Abstract
The mechanisms by which the activation of Smoothened (Smo), a protein essential to the actions of the Hedgehog family of secreted proteins, is translated into signals that converge on the Gli transcription factors are not fully understood. The seven-transmembrane structure of Smo has long implied the utilization of heterotrimeric GTP-binding regulatory proteins (G proteins); however, evidence in this regard has been indirect and contradictory. In the current study we evaluated the capacity of mammalian Smo to couple to G proteins directly. We found that Smo, by virtue of what appears to be constitutive activity, activates all members of the Gi family but does not activate members of the Gs, Gq, and G12 families. The activation is suppressed by cyclopamine and other inhibitors of Hedgehog signaling and is enhanced by the Smo agonist purmorphamine. Activation of Gi by Smo is essential in the activation of Gli in fibroblasts, because disruption of coupling to Gi with pertussis toxin inhibits the activation of Gli by Sonic hedgehog and a constitutively active form of Smo (SmoM2). However, Gi does not provide a sufficient signal because a truncated form of Smo, although capable of activating Gi, does not effect activation of Gli. Rescue of pertussis toxin-inhibited activation of Gli by Sonic hedgehog can be achieved with a constitutively active Gαi-subunit. The data suggest that Smo is in fact the source of two signals relevant to the activation of Gli: one involving Gi and the other involving events at Smo’s C-tail independent of Gi.
Keywords: GTP binding, Hedgehog, cyclopamine
Smoothened (Smo) is a seven-transmembrane (7-TM) protein that mediates all the known biological activities of the Hedgehog (Hh) family of secreted proteins, which are relevant to pattern formation in the embryo and tissue homeostasis in the adult (1). Deficiency in activation of the pathway leads to phenotypes ranging from severe malformations of the head and limbs to embryonic lethality (2), whereas unabated signaling contributes to several cancers (3). Substantial interest exists, therefore, in understanding and manipulating pathways relevant to Smo activity.
Genetic and biochemical studies imply that Smo can adopt an active conformation but that it is normally repressed by Patched (Ptch), a 12-transmembrane protein considered the receptor for Hh (4). Binding of Hh to Ptch relieves the repression of Smo, allowing Smo to signal. The activity of Smo is, therefore, evident in the presence of Hh or the absence of Ptch and by mutations in Smo that make it insensitive to repression by Ptch. The derepression of Smo results in the activation of transcription factors belonging to the Cubitus interruptus/Gli family and consequent induction of respective target genes. In the absence of Hh ligands, Cubitus interruptus (in Drosophila) and Gli2 and Gli3 (in vertebrates) are phosphorylated by protein kinase A and glycogen synthase kinase-3β and are proteolytically processed (5–8). Although several proteins have been implicated in Smo signaling (e.g., Fused kinase, Suppressor of Fused and, in Drosophila, Costal-2), little is known about the mechanism by which Smo increases Gli activity.
The 7-TM structure of Smo would suggest an ability to couple to one or more heterotrimeric GTP-binding regulatory proteins (G proteins). Efforts to uncover a G protein-based form of transduction, however, have been disappointing. A pertussis toxin (PTX) that ADP-ribosylates Gi inhibits Smo-elicited signaling in some, but reportedly few, instances (9, 10). Of course, effects of PTX do not preclude indirect activation of Gi through autocrine signaling, nor do they demonstrate G protein activation directly. Genetic studies of Drosophila, which provide a widely used platform for understanding Hh signaling, have not revealed a role for G proteins at all (1). Signals originating from the C-tail of Smo, for both Drosophila and vertebrate proteins, have instead captured the preponderance of attention (11–16).
We therefore sought to evaluate the coupling of Smo to G proteins using a direct assay of G protein activation documented previously to be sensitive to receptor constitutive activity, such as that anticipated for Smo, and to suppression of that activity by inverse agonists (17). Using this assay we determined that mouse Smo couples to all members of the Gi family but does not couple to those of other G protein families. Cyclopamine and other inhibitors of Hh signaling were found to inhibit Smo coupling to Gi in a manner consistent with inverse agonism. We further demonstrated that Gi is required for activation of Gli, as is the C-tail of Smo. Our findings argue that Smo is the source of two signals relevant to Hh signaling, one operating through Gi and the other originating with the C-tail, both of which are required for activation of Gli.
Results
Smo Activates Members of the Gi Family of G Proteins.
