Abstract
Background
The conventional solution-phase Chemical Cleavage of Mismatch (CCM) method is time-consuming, as the protocol requires purification of DNA after each reaction step. This paper describes a new version of CCM to overcome this problem by immobilizing DNA on silica solid supports.
Results
DNA test samples were loaded on to silica beads and the DNA bound to the solid supports underwent chemical modification reactions with KMnO4 (potassium permanganate) and hydroxylamine in 3M TEAC (tetraethylammonium chloride) solution. The resulting modified DNA was then simultaneously cleaved by piperidine and removed from the solid supports to afford DNA fragments without the requirement of DNA purification between reaction steps.
Conclusions
The new solid-phase version of CCM is a fast, cost-effective and sensitive method for detection of mismatches and mutations.
Keywords: solid phase, chemical cleavage reaction, mutation detection, chemical modification of DNA
Background
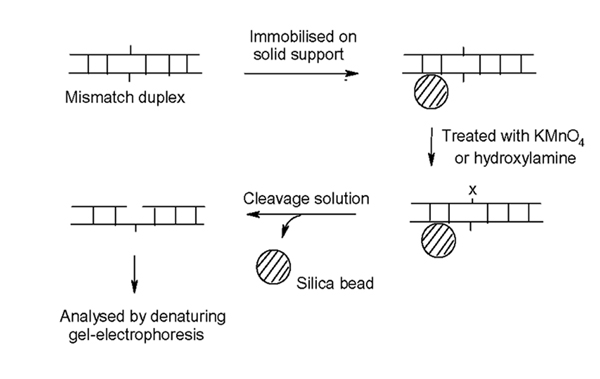
The solution phase Chemical Cleavage of Mismatch (CCM) is one of few methods capable of detecting nearly all single base mismatches [1]. This method was developed in 1988 by Cotton et al. [2] and has been widely used in research and diagnosis of many inherited diseases. The technology is based on selective reactions of mismatched thymine and cytosine with KMnO4 and hydroxylamine respectively [2-5]. The modified mismatched bases are subjected to piperidine cleavage reactions and the resulting fragments are separated and identified by gel-electrophoresis. The process is time-consuming, as the method requires purification of DNA by using the standard precipitation technique after each reaction step [2,4,5]. In our earlier work [6], attempts have been successfully made to attach biotinylated DNA samples onto streptavidin-coated magnetic beads for solid-phase chemical modification and cleavage reactions. The desire to simplify this approach further by reducing the use of the cumbersome biotin dependent assay led us to the development of a new solid-phase chemical cleavage of mismatched DNA (Figure 5). The method involves attachment of DNA on to the commercially available silica solid supports. The solid-bound DNA remained intact throughout the chemical reactions and the final product was removed from solid support by treatment with the piperidine cleavage solution at 90°C as described in our experimental section. This new version of the CCM method is simple, cost-effective and highly accurate by improving the quality of the cleavage fragments on sequencing gel traces.
Figure 5.
Solid-phase chemical cleavage of mismatch. X indicates the base modified by KMnO4 or hydroxylamine.
Methods
Chemicals and solvents were purchased from Aldrich Chemical Company (Castle Hill, Australia). The fluorescent primers (6-FAM and HEX for the 5' and 3' primers respectively) were purchased from Geneset corp, CA, USA. Test sample DNA (547-bp μ-globin mouse promoter) [7] was amplified as per the previously described conditions. The resulting PCR products (DNA) were purified by Strategene kit (Integrated Science, Inc. CA, USA). The silica solid support (Ultra-bind bead) and Ultrawash solution were purchased from MO BIO Laboratories Inc. (CA, USA).
DNA concentration: DNA concentrations were determined by the standard spectroscopic method (measurement at 260 nm) [8] using the Cary 300 UV-Visible spectrophotometer (Varian Inc., Australia).
Formation of heteroduplex DNA containing T.C mismatch
Equal amounts of the labeled wild-type (10 μg of 547 bp DNA containing C.G pair at the position 82 from 5' end) and mutant DNA (10 μg of 547 bp DNA containing A.T pair at the position 82 from 5' end) were mixed in TE buffer (100 μl of 1 M Tris-HCl, pH 8.0, 20 μl of 0.5 M ethylenediamine-tetraacetic acid and 9.88 ml distilled H2O). The mixture was heated to 95°C for 7 min, cooled down to 65°C and maintained at this temperature for 60 min and finally brought back to room temperature.
Formation of heteroduplex DNA containing C.C mismatch
The labeled wild-type DNA (10 μg of 540 bp DNA containing C.G pair at the position 83 from 5' end) and mutant DNA (10 μg of 540 bp DNA containing G.C pair at the position 83 from 5' end) were used for heteroduplex formation under the above conditions.
