Abstract
Nutrient sensing is critical for plant adaptation to the environment. Because of extensive farming and erosion, low content of mineral nutrients such as potassium (K+) in soils becomes a limiting factor for plant growth. In response to low-K conditions, plants enhance their capability of K+ uptake through an unknown signaling mechanism. Here we report the identification of a Ca2+-dependent pathway for low-K response in Arabidopsis. We are not aware of any other example of a molecular pathway for a nutrient response in plants. Earlier genetic analyses revealed three genes encoding two Ca2+ sensors (CBL1 and CBL9) and their target protein kinase (CIPK23) to be critical for plant growth on low-K media and for stomatal regulation, indicating that these calcium signaling components participate in the low-K response and turgor regulation. In this study, we show that the protein kinase CIPK23 interacted with, and phosphorylated, a voltage-gated inward K+ channel (AKT1) required for K+ acquisition in Arabidopsis. In the Xenopus oocyte system, our studies showed that interacting calcium sensors (CBL1 and CBL9) together with target kinase CIPK23, but not either component alone, activated the AKT1 channel in a Ca2+-dependent manner, connecting the Ca2+ signal to enhanced K+ uptake through activation of a K+ channel. Disruption of both CBL1 and CBL9 or CIPK23 gene in Arabidopsis reduced the AKT1 activity in the mutant roots, confirming that the Ca2+-CBL-CIPK pathway functions to orchestrate transporting activities in planta according to external K+ availability.
Keywords: calcium signaling, potassium channel, nutrient sensing, potassium uptake, protein kinase
Plants and microbes are able to adapt to widely varying environmental conditions, including the availability of inorganic nutrients. The response to K+ is especially relevant because it is the major inorganic osmoticum that contributes to turgor pressure and, hence, to cellular growth and development (1, 2). Both plants and fungi maintain internal K+ near 100 mM even when extracellular K+ varies between 1 μM and 100 mM (i.e., over five orders of magnitude in free concentration). A large collection of transporters is required for this homeostatic mechanism (3–6), yet little is known about the mechanism underlying regulation and integration of the transport activities in concert with the extracellular K+ levels. Some studies suggest that plants enhance their capability of K+ uptake by activating some K+ transporters in response to K+-deficient conditions (7–10). A signaling process exists for the plants to “monitor” external K+ concentration and “respond” to the low-K condition by enhancing the capability for K+ acquisition. This signaling pathway represents a typical nutrient sensing and response process in plants about which our knowledge is extremely limited. One recent study indicates that low-K status in the soil triggers elevated production of H2O2 that may serve as a signaling molecule to alter the expression of certain genes (11). Because H2O2 production leads to Ca2+ changes in plant cells (12, 13), it is conceivable that Ca2+ functions as a second messenger in plant response to low-K stress. How does a Ca2+ signal translate low-K stimulus into enhanced K+ uptake in plants?
Calcium signaling in both plants and animals involves Ca2+-binding proteins as initial sensors. Calcineurin B-like (CBL) proteins are a unique family of calcium sensors in plants (14, 15). The CBL proteins function by interacting with and regulating a unique family of plant protein kinases (called CIPKs for CBL-interacting protein kinases) (15–19). The presence of multigene families of CBLs and CIPKs in Arabidopsis suggests that CBL-CIPK network may be involved in a number of signaling processes in plants (20–22, ‡). Using a genetics approach to dissecting the function of the CBL and CIPK genes, we identified two CBLs (CBL1 and CBL9) that play specific roles in stress responses and response to abscisic acid (ABA) (20, 21). In addition, CBL1 and CBL9 also have redundant functions in the regulation of low-K response and stomatal movements. These two CBLs share a common interacting kinase, CIPK23, that is also required for low-K response and stomatal regulation in Arabidopsis.‡. However, it was not known how this CBL–CIPK pathway regulates K+ uptake or stomatal response. Here we report the identification of AKT1, a voltage-gated K channel, as a downstream target of the CBL1/9–CIPK23 pathway. The CBL–CIPK complex interacted with and activated AKT1 in a Ca2+-dependent manner, enhancing K+ uptake in Arabidopsis under low-K conditions.
Results and Discussion
CIPK23 Interacts with and Phosphorylates the AKT1 K+ Channel.