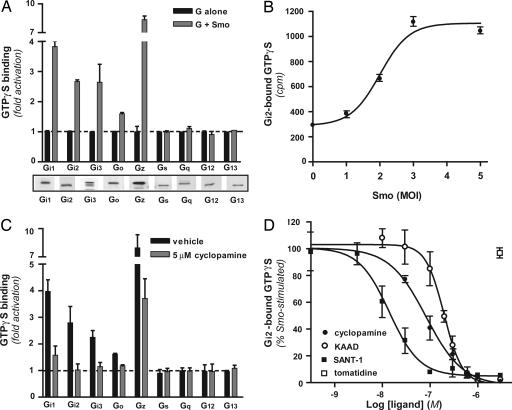
Our first objective was to evaluate the capacity of Smo to couple to any one or more heterotrimeric G proteins. We introduced selected Gα-subunits and Gβ1γ2, with or without Smo, into Sf9 cells by means of recombinant baculoviruses. Sf9 cells provide a background devoid of most receptors and G proteins and have been used extensively in studies of coupling (18). Coupling was evaluated in isolated membranes by means of guanine nucleotide exchange (19, 20) wherein the activation of a G protein would be evident as a Smo-promoted binding of [35S]guanosine 5′-(3-O-thio)triphosphate (GTPγS) to the Gα-subunit. We relied here on the probability that Smo would possess significant “constitutive” activity (21). We found that, depending on the G protein, expression of Smo promoted a 1.5- to 8-fold activation of various members of the Gi family, i.e., Gi1, Gi2, Gi3, Go, and Gz (Fig. 1A). Smo was inactive toward members of the other three families, represented in our assay by Gs, Gq, G12, and G13. The inactivity of Smo toward the latter G proteins is not attributable to some defect in the reconstitution, because all of the G proteins can be activated through one 7-TM receptor or another in the same assay (19, 22). Decreasing or increasing the multiplicity of infection for the recombinant Smo virus [a multiplicity of infection (MOI) of 2 was used in A and most other experiments] resulted in a corresponding decrease or increase in [35S]GTPγS binding (Fig. 1B).
Fig. 1.
Smo is coupled to G proteins of the Gi family. (A) Sf9 cells were infected with a mixture of baculoviruses encoding Gα-subunits (corresponding to the indicated G protein), Gβ1, and Gγ2, each at an MOI of 1, with (gray bars) or without (black bars) a baculovirus encoding Smo (MOI = 2). At 48 h, membranes were isolated, and [35S]GTPγS binding was evaluated for the Gα-subunits as described in Materials and Methods. [35S]GTPγS binding is expressed as fold stimulation of binding to each G protein alone. For reference, the maximal average cpm for each G protein were as follows: ≈500 (Gi1), ≈700 (Gi2), ≈50 (Gi3), ≈2,000 (Go), ≈1,000 (Gz), ≈10,000 (Gq), ≈250 (G12), ≈2,500 (G13), and ≈1,000 (Gs). Shown beneath the graph is a series of Western blots for Gα-subunits in the Sf9 membranes (10 μg of membrane protein per lane) using individual Gα-specific antisera. (B) Sf9 cells were infected with baculoviruses encoding Gαi2, Gβ1, and Gγ2, each at an MOI of 1, and Smo at MOIs ranging from 0 to 5. Membranes were isolated, and [35S]GTPγS binding was evaluated; binding is expressed as cpm. (C) Sf9 cells were infected with baculoviruses encoding Smo (MOI = 2), Gα, Gβ1, and Gγ2. Membranes were incubated for 10 min with vehicle (black bars) or 5 μM cyclopamine (gray bars) before the addition of [35S]GTPγS. [35S]GTPγS binding of 1-fold corresponds to each G protein alone. (D) Dose-dependent inhibition of Smo-promoted [35S]GTPγS binding to Gi2 was evaluated for cyclopamine (filled circles), KAAD-cyclopamine (open circles), SANT-1 (filled squares), and tomatidine (open square). Binding in the absence of any compound was set at 100%, and Smo-independent binding was set at 0%. Each value in the four graphs represents the average of three independent experiments performed in triplicate ± 1 SE.
Cyclopamine Suppresses Activation of Gi by Smo.
Cyclopamine is a plant alkaloid that inhibits mammalian Hh signaling consequent to binding to Smo (23). We found that cyclopamine completely inhibited Smo-stimulated [35S]GTPγS binding to all members of the Gi family, with the exception of Gz, which was only partly inhibited (Fig. 1C). Cyclopamine had no actions on the G proteins apart from Smo, or on coupling of the 5-hydroxytryptamine1A receptor to Gi2 (Fig. 5, which is published as supporting information on the PNAS web site), confirming that the original increase in binding in response to coexpression of Smo represented a Smo activity. In concentration–response experiments (Fig. 1D), the EC50 for cyclopamine was found to be ≈80 nM, in the range noted for inhibition of Sonic hedgehog (Shh)-effected Gli-dependent transcription (20–300 nM) (23, 24). KAAD-cyclopamine, reportedly more potent than cyclopamine in Gli-dependent transcriptional assays (23), was slightly less potent for inhibition of Smo–Gi2 coupling (EC50 ≈ 200 nM). SANT-1, an inhibitor of Shh signaling that is structurally dissimilar to the other inhibitors, inhibited coupling completely with the highest potency (EC50 ≈ 15 nM). Tomatidine, an alkaloid inactive with respect to Hh signaling, did not affect coupling at any tested concentration (up to 10 μM).