Solid-phase reaction of DNA with KMnO4
DNA sample (1 μl of the heteroduplex DNA contains ca. 0.1 μg DNA) was incubated with 2.5 μl of Ultra-bind bead suspension in an Eppendorf tube. The reaction mixture was gentlely mixed on shaker at 25°C for 2 h. The Eppendorf tube was centrifuged (2000 rpm) and supernatant was removed (by Pasteur pipette). The DNA bound solid supports were washed with Ultrawash solution (2 × 200 μl) and then treated with 30 μl of 1 mM KMnO4 in 3 M TEAC solution. The reaction mixture was incubated at 25°C for 10 min and the resulting DNA-bound beads were separated by centrifuge, washed with Ultrawash solution (2 × 200 μl). The beads were air-dried for 15 min and used for the next reaction step.
Solid-phase reaction of DNA with hydroxylamine
DNA-bound beads were prepared as described above (1 μl of homoduplex DNA for control or heteroduplex DNA for mismatch detection containing 0.1–0.2 μg DNA was incubated with 2.5 μl of Ultra-bind bead suspension in an Eppendorf tube). The DNA-bound beads were treated with 30 μl of 4.2 M hydroxylamine in 3 M TEAC solution. The reaction mixture was incubated at 37°C for 40 min and the resulting DNA bound beads were separated by centrifuge, washed with Ultra-wash solution (2 × 200 μl). The beads were air-dried for 15 min and used for the next step.
In the control experiments, the homoduplex DNA samples (wild type or mutant) were treated under identical reaction conditions.
Cleavage by piperidine
The DNA-bound beads obtained from the reactions with KMnO4 or hydroxylamine were treated with 10 μl of cleavage-dye solution at 90°C for 30 min. The cleavage dye solution was made from 20 μl of piperidine, 66.6 μl of formamide and 13.4 μl of dye solution (5 % w/v blue dextran in H2O). The solid beads were separated by centrifuge and the supernatant was loaded on to a denaturing polyacrilamide gel (acrylamide: bisacrylamide, 19:1). Electrophoresis was performed on ABI 377 DNA sequencer using TBE buffer (16.2 g Tris-base, 8.1 g boric acid and 1.12 g EDTA in 1500 ml distilled H2O, pH 8.0) at 3000 V for 3 h.
Results and Discussion
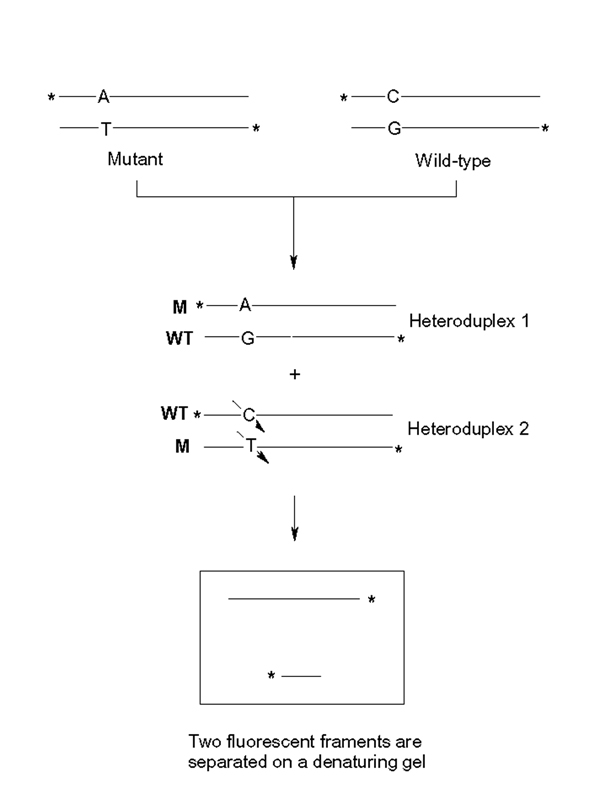
The chemical cleavage of mismatch has been widely used as a diagnostic tool for detection of mutations in many inherited diseases [1,9]. The method was based on the formation of heteroduplexes DNA containing a single base mismatch by mixing equal amounts of two homoduplexes (wild-type and mutant) DNA. At the mismatched site, nucleotide bases are extra-helical and susceptible to chemical modification reactions [10]. Therefore, KMnO4 and hydroxylamine have been employed in the CCM method to modify the mismatched thymine and cytosine respectively. The resulting modified DNA is then treated with piperidine before being loaded on to a sequencing polyacrylamide gel for fragment analysis (Figure 1). This process is time-consuming as the reactions are carried out in solution phases and the products require exhaustive purification procedures after each reaction step.