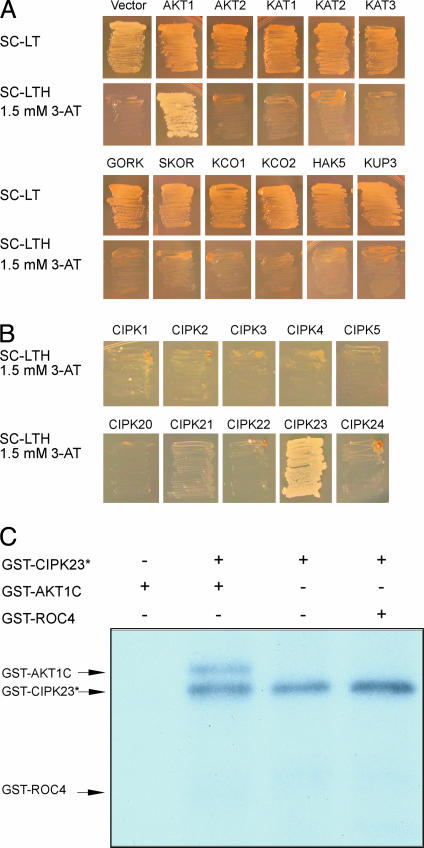
Genetic analysis of the CBL1/CBL9 and CIPK23 genes showed that they are important for K uptake in roots and regulation of stomatal movements.‡ We hypothesized that the CBL–CIPK pathway interacts with and regulates the activity of a transport protein or proteins involved in K+ uptake in the roots and in turgor regulation of stomatal guard cells. To test this possibility, we examined the potential physical interaction between CIPK23 and a number of K+ transporters by using a yeast two-hybrid assay. To avoid interference of the transmembrane domains in the transporters, we used the putative cytoplasmic hydrophilic domains of the transporters in the interaction assays. We chose representative members in different subfamilies of K transporters as described earlier (3). Among the transporters tested, we found that only the C-terminal domain of AKT1, a voltage-gated shaker-type K channel (24, 25), interacted with CIPK23 (Fig. 1A). The AKT1 protein did not interact with CIPK3, a CIPK involved in abscisic acid response (22) and several other CIPKs (Fig. 1B), indicating specific interaction of AKT1 with CIPK23.
Fig. 1.
CIPK23 specifically interacts with and phosphorylates AKT1. (A) Yeast two-hybrid assay of CIPK23 interaction with various K+ transporters. Yeast strain PJ69-4A was transformed with various K+ transporters in the pGAD.GH vector and CIPK23 cloned into pGBT9. The growth in selective medium indicates protein–protein interaction. SC, synthetic complete agar medium; SC-LT, SC minus leucine and tryptophan; SC-LTH, SC minus leucine, tryptophan, and histidine; 3-AT, 3-amino-1,2,4-aminotriazole. The K+ transporters are classified in Maser et al. (3), and the C-terminal regions of these transporters were defined by specific primers listed in Materials and Methods. (B) Yeast two-hybrid assays of AKT1 interaction with 10 CIPKs. The CIPKs were cloned into the pGBT9 and AKT1 was in pGAD.GH. (C) Autokinase activity and phosphorylation of AKT1 C-terminal region by CIPK23.
It is possible that CIPK23 interacts with AKT1 and targets the AKT1 protein as a substrate. We tested this possibility by in vitro phosphorylation assay. The CIPK23 protein was expressed in Escherichia coli (prokaryotic system) or Xenopus laevis oocytes (eukaryotic system). Although a large amount of CIPK23 protein was purified from E. coli, we detected little activity of CIPK23 in a typical kinase assay (data not shown). We purified the CIPK23 protein from oocytes after coexpression of CIPK23 and CBL1. This form of kinase was more active and showed both autokinase activity and phosphorylation against AKT1-GST fusion (Fig. 1C). The GST fusion with a cyclophilin protein ROC4 was not phosphorylated by CIPK23, indicating that CIPK23 uses AKT1 as a specific substrate.
Activation of AKT1 by the CBL–CIPK Pathway.
Earlier studies have shown that AKT1 plays a critical role in plant growth under low-K conditions (24). The akt1 mutant, like cipk23 or cbl1cbl9 double mutant,‡ displayed hypersensitivity to low K+. Together with our findings on CIPK23–AKT1 interaction and CBL1/9–CIPK23 function in K+ uptake, we proposed a model signaling pathway for plant response to low-K stress: calcium changes under low-K condition activate CBL1 or CBL9 calcium sensor that interacts and activates the target kinase CIPK23. The active CIPK23 interacts and phosphorylates inward K-channel AKT1, leading to activation of AKT1 and enhanced K+ uptake in plants.
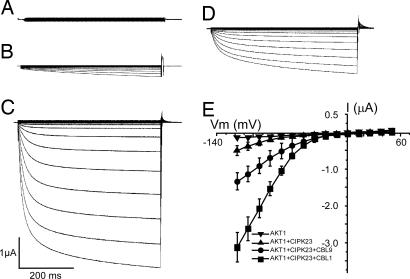
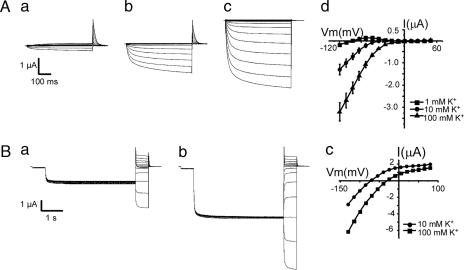
To test this model, we reconstituted the Ca2+–CBL–CIPK–AKT1 pathway in Xenopus oocytes, where ion channel activity can be readily studied by the electrophysiological approach (26–28). When AKT1 was expressed alone in the oocytes, no inward K+ current was recorded by using the two-electrode voltage-clamp procedure (Fig. 2A), consistent with an earlier report that AKT1 channel was not active in oocytes (25). Upon coexpression of AKT1 and CIPK23, a low level of AKT1 activity was detected (Fig. 2B), suggesting that CIPK23 alone was active to some extent in the activation of the AKT1 channel. After coexpression of AKT1, CIPK23, and CBL1, AKT1 activity was dramatically enhanced (Fig. 2C), reaching a magnitude of > 10-fold the current recorded from AKT1–CIPK23 coexpressed oocytes. The activation pattern of the AKT1 channel was analyzed with the I–V curves shown in Fig. 2E. The current traces in Fig. 2C depict time-dependent channel opening kinetics at different voltages and the I–V curves in Fig. 2E summarize the current values at the end of each voltage episode. Coexpression of CBL9 (instead of CBL1) with CIPK23 and AKT1 also enhanced the AKT1 current (Fig. 2D) although to a lesser extent, indicating that both CBL1 and CBL9 are capable of activating CIPK23. Coexpression of other combinations (AKT1 with CBL1 or CBL9; CBL1 with CIPK23) did not produce significant current (data not shown), confirming that the current is produced by AKT1, whose activity requires the CBL–CIPK complex. To confirm that the current produced by AKT1 channel is K+-dependent as it should be (25), we determined the channel activity with various K+ concentrations in the bath solution, and we found that the current was proportional to [K+] and showed a kinetics of activation with AKT1 similar to that reported earlier (Fig. 3A) (25). In addition, tail current analysis at various K+ concentrations (Fig. 3B) also indicated that the shift in the reverse potential is consistent with the fact that AKT1 is an inward K+ channel.