Coupling of Smo to Gi in Mammalian Cells.
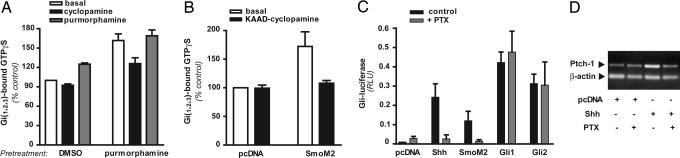
We next extended our findings to mammalian cells. Because the presence of Ptch represses Smo activity in its normal environment, we first determined whether an “agonist” for Smo, purmorphamine (25), was able to stimulate coupling of Smo to Gi. In the Sf9 system purmorphamine elicited a dose-responsive increase in Smo-dependent [35S]GTPγS binding to Gi2, with an EC50 = 1.4 ± 0.4 μM (Fig. 6, which is published as supporting information on the PNAS web site). In membranes from mouse embryonic fibroblasts, addition of 10 μM purmorphamine increased by ≈20% the [35S]GTPγS binding to Gi (i.e., Gi1, Gi2, and/or Gi3) (Fig. 2A). This value, which was significant, would reflect the contribution of Smo among many other 7-TM receptors that have some degree of constitutive activity toward Gi. The constitutive [35S]GTPγS binding to Gi was increased by 60% in membranes from cells pretreated with purmorphamine. This increase was likely attributable to stabilization of Smo at the plasma membrane in the intact cell (26). Purmorphamine had no further effect in the assay itself, but cyclopamine depressed the activity by 25%. Moreover, overexpression of the oncogenic mutant SmoM2, which is insensitive to repression by Ptch and also couples to Gi2 in Sf9 membranes (Fig. 7, which is published as supporting information on the PNAS web site), produced a 70% increase in total coupling to Gi compared with the empty vector in HEK 293 cells (Fig. 2B), an increase that was also inhibited by KAAD-cyclopamine.
Fig. 2.
Smo both activates and utilizes Gi in mammalian cells. (A) Mouse embryonic fibroblasts were treated for 45 min with vehicle (DMSO) or 10 μM purmorphamine in serum-free medium. Activation of Gi was determined in subsequently prepared membranes as described in Fig. 1 by using vehicle (white bars), 5 μM cyclopamine (black bars), or 10 μM purmorphamine (gray bars). (B) HEK 293 cells were transfected with empty pcDNA vector or SmoM2. After 48 h, membranes were prepared and used to determine the activation of Gi in the absence (white bars) or presence (black bars) of KAAD-cyclopamine. (C) NIH 3T3 cells were transiently transfected with expression vectors encoding Shh, oncogenic Smo (SmoM2), Gli1, Gli2, or empty pcDNA vector, together with reporters encoding Gli-luciferase and a control Renilla luciferase. After the cells reached confluency, the medium was replaced with 0.5% FCS (black bars) or 0.5% FCS plus 100 ng/ml PTX (gray bars). Luciferase activities were determined 24 h later. Results are expressed as the ratio of firefly/Renilla luciferase activities and represent an average of three independent experiments performed in triplicate ± 1 SE. (D) NIH 3T3 cells were transfected with an expression vector encoding Shh or with empty pcDNA vector. Cells treated subsequently with or without PTX were lysed after 24 h to extract total RNA. Semiquantitative RT-PCR was performed for Ptch-1 and β-actin. The result is representative of three independent experiments.
A PTX Blocks Signaling Through Smo to Gli.
Having shown that Smo is active toward the Gi family of G proteins, we next sought to determine whether activation of Gi is required for Gli-mediated transcriptional responses. We transfected NIH 3T3 cells with Shh or SmoM2, together with a Gli-luciferase reporter. Shh activates Gli through Ptch and Smo endogenous to these cells. The Gli reporter responded, as expected, to both Shh and SmoM2 (Fig. 2C). However, treatment with PTX (100 ng/ml) 24 h before assay of the reporter blocked this response. PTX had no effect on receptor-independent activation of the reporter achieved, as a control, by overexpression of Gli1 or Gli2. Ptch-1 is an endogenous target for Gli. Ptch-1 up-regulation by Shh, evaluated by RT-PCR, was also blocked by PTX (Fig. 2D).
