Abstract
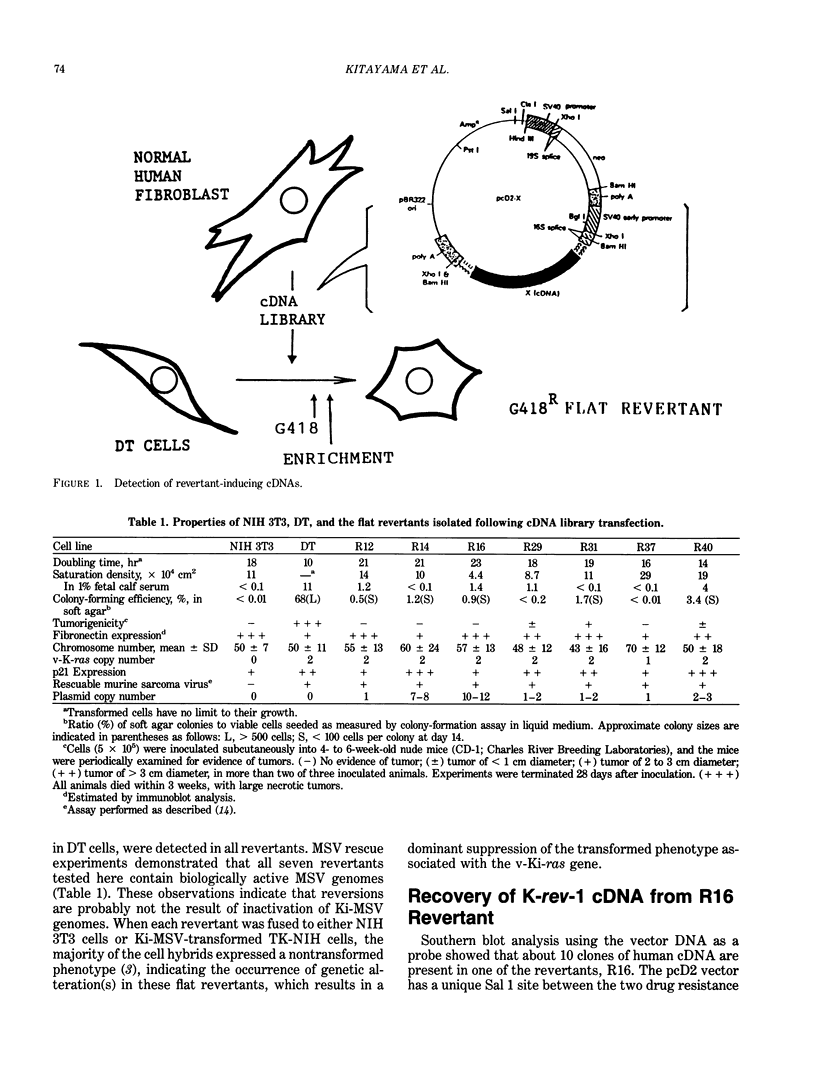
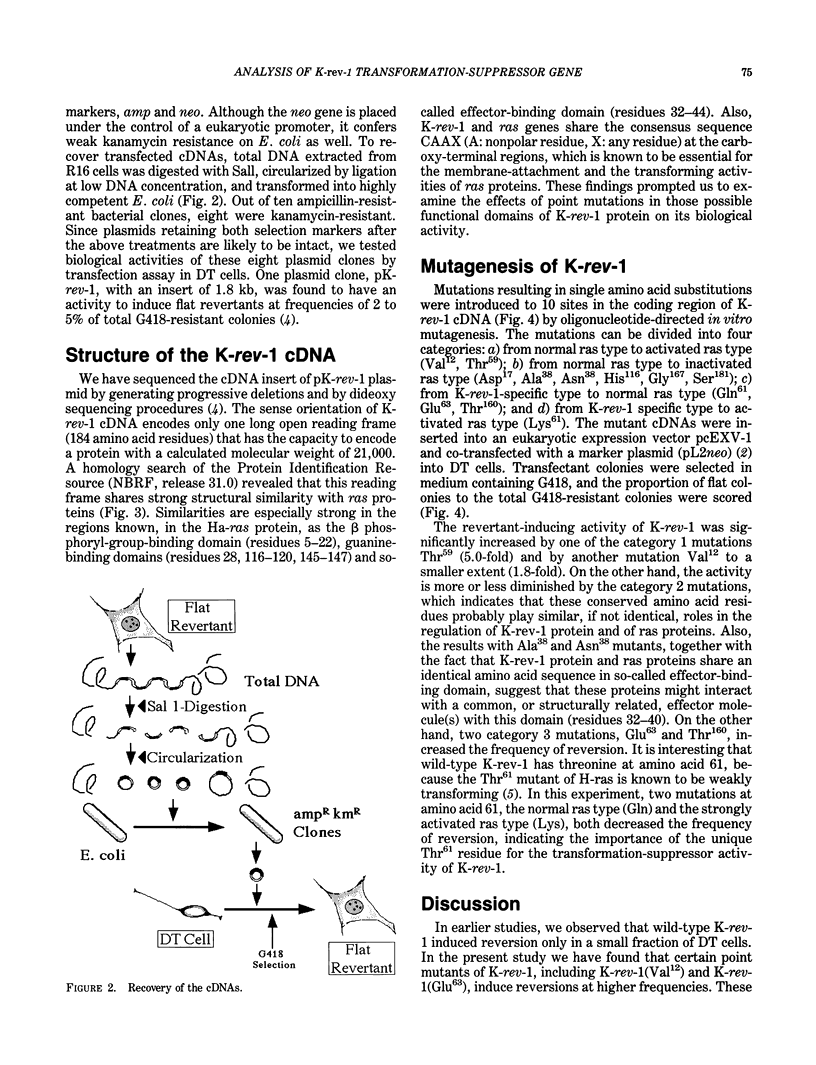
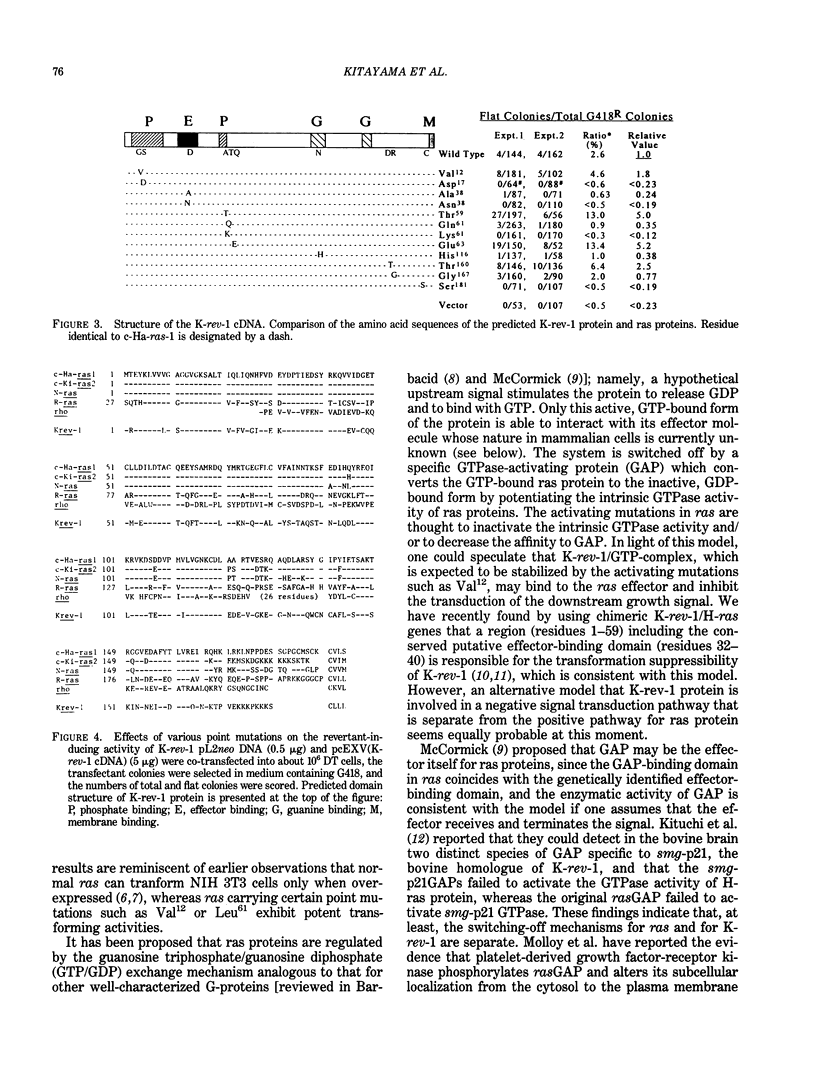
Flat revertants with reduced malignancy in vivo can be isolated from Kirsten sarcoma virus-transformed NIH 3T3 cells (DT line) following transfection with a normal human fibroblast cDNA expression library. We have recovered from one such revertant a 1.8-kb cDNA clone, K-rev-1, that exhibits an activity of inducing flat revertants at certain frequencies (2-5% of total transfectants) when transfected into DT cells. The K-rev-1 cDNA has the capacity to encode a protein with a calculated molecular weight of 21,000, having strong structural similarity to ras proteins (approximately 50% homology), especially in their guanosine triphosphate/guanosine diphosphate-binding, effector-binding, and membrane-attachment domains. Toward understanding the mechanism of action of K-rev-1 protein, we constructed a series of point mutants of K-rev-1 cDNA and tested their biological activities. Substitutions of the amino acid residues in the putative guanine nucleotide-binding regions (Asp17 and Asn116), in the putative effector-binding domain (residue 38), at the putative acylation site (Cys181), and at the unique Thr61 all decreased the transformation-suppressor activity. On the other hand, substitutions including Gly12 to Val12, Ala59 