Fig. 2.
Reconstitution of the CBL–CIPK–AKT1 pathway in Xenopus oocytes. Whole-cell currents were recorded in oocytes expressing AKT1 (A), AKT1 + CIPK23 (B), AKT1 + CIPK23 + CBL1 (C), or AKT1 + CIPK23 + CBL9 (D). The current traces depict time-dependent channel opening kinetics at different voltages. The current–voltage (I–V) curves in E plot current values at the end of each voltage-clamp episode (t = 0.5 s, n = 5 for each group). The bath solution contained 100 mM KCl. Holding potential was −20 mV. Clamp potentials ranged from +48 to −120 mV with a −12-mV step.
Fig. 3.
The AKT1 activity was K dependent. (A) Whole-cell currents recorded from oocytes expressing AKT1 + CIPK23 + CBL1 with bath solution containing 1 mM (a), 10 mM (b), or 100 mM (c) KCl. The I–V curves shown in d are derived from multiple recordings (n = 5). Holding potential was −20 mV. Clamp potential ranged from +48 to −120 with a −12-mV step. (B) Tail currents were obtained by activating the time-dependent currents with a 4-s pulse to −100 mV from holding potential of −40 mV followed by a step from +80 to −115 mV (a, 10 mM K+; b, 100 mM K+). Representative I–V plots of steady-state currents under the same recording condition are shown in c.
Regulation of AKT1 by CBL–CIPK Involves Modulation of the Channel Activity and Requires Kinase Activity of CIPK23.
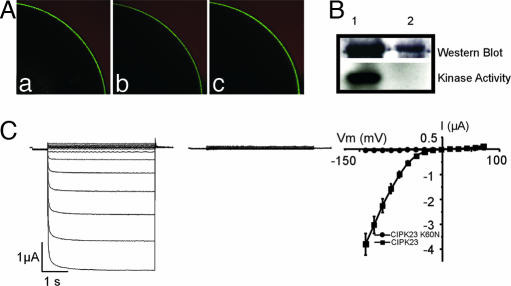
Activation of AKT1 channel by the CBL–CIPK regime may be achieved through two possible mechanisms. The first one is that CBL–CIPK facilitates the expression and plasma membrane localization of AKT1 and the second is the activation of the channel already assembled into the membrane. Our studies using AKT1-GFP fusion showed that AKT1 protein can be expressed and targeted to the plasma membrane without the need of the CBL–CIPK complex. Coexpression of CBL1–CIPK23 with AKT1-GFP did not have a significant effect on the level of AKT1-GFP fusion in the plasma membrane (Fig. 4A). It is therefore likely that the CBL–CIPK “complex” phosphorylates the AKT1 protein, leading to activation of its channel activity. In this regard, we tested whether the kinase activity is essential for the activation of AKT1. We mutated the ATP-binding lysine in the CIPK23 protein resulting in loss of its kinase activity (Fig. 4B). We coexpressed this form of “dead” kinase with CBL1 and AKT1 in the oocytes and found that AKT1 was no longer active (Fig. 4C), demonstrating that AKT1 phosphorylation by CIPK23 is required for AKT1 activation.
Fig. 4.
AKT1 activation by the CBL–CIPK regime involves modification of the channel activity and requires kinase activity of CIPK23. (A) Expression and localization of AKT1-GFP fusion in the oocytes injected with cRNA of AKT1-GFP (a), CIKP23 + AKT1-GFP (b), or CBL1 + CIPK23 + AKT1-GFP (c). A quarter of an oocyte is shown in each case. Similar GFP fluorescence was localized to the plasma membrane. (B) Expression of the wild-type and mutant version of CIPK23 (CIPK23K60N) in oocytes. Expression of GST-CIPK23 and GST-CIPK23K60N was confirmed by Western blot using anti-GST antibody. The mutant version was inactive and did not autophosphorylate in the kinase assay. Lane 1, GST-CIPK23; lane 2, GST-CIPK23K60N. (C) The CIPK23K60N mutant failed to activate AKT1. Whole-cell currents were recorded by two-electrode voltage-clamp from oocytes expressing AKT1 + CBL1 + GST-CIPK23 (Left) or AKT1 + CBL1 + GST-CIPK23K60N (Center). (Right) The I–V curves plot the current values at the end of the voltage-clamp episodes (t = 4 s, n = 5 for each group). Holding potential was −40 mV. Clamp potential ranged from +55 to −140 with a −15-mV step.