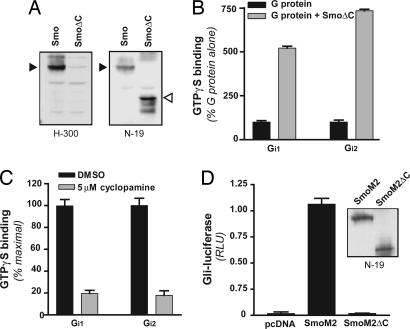
The C-Tail of Smo Is Required for Signaling to Gli but Not Activation of Gi.
The C-tail of Smo is known to be required in Drosophila for activation of Cubitus interruptus (11), and recent data imply that the C-tail of mammalian Smo may be required as well for activation of Gli (16). We therefore performed experiments to determine whether the C-tail was in any way relevant to the activation of Gi. A truncated version of Smo (residues 1–566), having only several residues distal to the seventh transmembrane domain, was expressed in Sf9 cells (Fig. 3A). In [35S]GTPγS-binding assays, SmoΔC displayed an activity toward members of the Gi family similar to that displayed by full-length Smo (Fig. 3B). Cyclopamine completely inhibited this activity (Fig. 3C). The activation of Gi by Smo, therefore, does not require the C-tail. We then generated a truncated form of SmoM2 (SmoM2ΔC) to evaluate the potential for Gli activation in NIH 3T3 cells; the use of SmoM2/SmoM2ΔC permitted circumvention of Ptch and Smo endogenous to the NIH 3T3 cells. SmoM2ΔC, in contrast to SmoM2, was completely inactive with regard to stimulation of Gli-dependent transcriptional events (Fig. 3D). Therefore, it would seem that, although Gi is required for activation of Gli, so also is the C-tail of Smo independent of Gi activation.
Fig. 3.
The C-tail of Smo is required for activation of Gli but not Gi. (A) Membranes from Sf9 cells infected with recombinant baculoviruses for Smo and a C-tail-deleted mutant of Smo (SmoΔC) were subjected to SDS/PAGE (10 μg of protein per lane) and then, after transfer to nitrocellulose, probed with antibodies directed against the C-terminal (H300) or N-terminal (N19) aspects of Smo. Solid arrowheads denote the position of full-length Smo (86 kDa), and the empty arrowhead denotes the position of truncated Smo (62 kDa). (B) [35S]GTPγS binding was evaluated as described in Fig. 1 for two members of the Gi family, Gi1 and Gi2, expressed with (gray bars) or without (black bars) SmoΔC in Sf9 cells. Results shown are representative of three independent experiments performed in triplicate. (C) SmoΔC-stimulated binding of [35S]GTPγS to the G proteins was evaluated in the presence or absence of 5 μM cyclopamine as described in Fig. 1 for Smo. (D) NIH 3T3 cells were transfected with empty vector (pcDNA), SmoM2, or a C-tail-deleted mutant of SmoM2 (SmoM2ΔC) together with Gli (and Renilla) luciferase reporters, then assayed for luciferase activities 24 h thereafter, as described in Fig. 2. (Inset) SmoM2 and SmoM2ΔC express equivalently in HEK 293 cells (Western blot with N-19); expression in 3T3 cells was not high enough for detection.
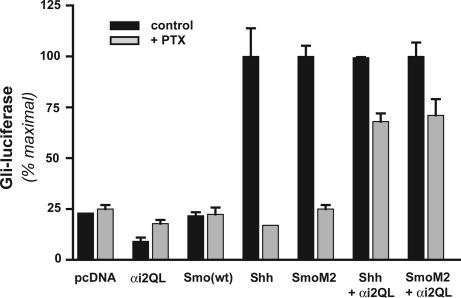
A Constitutively Active Gαi Rescues PTX-Inhibited Activation of Gli Through Smo.
Might two signals originate from Smo, then, one relating to activation of Gi and the other involving the C-tail, both of which are required for activation of Gli? We introduced a constitutively active form of a Gαi-subunit, Gαi2Q205L, into NIH 3T3 cells, expecting little activity toward the Gli reporter if indeed two signals are required. As anticipated, no activation of Gli was observed (Fig. 4). Noting that the activation state of the Gα mutant would not be affected by PTX, which influences only the interaction of the Gi protein with a receptor, we devised a protocol in which cells expressing Gαi2Q205L and either Shh or SmoM2 were treated with PTX to inactivate endogenous Gi. This strategy allowed us to test whether the Gαi2 mutant could complement signaling relevant to Gli by an activated Smo unable to communicate with Gi. In fact, Gαi2Q205L restored 60–70% of Gli-luciferase reporter activity normally inhibited by PTX. The reason for a less than complete complementation was not clear, but it possibly involves cotransfection efficiencies, contributions by Gβγ, or the need for precise colocalization of signals within the cells. Regardless, the result demonstrates that Gαi and another signal from Smo are, together, largely sufficient for activation of Gli.