to Thr59, and Gln63 to Glu63 were found to significantly increase the transformation-suppressor activity of K-rev-1. These findings are consistent with the idea that K-rev-1 protein is regulated like many other G-proteins by guanine triphosphate/guanine diphosphate-exchange mechanism probably in response to certain negative growth-regulatory signals.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Chang E. H., Furth M. E., Scolnick E. M., Lowy D. R. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982 Jun 10;297(5866):479–483. doi: 10.1038/297479a0. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der C. J., Finkel T., Cooper G. M. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986 Jan 17;44(1):167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Kikuchi A., Sasaki T., Araki S., Hata Y., Takai Y. Purification and characterization from bovine brain cytosol of two GTPase-activating proteins specific for smg p21, a GTP-binding protein having the same effector domain as c-ras p21s. J Biol Chem. 1989 Jun 5;264(16):9133–9136. [PubMed] [Google Scholar]
- Kitayama H., Matsuzaki T., Ikawa Y., Noda M. A domain responsible for the transformation suppressor activity in Krev-1 protein. Jpn J Cancer Res. 1990 May;81(5):445–448. doi: 10.1111/j.1349-7006.1990.tb02589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy C. J., Bottaro D. P., Fleming T. P., Marshall M. S., Gibbs J. B., Aaronson S. A. PDGF induction of tyrosine phosphorylation of GTPase activating protein. Nature. 1989 Dec 7;342(6250):711–714. doi: 10.1038/342711a0. [DOI] [PubMed] [Google Scholar]
- Noda M., Kitayama H., Matsuzaki T., Sugimoto Y., Okayama H., Bassin R. H., Ikawa Y. Detection of genes with a potential for suppressing the transformed phenotype associated with activated ras genes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):162–166. doi: 10.1073/pnas.86.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Selinger Z., Scolnick E. M., Bassin R. H. Flat revertants isolated from Kirsten sarcoma virus-transformed cells are resistant to the action of specific oncogenes. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5602–5606. doi: 10.1073/pnas.80.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulciani S., Santos E., Long L. K., Sorrentino V., Barbacid M. ras gene Amplification and malignant transformation. Mol Cell Biol. 1985 Oct;5(10):2836–2841. doi: 10.1128/mcb.5.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Noda M., Vass W. C., Papageorge A. G., Lowy D. R. Identification of small clusters of divergent amino acids that mediate the opposing effects of ras and Krev-1. Science. 1990 Jul 13;249(4965):162–165. doi: 10.1126/science.2115210. [DOI] [PubMed] [Google Scholar]