Activation of AKT1 by CBL-CIPK Is Ca2+-Dependent.
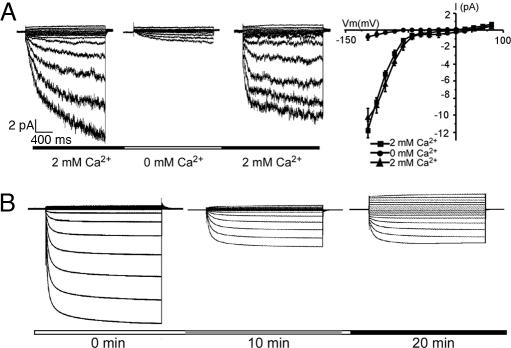
Because CBLs are calcium sensors, we tested whether Ca2+ is required for the CBL–CIPK activation of the AKT1 channel by a giant-patch recording procedure (26). In this procedure, a patch of membrane was excised from an oocyte cell surface so that the membrane retained an inside-out configuration. Although the cytoplasmic side of the membrane was exposed to the bath solution and was detached from the cytoplasm of the oocyte, the CBL–CIPK complex should be associated with the membrane through interaction with the C-terminal domain of the AKT1 channel. If this is true, the AKT1 channel in the excised membrane should be active in the presence of MgATP and Ca2+ to maintain the activation of the AKT1 by the Ca2+–CBL–CIPK pathway. This was indeed the case. After the patch of membrane was excised, the AKT1 current was retained in the bath solution containing MgATP and Ca2+ (Fig. 5A). If CBL–CIPK regulation of AKT1 depends on Ca2+, the channel activity should go down when the bath is perfused with the solution containing MgATP but not Ca2+. As shown in Fig. 5A, bath perfusion with solution lacking Ca2+ almost abolished the AKT1 currents. Further perfusion with Ca2+-containing solution restored the AKT1 current, demonstrating that the activation of AKT1 by the CBL–CIPK complex is reversible and requires Ca2+. As an additional test, we also injected EGTA into the oocyte by using a third pipette in a two-electrode voltage-clamp configuration and found that EGTA significantly reduced the AKT1 currents in the oocytes coexpressing CBL1–CIPK23–AKT1 (Fig. 5B). Injection of water instead of EDTA did not have any effect (data not shown).
Fig. 5.
Calcium-dependent activation of the CBL–CIPK–AKT1 pathway. (A) Giant-patch recordings of AKT1 activity with or without 2 mM calcium gluconate (shown as 2 mM or 0 mM in the perfusion scheme) in the perfusion solution. (Right) I–V curves of AKT1 activity. The voltage protocol is from +65 to −130 mV with a −13-mV step. (B) Inhibition of the AKT1 activity by injection of EGTA into the oocytes. The third pipette (besides the two electrodes for voltage clamping) was introduced to inject EGTA (23 nl, 500 mM) into the oocytes. The currents before injection and 10 min and 20 min after injection were recorded. The bath solution contained 100 mM KCl. Holding potential was −20 mV. Clamp potentials ranged from +48 to −132 mV with a −12-mV step.
CBL–CIPK Pathway Regulates AKT1 Activity in Planta.
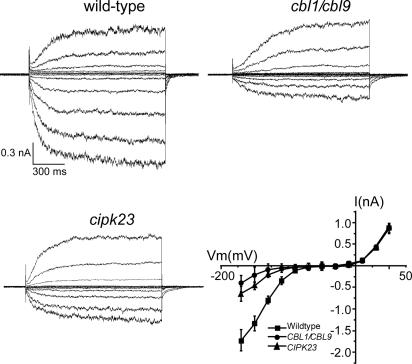
The Ca2+–CBL–CIPK–AKT1 pathway was reconstituted in the oocyte system, confirming that AKT1 channel is the target for the Ca2+-dependent signaling pathway involving two calcium sensors and their interacting kinase. If this pathway operates in planta, disruption of this pathway by mutation of CIPK23 or the CBLs should reduce the activity of the AKT1 channel in Arabidopsis roots. We examined the AKT1 activity in the roots from the wild-type, cipk23 mutant, and cbl1cbl9 double mutant by patch-clamp recording using isolated protoplasts. As described earlier (24, 29, 30), the root hair protoplasts displayed both inward and outward K+ currents and the AKT1 activity is the predominant contributor to the inward K+ current in root hair cells (24, 30). We show here that the same inward current in the root hair cells was significantly reduced in the cipk23 mutant or in the cbl1cbl9 double mutant (Fig. 6), confirming that the CBL–CIPK pathway is essential for the optimal function of AKT1 in vivo. The outward K+ currents in the same protoplasts were not altered in the mutants, consistent with the idea that CIPK23 specifically interacts with and regulates the activity of AKT1 but not other channels.