Fig. 4.
Partial reversal of PTX inhibition of Smo signaling by a constitutively active mutant of Gαi. NIH 3T3 cells were cotransfected with the indicated combination of vectors and, after reaching confluency, were incubated in 0.5% FCS with or without 100 ng/ml PTX, as described in Fig. 2. Gli-dependent luciferase activity was determined 24 h later as described; activities for Shh, SmoM2, Shh plus Gαi2Q205L, and SmoM2 plus Gαi2Q205L without PTX were each set to 100%. Results are one of three equivalent experiments performed in triplicate.
Discussion
Information regarding the mechanisms by which the derepression of Smo is translated into signals that culminate in the regulation of the Gli family of transcription factors is limited. The finding that Smo has a 7-TM structure elicited early speculation that Smo utilizes heterotrimeric G proteins (21, 27). A structure of this nature does not guarantee the capacity to couple to G proteins, however, and data in support of such signaling for Smo have been indirect at best and superceded to some extent by interest in events originating at the C-tail of Smo. In the current study we provide proof that vertebrate Smo indeed has the capacity to activate heterotrimeric G proteins, specifically those of the Gi family. We also demonstrate that this capacity is realized within the context of Shh signaling in the intact cell. We show that the signaling of Smo through G proteins is insufficient to activate Gli but is nonetheless required. We confirm that the C-tail of Smo is required as well, but note its actions apart from G protein activation, supporting the relevance of two signals that originate with Smo.
Smo signaling in Drosophila provides a widely referenced platform for understanding the actions of Hh in general. Surprisingly, given our results, studies with Drosophila have provided no support for the utilization by Smo of heterotrimeric G proteins. One study to our knowledge addresses the question directly, that of Lum et al. (28), which notes that the response to Hh of a ptch-luciferase reporter in wing imaginal disc-derived cl-8 cells was unaffected by RNAi targeting individual or multiple Gα and Gβγ genes. Ingham and McMahon (1) cite the absence of any report of a G protein mutation that disrupts Hh signaling. The lack of positive data in Drosophila for the interaction of Smo with G proteins is difficult to ignore. Quite possibly, the utilization of Gi by Smo in vertebrates has evolved in response to other functions or pressures on Smo to incorporate G proteins.
Previous data for the utilization of G proteins by Smo in vertebrates has been more substantial, but not unequivocal. Human Smo, when transfected into Xenopus melanophores, caused pigment aggregation, which was reversed with PTX (9), and PTX also inhibited Shh-induced capillary morphogenesis (10). However, the toxin effected a phenotype in zebrafish embryos that only partly mimicked that of Shh deficiency (29), and the induction of slow muscle by Shh in vitro was not affected (30). The lack of more data regarding an inhibitory action of PTX is perplexing given our results. Nonetheless, the lack of an action of PTX must be viewed cautiously: not all cells have a mechanism for internalizing the S1-subunit of PTX (31); a toxin-insensitive member of the Gi family (Gz) exists in neurons, platelets, and conceivably other tissues (32); and, perhaps most importantly for in vivo studies, anything short of complete intoxication may not be effective (33). It is conceivable, still, that not all of the actions of Smo require Gi or that in some instances this requirement is masked by coincident signaling by other agonists, e.g., growth factors in culture medium or in vivo, that in some way achieve the actions otherwise achieved by Smo through Gi. We note that no previous study with PTX deals specifically with the relevance of Gi to Gli, nor do any demonstrate, in fact, the activation of Gi explicitly. Except for one study (34), the possible relevance of other G proteins was not evaluated either. In the latter study, p115RGS, a dominant inhibitor of G12 and G13, suppressed Shh- and Smo-effected Gli activation in transfected HEK 293 cells. The extent to which Gli is activated by Shh and Smo, however, is unusually small in these cells, and the suppression by p115RGS is partial.
Our work shows that Smo communicates with members of the Gi family and appears to do so exclusively. The assay used to achieve this conclusion is based on a basic property of receptor-mediated activation, the exchange by Gα-subunits of GDP for GTP, in this case mimicked by [35S]GTPγS, and relies here on the activity of Smo. We believe that the latter activity is intrinsic to Smo, i.e., that it represents receptor constitutive activity, which is widely believed to be the physiological basis of Smo’s action. The inability to detect activation of G12 or G13 is not a failure of the assay, because the activation of G12 and G13 by other 7-TM receptors is readily detected. We believe instead that the reported effects of the p115RGS are indirect or that G12 and G13 are engaged indirectly by Smo-effected synthesis of autocrine factors. However, it is also conceivable that G proteins beyond the Gi family can be activated if the activity of Smo exceeds that of constitutive activity, e.g., by action of a direct agonist. The range of G proteins could be broadened as a result of increasing strength of stimulus or changed by an agonist-induced change in activating conformation (35). Physiological ligands of this nature are just now being proposed (36).