Figure 1.
Chemical Cleavage of Mismatch. * DNA strand was labeled with FAM or HEX at 5' and 3' ends respectively. Heteroduplex was formed by mixing equal amounts of wild-type and mutant DNA. Mismatched T.C on the heteroduplex was modified by KMnO4 and hydroylamine and then cleaved by piperidine. A denaturing gel displays two cleavage fragments derived from both strands. Note: When only one of two reagents (either KMnO4 or hydroxylamine) was applied on T.C mismatch, only one labeled cleavage fragment was detected by gel-electrophoresis – See Fig. 2 &3).
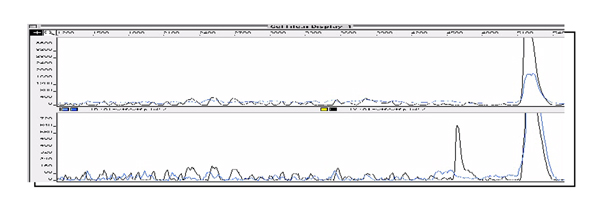
To overcome this problem, our initial target was focussed on development of the solid-phase CCM method using silica beads as a solid. Attachment of DNA on silica in high salt solution has been well established and widely used as an effective purification technique [11]. By this way, the commercially available silica beads have been adapted for improving the current CCM protocol. Figure 5 describes the new solid-phase version of CCM, in which DNA fragments are immobilised on silica (Ultra-bind) beads by the standard method. The DNA-attached beads are then treated with KMnO4 or hydroxylamine as usual. After reactions, the beads are simply washed with the Ultra-wash solution and used for the next step without further purification. This solid-phase CCM protocol was successfully applied for detection of T.C & C.C mismatches on the previously published 547 bp DNA sequences [6]. In one typical example, the heteroduplex 2 containing T.C mismatch was formed (the location of a T.C mismatch was close to one end, ie. 82 bp from 5' end, Fig. 1) and allowed to react with KMnO4 and hydroxylamine on solid supports in two separate reactions. After cleavage with piperidine, the reactions gave rise to two fluorescent-labeled DNA fragments of 82 bp and 466 bp as identified by denaturing gel-electrophoresis. Figure 2 and 3 show the strong cleavage peaks derived from the KMnO4/piperidine and the hydroxylamine/piperidine assays respectively while the control reactions show no reactions of homoduplexes (wild type or mutant) with both KMnO4 and hydroxylamine.
Figure 2.
Detection of a T.C mismatch in a 547 bp DNA fragment by using KMnO4 / piperidine assay. The control trace (homoduplex, top) shows no cleavage peak and the mismatched DNA trace (heteroduplex, bottom) displays a strong cleavage peak of the mismatched T base in the 3'HEX sequence. Note consistent background in both traces which is essentially a chemical sequencing trace of T bases allowing confidence that reaction has occurred and a position reference.
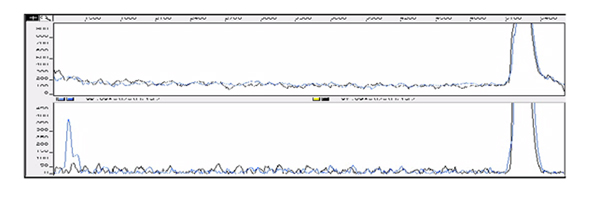
Figure 3.
Detection of T.C mismatch of 547 bp DNA fragment by using hydroxylamine / piperidine assay. The control trace (homoduplex, top) shows no cleavage peak and the mismatched DNA trace (heteroduplex, bottom) displays a strong cleavage peak of 5'FAM sequence at the mismatched C base. Note this is the second chance of detecting the mutation, the first being in Fig 2.
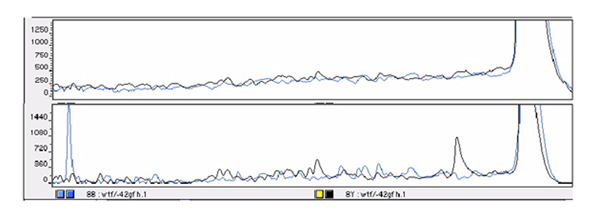
In another example, a DNA fragment containing C.C mismatch was treated with hydroxylamine/piperidine under similar solid-phase reaction conditions. The results showed two cleavage peaks on the same gel which correspond to two fragments (83 bp and 465 bp), each from 5'- and 3'-labeled DNA fragments derived from the 547 bp DNA sample (Figure 4).
Figure 4.
Detection of C.C mismatch of 547 bp DNA fragment by using hydroxylamine / piperidine assay. The control trace (homoduplex, top) shows no cleavage peak and the mismatched DNA trace (heteroduplex, bottom) displays two strong cleavage peaks of 5'FAM sequence and 3'HEX sequence at the mismatched C bases.