Fig. 6.
Reduced AKT1 activity in the cipk23 and cbl1cbl9 mutants. Voltage- and time-dependent activity of inward and outward K+ currents in the root hair protoplasts from wild type, cbl1cbl9 double mutant, and cipk23 mutant were recorded in a whole-cell configuration. Current traces represent typical recordings obtained in five independent experiments. The I–V curves of K+ steady-state current values were derived as mean ± SE from five independent experiments (each experiment recorded three cells for each sample; n = 15). The bath and pipette solutions were the same as described in ref. 29. Voltage-clamp protocols were applied from a holding potential of −40 mV in the range from +65 mV to −188 mV with a −23-mV step.
K channels are involved in numerous other processes (besides K uptake in roots) such as turgor regulation in guard cells (4, 31). In the context of AKT1 regulation by the CBL–CIPK pathway, it is important to note that the same CBL–CIPK pathway was also shown to regulate stomatal movement in Arabidopsis.‡ However, it is not known whether AKT1 or other transporter(s) serves as the downstream target of CBL–CIPK in turgor regulation of guard cells. Further analysis of ion channel activities in guard cells in the cbl1cbl9 and cipk23 mutants will resolve the mechanism of CBL–CIPK action in stomatal regulation. We speculate that the CBL–CIPK network constitutes a critical regulatory circuit for the regulation of ionic transports in plants.
Concluding Remarks.
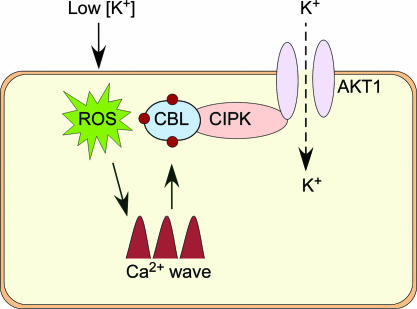
Nutrient sensing is an ancient mechanism for environmental adaptation of organisms such as plants and bacteria (6, 32). A low-K condition in the environment is a rate-limiting factor for plant growth. In response to such a condition, plants initiate mechanisms to enhance their K+-uptake capability (6), which represents an effective strategy to survive and adapt to the low-K environment. Using a combination of genetic, biochemical, and electrophysiological approaches, we have uncovered a comprehensive signaling pathway for the low-K response in Arabidopsis, providing an example of nutrient response mechanism in plants. This pathway involves two calcium sensors (CBL1 and CBL9) physically interacting with their target protein kinase CIPK23, which, in turn, interacts with and phosphorylates the voltage-gated K channel, AKT1. Activation of the CBL–CIPK pathway by Ca2+ triggers the activation of the AKT1 channel, enhancing K+ uptake by roots. As a result, disruption of the genes encoding the calcium sensors or the kinase in Arabidopsis plants leads to reduced AKT1 channel activity in root cells (this study), lowers K+ uptake, and results in a hypersensitive response to low-K.‡ Fig. 7 presents a schematic model of this pathway.
Fig. 7.
A schematic model of the Ca2+-dependent pathway for low-K response in Arabidopsis (see details in Concluding Remarks). ROS, reactive oxygen species.
Potassium is the most abundant inorganic cation in plants, constituting up to 5–10% of a plant’s dry weight (1, 2). The essential role of K+ in plant growth has long been recognized, and early physiological studies identified a number of symptoms associated with K+ deficiency (2). Because low K in the soil has been an ongoing condition, plants have evolved mechanisms to adapt by enhancing K+ uptake in the roots (6). Strong evidence supports that the voltage-gated K channel AKT1 participates in low-K response as a downstream effector molecule (10, 24, 30). Because AKT1 mRNA is not elevated by low-K treatment (33), activation of AKT1 by low-K signal may occur at the posttranslational level. Our study here has demonstrated that AKT1 is activated by a Ca2+-dependent signaling mechanism involving a protein kinase directly interacting with and phosphorylating the AKT1 protein. To our knowledge, this is the first case in which plant ion channel is activated by a protein kinase through direct protein–protein interaction and phosphorylation, setting the stage for further mechanistic analysis of AKT1 regulation by CIPK23 or other CIPKs and for the regulation of other ion channels by protein phosphorylation in plants.
Materials and Methods
Yeast Two-Hybrid Assays.