The effects of cyclopamine, KAAD-cyclopamine, and SANT-1 confirm that the increased [35S]GTPγS binding to members of the Gi family upon coexpression of Smo is in fact attributable to Smo activity. These effects, in that they represent a reversal of activity, are consistent with inverse agonism. However, proof of inverse agonism requires that the actions of an endogenous agonist be ruled out as a cause of Smo activity in the first place. The definitive experiment for inverse agonism will therefore require a neutral antagonist as a control, which has yet to be identified for Smo. The rank-ordering of potencies for cyclopamine, KAAD-cyclopamine, and SANT-1 in suppressing [35S]GTPγS binding to Gi (SANT-1>cyclopamine>KAAD-cyclopamine) differed from that reported for inhibition of Shh signaling in intact cells (KAAD-cyclopamine≈SANT-1>cyclopamine) (23, 37). We do not know the basis for this difference.
The results here strongly argue that vertebrate Smo is the source of two signals relevant to the activation of Gli, one involving activation of Gi and the other involving events specific to the C-tail of Smo. Specifically, the requirement for Gi was demonstrated by the inhibition by PTX of Shh- and SmoM2-effected activation of Gli, whereas the requirement for the C-tail was demonstrated by the inactivity toward Gli of the truncated Smo, which can nonetheless effectively activate Gi. With regard to the C-tail, vertebrate Smo is phosphorylated by G protein-coupled receptor kinase 2, leading to interaction of Smo with arrestin (15), which is relevant to internalization and various forms of signaling (38). It is possible that the activation of Gi is a prerequisite to phosphorylation of vertebrate Smo in NIH 3T3 cells, because Gβγ once released from Gα can serve as an anchor for G protein-coupled receptor kinase 2/3 (39). Given the partial reconstitution of Gli activation by constitutively active Gαi in PTX-treated cells, however, a more direct form of signaling by Gi must exist.
Effectors modulated by Gi in the context of Gli activation remain to be determined. A reduction in cAMP through inhibition of adenylyl cyclase is attractive, because basal levels of cAMP may support the small degree of protein kinase A activity necessary to inhibit Gli activation (40). We note, however, that Shh did not reduce forskolin-elevated concentrations of cAMP at least in one cell type (41) and that, based on work with Drosophila Smo, the effects of protein kinase A can be stimulatory as well as inhibitory (12, 14, 42, 43). Other potential targets for Gi include phosphatidylinositol 3-kinase, MEK-1, and protein kinase C, all of which serve roles in Shh signaling to Gli (7, 44). We note also the relevance of Gi in transactivation of receptor tyrosine kinases (45) and in the production of autocrine factors (46). The activation of Gi by Smo might also account for events that occur simply too rapidly to be achieved through transcriptional activation. One such event is the negative regulation of growth cone movement (47). Regardless, the demonstration of Gi as a transducer for Smo provides now the rationale to evaluate more systematically the relevance of Gi-based effector pathways.
Materials and Methods
Materials.
Cyclopamine was obtained from Toronto Research Chemicals (Toronto, Canada). KAAD-cyclopamine, SANT-1, tomatidine, Pansorbin cells, and Nonidet P-40 were from EMD Biosciences (La Jolla, CA). [35S]GTPγS [1,300 Ci/mmol (1 Ci = 37 GBq)] was obtained from PerkinElmer Life Science Products (Boston, MA). Anti-Smo (H-300), anti-Smo (N-19), and horseradish peroxidase-conjugated anti-rabbit and anti-goat antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit antisera specific for individual Gα-subunits were already reported (19, 22, 48).
Baculoviruses and Plasmid Constructs.
Recombinant baculoviruses encoding Gα, Gβ1, and Gγ2 have been described (19, 22). Baculoviruses encoding full-length and truncated (residues 1–566) mouse Smo were generated by using pFASTBAC and the Bac-To-Bac expression system (Invitrogen, Carlsbad, CA) . The Smo WT, Shh, and SmoM2 expression vectors were provided by Philip Beachy (Johns Hopkins University, Baltimore, MD). SmoM2ΔC was generated by removal of a BglII-XbaI fragment of SmoM2 corresponding to the last 242 amino acid residues, ligation, and subcloning of the HindIII-XbaI coding sequence into pcDNA3.1+ and was sequence-verified. Full-length mGli1 and mGli2 expression constructs and the reporter vectors 8xGBSwt-luc and 8xGBSmut-luc were a gift from H. Sasaki (Osaka University, Osaka, Japan). pRL-TK was obtained from Promega (Madison, WI).