The identities of all cleavage fragments were confirmed with the previously published data obtained from the standard CCM and sequencing techniques [6].
Conclusions
The new solid-phase chemical cleavage of mismatched DNA has been successfully carried out on silica solid supports. The following results have been obtained: (i) The commercially available silica beads which was originally developed for DNA purification of small DNA fragments is now suitable for solid-phase chemical reactions. (ii) Typical reactions with KMnO4 and hydroxylamine [12,13] have been successfully demonstrated on DNA bound silica support. (iii) DNA bound silica support remains intact during reactions and purification steps. Cleavage of DNA product from silica can be done as per the instructions of the commercial silica product or our described conditions. As the solid-phase chemical cleavage reactions are very convenient in terms of manipulation and purity of the products, the silica solid support is highly recommended for any chemical modification reactions on DNA.
Scheme 1: Solid-phase chemical cleavage of mismatch. X indicates the base modified by KMnO4 or hydroxylamine. (See Figure 5 for schematic)
Contributor Information
Chinh T Bui, Email: chinhbui@mail.medstv.unimelb.edu.au.
Andreana Lambrinakos, Email: andyl@ariel.ucs.unimelb.edu.au.
Jeffrey J Babon, Email: babon@hotmail.com.
Richard GH Cotton, Email: cotton@ariel.ucs.unimelb.edu.au.
References
- Ellis TP, Humphrey KE, Smith MJ, Cotton RGH. Chemical cleavage of mismatch: a new look at an established method/recent developments. Hum Mutat. 1998;11:345–353. doi: 10.1002/(SICI)1098-1004(1998)11:5<345::AID-HUMU1>3.3.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Cotton RGH, Rodrigues NR, Campbell RD. Reactivity of cytosine and thymine in single base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci USA. 1988;85:4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Karayiorgou M, Aburatani H, Kafatos FC. Detection of single base mismatches of thymine and cytosine residues by potassium permanganate and hydroxylamine in the present of tetralkylammonium salts. Nucleic Acids Res. 1990;18:6807–6814. doi: 10.1093/nar/18.23.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton RGH, Grompe M. In: Chemical cleavage of heteroduplex DNA to identify mutations in Current Protocols in Human Genetic. Dracopoli NC, Haines JL, Korf BR, Morton CC, Seidman CE, Seidman JG, Smith DR, editor. John Wiley & Sons, Inc. Unit 7.6; 1994. [DOI] [PubMed] [Google Scholar]
- Cotton RGH. In: Detection of mutations in DNA and RNA by chemical cleavage in The Nucleic Acid Protocols handbook. Rapley R, editor. Humana Press; 1999. pp. 685–693. [Google Scholar]
- Lambrinakos A, Humphrey KE, Babon JJ, Ellis TP, Cotton RGH. Reactivity of potassium permanganate and tetraethylammonium chloride with mismatched bases and a simple mutation detection protocol. Nucleic Acids Res. 1999;27:1866–1874. doi: 10.1093/nar/27.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youil R, Kemper B, Cotton RGH. Detection of 81 of 81 known mouse b-globin promoter mutations with T4 endonuclease VII – the EMC method. Genomics. 1996;32:431–435. doi: 10.1006/geno.1996.0138. [DOI] [PubMed] [Google Scholar]
- Manchester KL. Value of A260/A280 ratios for measurement of purity of nucleic acids. Biotechniques. 1995;19:208–209. [PubMed] [Google Scholar]
- Cotton RGH. Slowly, but surely towards better scanning for mutations. Trends in genetics. 1997;13:43–46. doi: 10.1016/S0168-9525(97)01011-1. [DOI] [PubMed] [Google Scholar]
- Bui CT, Rees K, Lambrinakos A, Bedir A, Cotton RGH. Site-selective reactions of imperfectly matched DNA with small chemical molecules: applications in mutation detection. Bioorganic Chemistry. 2002;30:216–232. doi: 10.1016/S0045-2068(02)00019-6. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Gillesoie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman F, Fuselier CO, Karchefski EM. Permanganate ion oxidation of thymine: spectrophotometric detection of a stable organomanganese intermediate. Tet Letters. 1975;25:2133–2136. doi: 10.1016/S0040-4039(00)75315-9. [DOI] [Google Scholar]
- Stolarski R, Kierdaszuk B, Hagberg CE, Shugar D. Mechanism of hydroxylamine mutagenesis: tautomeric shifts and proton exchange between the promutagen N6-methoxyadenosine and cytidine. Biochemistry. 1987;26:4332–4337. doi: 10.1021/bi00388a022. [DOI] [PubMed] [Google Scholar]