Hodgkinson, L. Schwarzer, D. Stabat, S. F. Zenk, and A.J.B., unpublished data).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2CA5).
References
- 1.Kotloff K. L., Winickoff J. P., Ivanoff B., Clemens J. D., Swerdlow D. L., Sansonetti P. J., Adak G. K., Levine M. M. Bull. W. H. O. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 2.Sansonetti P. J., Kopecko D. J., Formal S. B. Infect. Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasakawa C., Kamata K., Sakai T., Makino S., Yamada M., Okada N., Yoshikawa M. J. Bacteriol. 1988;170:2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonetti P. J. FEMS Microbiol. Rev. 2001;25:3–14. doi: 10.1111/j.1574-6976.2001.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 5.Blocker A., Komoriya K., Aizawa S. Proc. Natl. Acad. Sci. USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minamino T., Namba K. J. Mol. Microbiol. Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- 7.Blocker A., Gounon P., Larquet E., Niebuhr K., Cabiaux V., Parsot C., Sansonetti P. J. Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mota L. J., Journet L., Sorg I., Agrain C., Cornelis G. R. Science. 2005;307:1278. doi: 10.1126/science.1107679. [DOI] [PubMed] [Google Scholar]
- 9.West N. P., Sansonetti P., Mounier J., Exley R. M., Parsot C., Guadagnini S., Prevost M. C., Prochnicka-Chalufour A., Delepierre M., Tanguy M., Tang C. M. Science. 2005;307:1313–1317. doi: 10.1126/science.1108472. [DOI] [PubMed] [Google Scholar]
- 10.Torruellas J., Jackson M. W., Pennock J. W., Plano G. V. Mol. Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 11.Kenjale R., Wilson J., Zenk S. F., Saurya S., Picking W. L., Picking W. D., Blocker A. J. Biol. Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 12.Tamano K., Aizawa S., Katayama E., Nonaka T., Imajoh-Ohmi S., Kuwae A., Nagai S., Sasakawa C. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blocker A., Jouihri N., Larquet E., Gounon P., Ebel F., Parsot C., Sansonetti P., Allaoui A. Mol. Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 14.Kimbrough T. G., Miller S. I. Proc. Natl. Acad. Sci. USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubori T., Sukhan A., Aizawa S., Galan J. E. Proc. Natl. Acad. Sci. USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordes F. S., Komoriya K., Larquet E., Yang S., Egelman E. H., Blocker A., Lea S. M. J. Biol. Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 17.Johnson S., Deane J. E., Lea S. M. Curr. Opin. Struct. Biol. 2005;15:700–707. doi: 10.1016/j.sbi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Deane J. E., Cordes F. S., Roversi P., Johnson S., Kenjale R., Picking W. D., Picking W. L., Lea S. M., Blocker A. Acta Crystallogr. F. 2006;62:302–305. doi: 10.1107/S1744309106006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yonekura K., Maki-Yonekura S., Namba K. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 20.Yip C. K., Finlay B. B., Strynadka N. C. Nat. Struct. Mol. Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 21.Phan J., Austin B. P., Waugh D. S. Protein Sci. 2005;14:2759–2763. doi: 10.1110/ps.051706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day J. B., Guller I., Plano G. V. Infect. Immun. 2000;68:6466–6471. doi: 10.1128/iai.68.11.6466-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinaud M., Chabert J., Faudry E., Neumann E., Lemaire D., Pastor A., Elsen S., Dessen A., Attree I. J. Biol. Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 24.Samatey F. A., Matsunami H., Imada K., Nagashima S., Shaikh T. R., Thomas D. R., Chen J. Z., Derosier D. J., Kitao A., Namba K. Nature. 2004;431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- 25.Cordes F. S., Daniell S., Kenjale R., Saurya S., Picking W. L., Picking W. D., Booy F., Lea S. M., Blocker A. J. Mol. Biol. 2005;354:206–211. doi: 10.1016/j.jmb.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 26.Vonderviszt F., Kanto S., Aizawa S., Namba K. J. Mol. Biol. 1989;209:127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- 27.Aizawa S. I., Vonderviszt F., Ishima R., Akasaka K. J. Mol. Biol. 1990;211:673–677. doi: 10.1016/0022-2836(90)90064-S. [DOI] [PubMed] [Google Scholar]
- 28.Namba K. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 29.Mueller C. A., Broz P., Muller S. A., Ringler P., Erne-Brand F., Sorg I., Kuhn M., Engel A., Cornelis G. R. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 30.Derewenda U., Mateja A., Devedjiev Y., Routzahn K. M., Evdokimov A. G., Derewenda Z. S., Waugh D. S. Structure (London) 2004;12:301–306. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 31.DeLano W. L. The PyMOL Molecular Graphic System. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- 32.Zhang L., Wang Y., Picking W. L., Picking W. D., De Guzman R. N. J. Mol. Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 33.Espina M., Olive A. J., Kenjale R., Moore D. S., Ausar S. F., Kaminski R. W., Oaks E. V., Middaugh C. R., Picking W. D., Picking W. L. Infect. Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.