Cell Culture, Infection, and Transfection.
Sf9 cells (Invitrogen) were maintained in Grace’s insect medium plus 10% heat-inactivated FBS as described (48). The cells were infected with combinations of baculoviruses at an MOI of one for G protein subunits and two (unless otherwise indicated) for Smo. NIH 3T3 cells (American Type Culture Collection, Manassas, VA) were maintained in DMEM with 10% FCS and penicillin/streptomycin at 37°C with 5% CO2. For transfections, NIH 3T3 cells were seeded in 24-well plates at ≈70% confluence and transfected with different mammalian expression vectors using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s protocol. After reaching 100% confluency, the medium was changed to 0.5% FCS with or without 100 ng/ml PTX and left for an additional 24 h. Firefly and Renilla luciferase activities were determined in lysates with the Dual Luciferase Reporter Assay System (Promega).
[35S]GTPγS Binding Assay.
[35S]GTPγS binding was assayed for various cell membranes essentially as described previously for those of Sf9 cells (19). Briefly, membranes (20 μg of protein per assay point) were incubated with vehicle or ligands for 10 min at 30°C in the presence of 20 mM Mg2+ and 0.1–30 μM GDP, depending on the G protein. [35S]GTPγS (final concentration 1 or 5 nM) was subsequently added, and membranes were incubated for an additional 10 min at 30°C. The membranes were solubilized under nondenaturing conditions, and the Gα-subunits were immunoprecipitated by using subunit-selective rabbit antisera. Bound radioactivity was quantitated by scintillation spectrometry.
Semiquantitative RT-PCR.
Total RNA was isolated from NIH 3T3 cells with the RNeasy kit (Qiagen, Valencia, CA) as directed. An aliquot of 2 μg was used for reverse transcription and PCR as described elsewhere (44).
Analysis of Data.
All differences stated in the text were statistically significant at P < 0.05 using Student’s t test.
Supplementary Material
Acknowledgments
Support for this study was provided by National Institutes of Health Grant GM066892.
Glossary
Abbreviations
- GTPγS
guanosine 5′-(3-O-thio)triphosphate
- Hh
Hedgehog
- MOI
multiplicity of infection
- Ptch
Patched
- PTX
pertussis toxin
- Shh
Sonic hedgehog
- Smo
Smoothened
- 7-TM
seven-transmembrane.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ingham P. W., McMahon A. P. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwenhuis E., Hui C.-c. Clin. Genet. 2005;67:193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 3.di Magliano M. P., Hebrok M. Nat. Rev. Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 4.Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., et al. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang B., Fallon J. F., Beachy P. A. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 6.Price M. A., Kalderon D. Cell. 2002;108:823–836. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 7.Riobo N. A., Lu K., Ai X., Haines G. M., Emerson C. P. Proc. Natl. Acad. Sci. USA. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Bai C. B., Joyner A. L., Wang B. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCamp D. L., Thompson T. M., de Sauvage F. J., Lerner M. J. Biol. Chem. 2000;275:26322–26327. doi: 10.1074/jbc.M004055200. [DOI] [PubMed] [Google Scholar]
- 10.Kanda S., Mochizuki Y., Suematsu T., Miyata Y., Nomata K., Kanetake H. J. Biol. Chem. 2003;278:8244–8249. doi: 10.1074/jbc.M210635200. [DOI] [PubMed] [Google Scholar]
- 11.Jia J., Tong C., Jiang J. Genes Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C., Williams E. H., Guo Y., Lum L., Beachy P. A. Proc. Natl. Acad. Sci. USA. 2004;101:17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apionishev S., Katanayeva N., Marks S. A., Kalderon D., Tomlinson A. Nat. Cell Biol. 2004;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- 14.Jia J., Tong C., Wang B., Luo L., Jiang J. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 15.Chen W., Ren X.-R., Nelson C. D., Barak L. S., Chen J. K., Beachy P. A., de Sauvage F. J., Lefkowitz R. J. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 16.Varjosalo M., Li S.-P., Taipale J. Dev. Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Barr A. J., Manning D. R. J. Biol. Chem. 1997;272:32979–32987. doi: 10.1074/jbc.272.52.32979. [DOI] [PubMed] [Google Scholar]