Cytoplamic C-terminal regions of 11 K+ transporters were cloned into the activation domain vector (pGAD.GH), and the CIPK23 full-length cDNA was cloned into the DNA-binding domain vector (pGBT9.BS). Primers used in these clonings are listed below, with restriction sites underlined. The “F” following the gene name indicates forward and “R” indicates reverse primers. SKORF, 5′-GTAAAGGATCCGAAAACAGAGAGATTCCGG-3′; SKORR, 5′-TATGTGTCGACAGCCAAATACAGTTTTTGTCC-3′; GORKF, 5′-GATGGGGGAATTCTCCGCTTGATGAAGCCCGA-3′; GORKR, 5′-GAAGAGTCGACCATTATGTTTGATCAGTAGTATCAC-3′; KCO1F, 5′-GAGCTGAATTCAGAAAACAAACAGAGGGCG-3′; KCO1R, 5′-GATGATGTCGACGATGAGGCTTACCTTTGAA-3′; KCO2F, 5′-GGCGAATTCAAGAGTTGATAAGAGGAATAGG-3′; KCO2R, 5′-CATGTGTCGACGTGTGTTACTAAATAGAAGTTG-3′; KAT2F, 5′-ACCAGGAATTCTAGAGATACTGTAAGAGCTG-3′; KAT2R, 5′-GGTTACTCGAGTCTTGTTAAGAGTTTTCATTG-3′; KAT3F, 5′-CGTACGAATTCCATGAGGAGTGCGATCAAT-3′; KAT3R, 5′-GAGCCTCGAGTTGTTCAACATGTGCACCATC-3′; AKT1F, 5′-GACTAGGAATTCTAGAGATACAATCCAAGC-3′; AKT1R, 5′-GTTTCTGTCGACGTTAAGAATCAGTTGCAAAG-3′; AKT2F, 5′-GGGACACTAGTTACCATGGAATTTAGGAAT-3′; AKT2R, 5′-AATTTCTCGAGTTCTAAATTATCTTGTTTACG-3′; HAK5F, 5′-CTCAAGGATCCGTACGAGCTGAGGGAGAAG-3′; HAK5R, 5′-ACACGTCGACGAAATAGAGAATCTTATAAC-3′; KUP3F, 5′-GCAATGAATTCTCACTTTGTGACCAACCTG-3′; KUP3R, 5′-TGAAGTCGACTCATATTAGTGGCTTAAACAT-3′; CIPK23F, 5′-ATATATATCTAGAGAGAGAGATGGCTTCTC-3′; and CIPK23R, 5′-GAACAAACTCGAGATTAGAAAAAAGTAAAC-3′.
The CIPK23 BD and each of potassium transporter AD plasmids were introduced into yeast strain PJ69–4A by the lithium acetate method (23). After selection of transformants on SC minus leucine minus tryptophan agar medium, three colonies were streaked on SC minus leucine minus tryptophan minus histidine agar medium supplemented with 1.5 mM 3-amino-1,2,4-aminotriazole to score growth as an indicator of protein–protein interaction. To test the specificity of CIPK23–AKT1 interaction, nine other CIPK full-length cDNAs were cloned into DNA-binding domain vector (pGBT9.BS) and included in the yeast two-hybrid interaction assays with AKT1. The following primers (with restriction sites underlined) were used for the cloning process of CIPKs. CIPK1F, 5′-AATTGAATTCAATGGTGAGAAGGCAAGAGGA-3′; CIPK1R, 5′-AATTGTCGACCTAAGTTACTATCTCTTGCT-3; CIPK2F, 5′-TATGGATCCGATGGAGAACAAACCAAGTG; CIPK2R, 5′-CTAGTCGACCTATGATGGTTCTTGCTCTC-3′; CIPK3F, 5′-AGTGGATCCAATGAATCGGAGACAGCAAG-3′; CIPK3R, 5′-AATGTCGACTCATACATCACTTTGCTGTT-3′; CIPK4F, 5′-AATTGGATCCAATGGAATCTCCATATCCAAAATC-3′; CIPK4R, 5-AATTGAATTCTCAATTGTGCCATGAGAGCACAA-3′; CIPK5F, 5′-GAGGGATCCGATGGGAAGGTTATTAGGCA-3′; CIPK5R, 5′-AATGTCGACTCAACAATCCTCGGAAGAAG-3′; CIPK20F, 5′-AAGCGGGATCCAATGGATAAAAACGGC-3′; CIPK20R, 5′-CTCATCGTCGACACGGAGATCTAAAGAAAAAC-3′; CIPK21F, 5-AATTGAATTCAATGGGTTTGTTTGGAACGAAG-3′; CIPK21R, 5′-AATTGTCGACTTAGCTTACTTCCGCGGTAA-3′; CIPK22F, 5′-GTTTGGAATTCAATGGCCGAAGACTCTAA-3′; CIPK22R, 5′-TTAGTGTCGACACCGTTTTACGGTTTGTC-3′; CIPK24F, 5′-AATTGAATTCAATGACAAAGAAAATGAGAAG-3′; and CIPK24R, 5′-AATTGTCGACTCAAAACGTGATTGTTCTGA-3′.
Production of Recombinant Proteins and Protein Kinase Activity Assay.
To produce the CIPK23-GST protein in Xenopus oocytes, the CIPK23 cDNA was cloned into pGEX4T-3 vector and then the CIPK23-GST cassette was cloned into pGEMHE vector for expression in Xenopus oocytes. The complementary RNA (cRNA) production and injection procedure was described previously (26). After injection, oocytes were incubated for 2 days and harvested to purify the GST-CIPK23 protein as follows. Oocytes were washed in 1× TBS (50 mM Tris·HCl, pH 7.6/0.15 M NaCl) and lysed by pipetting up and down in extraction buffer (1× TBS/protease inhibitor mixture/0.1% Triton X-100/50 mM 2-glycerophosphate/1 mM sodium orthovanadate/1 mM sodium fluoride). The cytosolic fraction was separated by centrifugation at 13,800 × g for 10 min. Lysates were incubated with glutathione-Sepharose beads for 2 h at 4°C with moderate mixing. The beads were washed four times with extraction buffer and eluted by 10 mM glutathione in 1× TBS. The protein purity and quantity were determined by SDS/PAGE and silver stain.
