Abstract
Kit receptor-activating mutations are critical in the pathogenesis of gastrointestinal stromal tumors (GIST). We investigated mechanisms of oncogenic Kit signaling and the consequences of therapeutic intervention in a mouse model of human GIST. Treatment of GIST mice with imatinib decreased cell proliferation and increased apoptosis in the tumor. Analysis of tumor tissue from imatinib-treated mice showed diminished phosphatidylinositol 3-kinase (PI3-kinase) and mammalian target of rapamycin (mTOR) signaling suggesting that oncogenic Kit signaling critically contributes to the translational response in GIST. Treatment with RAD001 (everolimus), an mTOR inhibitor, diminished the translational response and cell proliferation in tumor lesions, pointing to mTOR inhibition as a therapeutic approach for imatinib-resistant GIST. Analysis of RNA expression profiles in GIST lesions with and without imatinib treatment showed changes in expression of IFN-inducible genes and cell cycle regulators. These results convincingly show that KitV558Δ/+ mice represent a unique faithful mouse model of human familial GIST, and they demonstrate the utility of these mice for preclinical investigations and to elucidate oncogenic signaling mechanisms by using genetic approaches and targeted pharmacological intervention.
Keywords: imatinib, Kit receptor tyrosine kinase, signal transduction
The Kit receptor tyrosine kinase has a critical role in the normal development and function of the interstitial cells of Cajal (ICC), as well as in hematopoietic cell populations, gametogenesis, and melanogenesis during embryonic development and in the postnatal organism (1–3). The finding of Kit receptor-activating mutations in human tumors, including gastrointestinal stromal tumor (GIST), seminomas, and mastocytosis, as well as some acute myelogenous leukemias, suggested a role for Kit in oncogenesis. GIST is the most common mesenchymal tumor of the gastrointestinal tract. GISTs express Kit and are thought to derive from a Kit+ or Kitlow ICC progenitor or ICC. The vast majority of GISTs contain Kit receptor-activating mutations (4, 5). Kit-activating mutations in GIST are found predominantly in the juxtamembrane domain of the Kit receptor, but mutations in the extracellular and kinase domains of Kit have been described as well (6, 7). Imatinib mesylate (Gleevec, STI571), an inhibitor of the Kit, PDGFR, and BCR-ABL tyrosine kinases, is used to treat patients with GIST and chronic myelogenous leukemia. In GIST, it elicits a partial response or stable disease in a majority of patients with metastatic or recurrent disease. The clinical response correlates with a decrease in tumor cellularity and myxoid degeneration of the tumor. Although the clinical response to imatinib is quite well described, the molecular response of Kit inhibition by imatinib in GIST is poorly understood. Imatinib is most effective in GISTs with Kit-activating mutations in the juxtamembrane domain, some kinase domain mutations, or extracellular domain mutations. But Kit mutations that destabilize the inactive form of the kinase are resistant to inhibition by imatinib. Unfortunately, long-term treatment with imatinib is associated with the development of drug resistance, and in some cases, resistance appears to derive from second site mutations in the Kit receptor (8, 9). Therefore, the development of new strategies for the treatment of GIST is highly relevant.
Several cases of human familial GIST syndrome with associated interstitial cells of Cajal hyperplasia, hyperpigmentation, and/or urticaria pigmentosa with germ-line Kit mutations have been reported (10, 11). Based on these findings, we produced a mouse carrying a Kit-activating mutation, KitV558Δ/+, in the germ line (12). The KitV558Δ/+ mutation is located in the juxtamembrane domain of Kit (exon 11), a negative regulatory region of the receptor where the majority of the somatic GIST mutations in patients occur. Heterozygous mutant KitV558Δ/+ mice develop symptoms of disease and eventually die from pathology in the gastrointestinal (GI) tract. Patchy hyperplasia of Kit-positive cells is observed within the myenteric plexus of the GI tract, and neoplastic lesions indistinguishable from human GIST are found with complete penetrance in the cecum of mutant mice (12). Thus the KitV558Δ/+ mice provide a unique opportunity to investigate the development of GIST and the consequences of therapeutic intervention.