- 18.Windh R. T., Barr A. J., Manning D. R. In: Handbook of Experimental Pharmacology, Kenakin T., Angus J. A., editors. Heidelberg, Germany: Springer; 2000. pp. 335–362. [Google Scholar]
- 19.Barr A. J., Brass L. F., Manning D. R. J. Biol. Chem. 1997;272:2223–2229. doi: 10.1074/jbc.272.4.2223. [DOI] [PubMed] [Google Scholar]
- 20.Windh R. T., Manning D. R. Methods Enzymol. 2002;344:3–14. doi: 10.1016/s0076-6879(02)44702-7. [DOI] [PubMed] [Google Scholar]
- 21.van den Heuvel M., Ingham P. W. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 22.Windh R. T., Lee M.-J., Hla T., An S., Barr A. J., Manning D. R. J. Biol. Chem. 1999;274:27351–27358. doi: 10.1074/jbc.274.39.27351. [DOI] [PubMed] [Google Scholar]
- 23.Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 24.Incardona J. P., Gaffield W., Kapur R. P., Roelink H. Development (Cambridge, U.K.) 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- 25.Sinha S., Chen J. K. Nat. Chem. Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 26.Frank-Kamenetsky M., Zhang X., Bottega S., Guicherit O., Wichterle H., Dudek H., Bumcrot D., Wang F., Jones S., Shulok J., et al. J. Biol. 2002;1:10.1–10.19. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcedo J., Ayzenzon M., Von Ohlen T., Noll M., Hooper J. E. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 28.Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. Mol. Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- 29.Hammerschmidt M., McMahon A. P. Dev. Biol. 1998;194:166–171. doi: 10.1006/dbio.1997.8796. [DOI] [PubMed] [Google Scholar]
- 30.Norris W., Neyt C., Ingham P. W., Currie P. D. J. Cell Sci. 2000;113:2695–2703. doi: 10.1242/jcs.113.15.2695. [DOI] [PubMed] [Google Scholar]
- 31.Ui M. In: ADP-Ribosylating Toxins and G Proteins, Moss J., Vaughan M., editors. Washington, DC: Am. Soc. for Microbiol; 1990. [Google Scholar]
- 32.Yang J., Wu J., Kowalska M. A., Dalvi A., Prevost N., O’Brien P. J., Manning D., Poncz M., Lucki I., Blendy J. A., Brass L. F. Proc. Natl. Acad. Sci. USA. 2000;97:9984–9989. doi: 10.1073/pnas.180194597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pobiner B. F., Hewlett E. L., Garrison J. C. J. Biol. Chem. 1985;260:16200–16209. [PubMed] [Google Scholar]
- 34.Kasai K., Takahashi M., Osumi N., Sinnarajah S., Takeo T., Ikeda H., Kehrl J. H., Itoh G., Arnheiter H. Genes Cells. 2004;9:49–58. doi: 10.1111/j.1356-9597.2004.00701.x. [DOI] [PubMed] [Google Scholar]
- 35.Manning D. R. Mol. Pharmacol. 2002;62:451–452. doi: 10.1124/mol.62.3.451. [DOI] [PubMed] [Google Scholar]
- 36.Corcoran R. B., Scott M. P. Proc. Natl. Acad. Sci. USA. 2006;103:8408–8413. doi: 10.1073/pnas.0602852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J. K., Taipale J., Cooper M. K., Beachy P. A. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefkowitz R. J., Shenoy S. K. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 39.Pitcher J. A., Inglese J., Higgins J. B., Arriza J. L., Casey P. J., Kim C., Benovic J. L., Kwatra M. M., Caron M. G., Lefkowitz R. J. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 40.Epstein D. J., Marti E., Scott M. P., McMahon A. P. Development (Cambridge, U.K.) 1996;122:2885–2894. doi: 10.1242/dev.122.9.2885. [DOI] [PubMed] [Google Scholar]
- 41.Murone M., Rosenthal A., de Sauvage F. J. Curr. Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 42.Ohlmeyer J. T., Kalderon D. Genes Dev. 1997;11:2250–2258. doi: 10.1101/gad.11.17.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Gallaher N., Goodman R. H., Smolik S. M. Proc. Natl. Acad. Sci. USA. 1998;95:2349–2354. doi: 10.1073/pnas.95.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riobo N. A., Haines G. M., Emerson C. P. Cancer Res. 2006;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 45.Daub H., Wallasch C., Lankenau A., Herrlich A., Ullrich A. EMBO J. 1997;16:7032–7044. doi: 10.1093/emboj/16.23.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berk B. C. Physiol. Rev. 2001;81:999–1030. doi: 10.1152/physrev.2001.81.3.999. [DOI] [PubMed] [Google Scholar]
- 47.Trousse F., Marti E., Gruss P., Torres M., Bovolenta P. Development (Cambridge, U.K.) 2001;128:3927–3936. doi: 10.1242/dev.128.20.3927. [DOI] [PubMed] [Google Scholar]
- 48.Butkerait P., Zheng Y., Hallak H., Graham T. E., Miller H. A., Burris K. D., Molinoff P. B., Manning D. R. J. Biol. Chem. 1995;270:18691–18699. doi: 10.1074/jbc.270.31.18691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.