The GST-AKT1 fusion protein was produced in E. coli after the cytoplasmic C-terminal domain of AKT1 was cloned into pGEX4T-3 vector. The primers used for cloning were AKT1F, 5′-GACTAGGAATTCTAGAGATACAATCCAAGC-3′; AKT1R, 5′-GTTTCTGTCGACGTTAAGAATCAGTTGCAAAG-3′ with restriction sites underlined. Phosphorylation assays were performed as follows. The kinase buffer includes 20 mM Tris·HCl (pH 8.0), 5 mM MgCl2, 1 mM CaCl2, and 1 mM DTT. Kinase reactions were performed in 30 μl of kinase buffer supplemented with 15 μCi (1 μCi = 37 kBq) of [γ-32P]ATP for 20 min at 30°C. Reactions were stopped by addition of 8 μl of 6× Laemmli buffer, and half of the reaction mixture was separated by SDS/PAGE. The gel was dried and 32P was detected by autoradiography using a Typhoon 8600 imager (Molecular Dynamics, Piscataway, NJ).
Protein Expression in Xenopus Oocytes.
To express proteins in Xenopus oocytes for the electrophysiological procedures, CBL1, CBL9, CIPK23, and AKT1 genes were cloned into the pGEMHE oocyte expression vector (26). The cDNAs were amplified by PCR using the following gene-specific primers: CBL1F, 5′-AATTCCCGGGATGGGCTGCTTCCACTCAAAGGCAGC-3′; CBL1R, 5′-AATTGAATTCTCATGTGGCAATCTCATCGACC-3′; CBL9F, 5′-GTCATGAATTCATGGGTTGTTTCCATTCCACG-3′; CBL9R, 5′-CTTGTCTTCTAGACGTCGCAATCTCGTCCA-3′; AKT1F, 5′-GCCAAGATCTAAAAACGTCGGTGATGAGAG-3′; AKT1R, 5′-GGCAGAGAATTCCTATGGTTGTTAAGAATC-3′; CIPK23F, 5′-TATAGAGCCCGGGATGGCTTCTCGAACAAC-3′; CIPK23R, 5′-GTAAACTCTAGATGTCGACTGTTTTGCAATTG-3′; For making AKT1-GFP fusion, the GFP tag was amplified by PCR and inserted in-frame to the C terminus of the AKT1 into BamHI–XbaI sites of pGEMHE-AKT1. All of the PCR-generated constructs were verified by sequencing to avoid mutated clones.
To make the dead kinase CIPK23 mutant, the ATP-binding Lys at the 60th amino acid position was changed into Asn by using PCR site-directed mutagenesis. The CIPK23-GST clone in the pGEMHE vector was used as template and amplified by using the following primer pairs (underlined nucleotides encode the mutated residue): F5′-TGATAATGTCGCTATTAACGTTATTGATAAAGAGAAAG-3′; R5′-CTTTCTCTTTATCAATAACGTTAATAGCGACATTATCA-3′.
Electrophysiological Procedures.
Oocyte expression and voltage-clamp recording were performed as described in ref. 26. The capped cRNA was transcribed with T7 RNA polymerase by using the mMESSAGE mMACHINE T7 RNA transcription Kit (Ambion, Austin, TX). The cRNA quality was checked by agarose gel electrophoresis. The concentration was determined by A260/A280 and adjusted to final concentration of 0.5 μg/μl. Freshly isolated Xenopus oocytes were injected with 23 nl of cRNA and used for voltage-clamp experiments 2–3 days after injection. Electrophysiological experiments were performed by using a two-electrode voltage-clamp amplifier as described previously with some modifications. Unless otherwise indicated, the solutions used in two-electrode voltage clamp were as follows. The oocytes were bathed with a solution containing 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, and 10 mM Hepes (pH 7.3) and perfused with the same solution. The exact procedure described in Liu and Luan (26) was followed in giant-patch recordings except that the bath solution contained 1 mM potassium gluconate, 183 mM mannitol, 10 mM Hepes (pH 7.3), 2 mM MgATP, with or without 2 mM calcium gluconate. The pipette solution consisted of 140 mM potassium gluconate, 2 mM MgCl2, and 10 mM Hepes (pH 7.3).
For the measurement of inward currents in Arabidopsis root cells, both protoplast isolation and electrophysiological procedure were performed according to Ivashikina et al. (29). Patch-clamp recordings were conducted in the whole-cell mode and currents recorded from the wild type (Col-0), cipk23 mutant, and cbl1cbl9 double mutant were compared. The voltage protocols used for two-electrode, giant-patch, and patch-clamp recordings are described in figure legends.
Acknowledgments
We thank Dr. Bob B. Buchanan for critical reading of this manuscript and Drs. Joerg Kudla and Weihua Wu for helpful discussions on this work. B.-G.K. was partially supported by Korea Research Foundation Grant M01-2003-000-20328-0. This work was supported by grants from the National Science Foundation and the U.S. Department of Agriculture (to S.L.).