The Kit receptor has a role in distinct cellular responses, including cell proliferation, survival, adhesion, chemotaxis, and secretory responses, as well as the desensitization of the activated receptor in various cell types in vitro and in vivo. The downstream signaling cascades that are known to be activated by Kit include the Ras/MAP kinase, Rac/Rho-JNK, phosphatidylinositol 3-kinase (PI3-kinase)/AKT/PDK1/FOXO, and src family kinase (SFK)/STAT signaling networks. Cell-type-specific responses depend in part on the cellular context and the presence of signaling components, and therefore signaling cascades that may be activated by Kit vary in different cell types. Consequently, disruption of Kit-specific signaling pathways by knock-in mutations produces cell-specific effects, e.g., disruption of Kit-induced PI3-kinase signaling was shown to impair male fertility, whereas melanogenesis and hematopoiesis were not affected (13), and disruption of SFK binding shows mainly age-dependent lymphopoietic defects (14). We have used imatinib to investigate oncogenic Kit signaling in mouse GIST in vivo. We show that treatment of GIST mice with imatinib abolished cell cycle progression concomitant with an increase in apoptosis in the tumor lesions. Analysis of gene expression profiles in placebo- and imatinib-treated mice revealed roles for cell cycle regulators and IFN-inducible genes in GIST. Biochemical analysis of tumor tissue from imatinib-treated mice showed diminished PI3-kinase and mammalian target of rapamycin (mTOR) signaling, implying a role for the translational response in oncogenic Kit signaling in GIST. To investigate the role of the translational response in GIST, mice were treated with the mTOR inhibitor RAD001 (everolimus).
Results
Effects of Imatinib Treatment in the GIST Mouse Model.
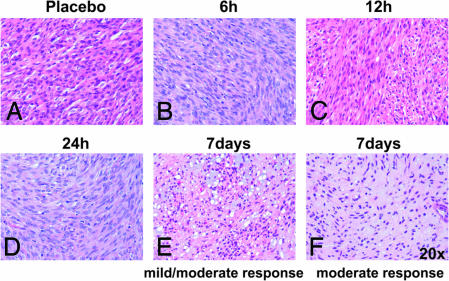
To determine whether GISTs in KitV558Δ/+ mice respond to imatinib treatment, heterozygous KitV558Δ/+ mice were treated with 45 mg/kg imatinib twice per day. After 7 days of treatment, a decrease in cellularity and an increase in myxoid stroma were observed microscopically in H&E-stained tumor sections. Whereas in 8 of 10 treated mice, the degree of response varied from a mild response with patchy acellular areas comprising 10–50% of the lesion, to a moderate response, where larger areas of tumor cells were replaced by myxoid stroma and necrosis, in two mice, the response was minimal, <10% (Fig. 1E and F; Table 1). However, no obvious changes in histology were observed for shorter time treatments, i.e., 6, 12, and 24 h (Fig. 1 A–D). These results showed that after 7 days of treatment, the murine GIST lesions responded to imatinib treatment, and that some variability was observed between mutant mice.
Fig. 1.
Histological response of GIST lesions in KitV558Δ/+ mice to imatinib treatment. H&E-stained tumor sections of placebo and imatinib-treated mice for the indicated time intervals (×20).
Table 1.
Individual response of GIST lesions in KitV558Δ/+ mice to 7-day imatinib treatment
| Animals | Imatinib treatment, 7 day | Histologic response | Ki67 staining | Cleaved Cas3 staining | P-S6 protein staining |
|---|---|---|---|---|---|
| Placebos (n = 10) | − | − | +++ | − | +++ |
| 1 | + | Minimal | − | 8.50 ± 1.29 | − |
| 2 | + | Moderate | − | 9.25 ± 2.21 | − |
| 3 | + | Mild | − | 15.75 ± 2.5 | − |
| 4 | + | Mild | − | 13.75 ± 1.70 | − |
| 5 | + | Mild | − | 4.20 ± 1.30 | − |
| 6 | + | Mild | − | 4.40 ± 2.50 | − |
| 7 | + | Mild to moderate | − | 23.4 ± 1.51 | − |
| 8 | + | Mild | − | 3.83 ± 1.47 | − |
| 9 | + | Minimal | − | 4.20 ± 3.70 | − |
| 10 | + | Mild | − | 17.4 ± 3.5 | − |
Histologic response based on microscopic findings of necrosis, increased stromal fibrosis, and myxoid changes, scored as: minimal or no (<10% response), mild (10–50% response), moderate (50–90% response), or very good (≥90% response). Cleaved caspase 3-positive cells were counted in 20 fields, and standard deviations are indicated (±). No correlation was found between histological response and number of cleaved caspase 3-stained cells. (−) and (+++) indicate no obvious staining or strong staining, respectively, for Ki67 (see Figs. 2 and 7) and P-S6 protein (see Fig. 11, which is published as supporting information on the PNAS web site).