Glossary
Abbreviations
- CBL
calcineurin B-like
- CIPK
CBL-interacting protein kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Leigh R. A., Jones R. G. A. New Phytol. 1984;97:1–13. [Google Scholar]
- 2.Kochian L. V. In: Biochemistry and Molecular Biology of Plants, Buchanan B., Gruissem W., Jones R., editors. Rockville, MD: Am. Soc. Plant Biologists; 2000. pp. 1204–1249. [Google Scholar]
- 3.Maser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., Sze H., Talke I. N., Amtmann A., Maathuis F. J., Sanders D., et al. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Very A. A., Sentenac H. Annu. Rev. Plant Biol. 2003;54:575–603. doi: 10.1146/annurev.arplant.54.031902.134831. [DOI] [PubMed] [Google Scholar]
- 5.Cherel I. J. Exp. Bot. 2004;55:337–351. doi: 10.1093/jxb/erh028. [DOI] [PubMed] [Google Scholar]
- 6.Ashley M. K., Grant M., Grabov A. J. Exp. Bot. 2006;57:425–436. doi: 10.1093/jxb/erj034. [DOI] [PubMed] [Google Scholar]
- 7.Armengaud P., Breitling R., Amtmann A. Plant Physiol. 2004;136:2556–2576. doi: 10.1104/pp.104.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gierth M., Maser P., Schroeder J. I. Plant Physiol. 2005;137:1105–1114. doi: 10.1104/pp.104.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pilot G., Gaymard F., Mouline K., Cherel I., Sentenac H. Plant Mol. Biol. 2003;51:773–787. doi: 10.1023/a:1022597102282. [DOI] [PubMed] [Google Scholar]
- 10.Maathuis F. J. M., Sanders D. Planta. 1995;197:456–464. doi: 10.1007/BF00196667. [DOI] [PubMed] [Google Scholar]
- 11.Shin R., Schachtman D. P. Proc. Natl. Acad. Sci. USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foreman J., Demidchik V., Bothwell J. H., Mylona P., Miedema H., Torres M. A., Linstead P., Costa S., Brownlee C., Jones J. D., et al. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- 13.Pei Z. M., Murata Y., Benning G., Thomine S., Klusener B., Allen G. J., Grill E., Schroeder J. I. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 14.Kudla J., Xu Q., Harter K., Gruissem W., Luan S. Proc. Natl. Acad. Sci. USA. 1999;96:4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. Plant Cell. 2002;14:S389–S400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J., Kim K. N., Ritz O., Albrecht V., Gupta R., Harter K., Luan S., Kudla J. Plant Cell. 1999;11:2393–2405. doi: 10.1105/tpc.11.12.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K. N., Cheong Y. H., Gupta R., Luan S. Plant Physiol. 2000;124:1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht V., Ritz O., Linder S., Harter K., Kudla J. EMBO J. 2001;20:1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batistic O., Kudla J. Planta. 2004;219:915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- 20.Cheong Y. H., Kim K. N., Pandey G. K., Gupta R., Grant J. J., Luan S. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey G. K., Cheong Y. H., Kim K. N., Grant J. J., Li L., Hung W., D’Angelo C., Weinl S., Kudla J., Luan S. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K. N., Cheong Y. H., Grant J. J., Pandey G. K., Luan S. Plant Cell. 2003;15:411–423. doi: 10.1105/tpc.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H., Fukuda Y., Murata K., Kimura A. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch R. E., Lewis B. D., Spalding E. P., Sussman M. R. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 25.Gaymard F., Cerutti M., Horeau C., Lemaillet G., Urbach S., Ravallec M., Devauchelle G., Sentenac H., Thibaud J. B. (1996) J. Biol. Chem. 1996;271:22863–22870. doi: 10.1074/jbc.271.37.22863. [DOI] [PubMed] [Google Scholar]
- 26.Liu K., Luan S. Plant Cell. 2001;13:1453–1465. doi: 10.1105/tpc.13.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K., Li L., Luan S. Plant J. 2005;42:433–443. doi: 10.1111/j.1365-313X.2005.02384.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu K., Li L., Luan S. Plant J. 2006;46:260–268. doi: 10.1111/j.1365-313X.2006.02689.x. [DOI] [PubMed] [Google Scholar]
- 29.Ivashikina N., Becker D., Ache P., Meyerhoff O., Felle H. H., Hedrich R. FEBS Lett. 2001;508:463–469. doi: 10.1016/s0014-5793(01)03114-3. [DOI] [PubMed] [Google Scholar]
- 30.Reintanz B., Szyroki A., Ivashikina N., Ache P., Godde M., Becker D., Palme K., Hedrich R. Proc. Natl. Acad. Sci. USA. 2002;99:4079–4084. doi: 10.1073/pnas.052677799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder J. I., Allen G. J., Hugouvieux V., Kwak J. M., Waner D. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 32.Heermann R., Fohrmann A., Altendorf K., Jung K. Mol. Microbiol. 2003;47:834–848. doi: 10.1046/j.1365-2958.2003.03348.x. [DOI] [PubMed] [Google Scholar]
- 33.Lagarde D., Basset M., Lepetit M., Conejero G., Gaymard F., Astruc S., Grignon C. Plant J. 1996;9:195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]