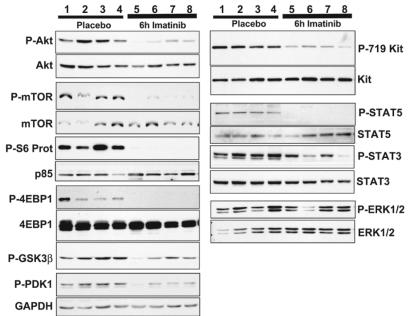
To confirm that imatinib inhibited Kit receptor activation and autophosphorylation in the murine GIST lesions, tumor protein extracts were prepared from mice treated with placebo or imatinib. To assess the range of individual variability in the molecular response to the drug after short treatment periods, tumor protein extracts were prepared from several mice and analyzed individually. We had previously shown that Kit is constitutively activated and autophosphorylated in tumor lesions of untreated mice using the antiphospho-Y719-Kit antibody (Fig. 3 and ref. 12). As anticipated, imatinib treatment significantly reduced Kit Y719 phosphorylation. The reduction in phosphorylation was observed after as early as 1 h of treatment and was maintained at 6 and 24 h of treatment (Fig. 3 and results not shown). Total Kit protein levels were not affected by the treatment. Similarly, immunohistochemical analysis of GISTs showed similar Kit staining patterns independent of treatment (Fig. 6 A and B, which is published as supporting information on the PNAS web site). These results indicated that imatinib treatment did not affect Kit receptor expression and/or turnover but only inhibited its kinase activity.
Fig. 3.
Imatinib down-regulates the translational response in GIST. Tumor protein lysates from four placebo (lanes 1–4) and four 6-h imatinib-treated mice (lanes 5–8) were fractionated by SDS/PAGE and subjected to Western blotting to assess protein activation using the following phospho-specific antibodies: P-Akt T308, P-mTOR S2448, P-S6 S235/236, P-4EBP1 S65, P-GSK3β S9, P-PDK1 S241, P-STAT5 Y694, P-STAT3 Y705, P-ERK Y202/204, Kit, and P-Y719-Kit. Protein extracts were prepared from tumors of several mice to assess individual variability in response to the drug.
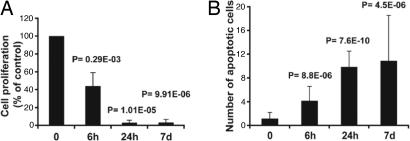
We then investigated the effect of imatinib treatment on GIST cell proliferation and apoptosis. GISTs from placebo-treated mice readily stained for Ki67, cell proliferation antigen, an indication that the tumors were viable and proliferating (Fig. 7A, which is published as supporting information on the PNAS web site); the mitotic index was quite low, ≈4%. Treated GISTs showed a reduction of Ki67-positive cells after a 6-h treatment time, and virtually no Ki67-positive staining was observed after 24 h or 7 days of imatinib treatment (Figs. 2A and 7 B–D), indicating an almost complete arrest of cell proliferation in tumor lesions as early as 24 h of treatment. We next determined whether there was an increase in apoptosis in the tumor by measuring cleaved caspase 3 levels by immunohistochemistry. GISTs from placebo-treated mice showed no obvious cleaved caspase 3 staining. However, positive staining was already seen after only 6 h of treatment. The number of caspase 3-positive staining cells increased with treatment time, and the most pronounced effect was observed after 7 days of treatment (Figs. 2B and Fig. 7 E–H). Taken together, these results show that GIST lesions in KitV558Δ/+ mice are sensitive to imatinib treatment.
Fig. 2.
Imatinib treatment decreases cell proliferation and increases apoptosis in tumor lesions of imatinib-treated KitV558Δ/+ mice. Quantification of proliferating (A) and apoptotic (B) cells (percent of control) in tumor sections of placebo and imatinib-treated KitV558Δ/+ mice. Groups were judged to differ significantly at P < 0.05 (see Materials and Methods).
Phosphorylation of Downstream Targets of Oncogenic Kit Signaling in GIST.
Because imatinib treatment abolishes Kit signaling in GIST lesions, a comparison of known signaling cascades that might be activated by Kit receptor signaling in tumor lysates before and after drug treatment should provide insights into the mechanisms of Kit-mediated responses in GIST. Our immunohistochemical analysis in imatinib-treated KitV558Δ/+ mice indicated that drug treatment induced both apoptosis and arrest of cell cycle progression in murine GISTs. To establish which signaling pathways are activated in GIST in vivo, we assessed the phosphorylation state of downstream substrates of the oncogenic Kit receptor. Kit is thought to mediate proliferation and survival by the PI3-kinase- and mitogen-activated protein kinase (ERK1 and -2) pathways and by signaling through STAT transcription factors (15). We prepared tumor lysates from placebo and 6-h-treated mice and evaluated protein activation by Western blotting with phospho-specific antibodies. Protein extracts from individual tumors of several mice were analyzed to assess potential variability. The ras/MAP kinase pathway is activated by many receptor tyrosine kinases, including Kit. Although ERK1/2 phosphorylation was observed in tumor lysates from untreated mice, there was no effect of imatinib treatment on ERK1/2 activation (Fig. 3). Thus ERK signaling is not critical for cell proliferation and survival response in tumor cells.
The PI3-kinase/Akt pathway, a key pathway in the control of apoptosis and the translational response, is known to play a role in malignancies (for reviews, see refs. 16 and 17). In tumor extracts from untreated control animals, PDK1 and Akt, as well as the downstream components GSK3β and mTOR, and ribosomal protein S6 were all strongly phosphorylated with minimal variability in all placebo-treated samples (Fig. 3). Phosphorylation of 4EBP1, a downstream target of mTOR, was more variable but also present in all placebo samples (Fig. 3). In tumor extracts from imatinib-treated mice, PDK1 and GSK3β phosphorylation was still visible but reduced compared to placebo controls (compare lanes 1–4 to 5–8; Fig. 3). Interestingly, Akt and its downstream targets (mTOR, the ribosomal protein S6, and 4EBP1) showed the strongest reduction of phosphorylation upon imatinib treatment. The Ser/Thr kinase mTOR is a regulator of the translational response, phosphorylating, among others, ribosomal protein S6 and 4EBP-1, components of the protein synthesis machinery. Phosphorylation of 4EBP-1 releases eIF4E to initiate translation. The decreased phosphorylation of mTOR, ribosomal protein S6, and 4EBP1 upon imatinib treatment indicates a strong down-regulation of the translational response upon imatinib treatment in GIST.
We next assessed the activation of the STAT transcription factors in tumor extracts from imatinib- and placebo-treated mice. STAT1 was expressed at very low levels in tumor extracts, and we could not reliably assess its expression or phosphorylation state. However, STAT3 and -5 were found to be expressed and consistently phosphorylated in placebo-treated GIST, and their phosphorylation appeared to be diminished upon treatment with imatinib. Interestingly, the decrease of phosphorylation of the smaller STAT3β isoform was more pronounced than that of the α isoform (Fig. 3). The reasons for this difference are not clear. Therefore, STAT5 and, to a lesser degree, STAT3 appear to be involved in the imatinib response of GIST tumor cells.
Gene Expression Profile of Kit Oncogenic Signaling in GIST.
To further elucidate the mechanisms underlying the response of murine GIST to imatinib, we compared gene expression profiles in RNA isolated from GISTs from placebo and imatinib-treated animals. Total RNA was isolated from the GISTs of five mice for each group: placebo and 6- and 24-h imatinib-treated and -labeled cRNA were synthesized and hybridized to an Affymetrix MOE430A murine expression array platform.
A one-way ANOVA was performed to determine which genes changed significantly between the three treatment groups, 0, 6, and 24 h. The genes found to be significantly different [68 and 76 probe identifications (IDs)] are listed in Table 2, which is published as supporting information on the PNAS web site. They can be assigned into 11 categories, including cell signaling, transcription, metabolism, and IFN response as the most represented categories. To determine which genes were differentially expressed after 6 or 24 h of imatinib treatment, a t test analysis between the placebo and the 6- or 24-h groups was performed. One hundred twenty-four (138 probe IDs) and 123 (132 probe IDs) genes were found differentially expressed after 6 and 24 h of treatment, respectively (Tables 3 and 4, which are published as supporting information on the PNAS web site), including 43 (6-h) and 47 (24-h) genes present in the ANOVA.
Genes belonging to the IFN response group were consistently down-regulated upon treatment with imatinib in all three types of analysis, indicating they were active in GIST. Quantitative PCR was used to validate that IFNγ-induced GTPase (Igtp) expression was reduced after 6 and 24 h of treatment by 2.3 and 3 times, respectively, consistent with the array data (Table 5, which is published as supporting information on the PNAS web site). Similarly, semiquantitative PCR showed that IFN-inducible protein 1 (Ifi1) expression was reduced in treated GIST (Fig. 8, which is published as supporting information on the PNAS web site).
Another observation from the ANOVA and t test analysis was that cyclins D1, D2, and/or D3 were down-regulated upon drug treatment. The down-regulation of cyclins D2 and D3 was confirmed by real-time and semiquantitative PCR, respectively (Table 5 and Fig. 8), and the results were consistent with the array data. Inversely, the cyclin-dependent kinase inhibitor p18 (Cdkn2c) was up-regulated almost 2-fold, a result confirmed by semiquantitative PCR (Fig. 8). Interestingly, down-regulation of the eukaryotic translation initiation factor 1A (Eif1a) was observed only after 24 h (Table 5). Taken together, the analysis of RNA expression profiles showed that IFN-responsive genes are expressed in GIST, and that their expression is diminished upon Kit inhibition. Furthermore, cyclin D proteins were also down-regulated by imatinib treatment, consistent with the cell cycle arrest observed in GIST lesions of imatinib-treated KitV558Δ/+ mice (Fig. 2A).
The ATP-binding cassette (ABC) transporters are a family of transporters involved in the transport of drugs and drug conjugates. At present, 10 ABC transporters have been implicated in the development of drug resistance (see refs. 18 and 19 for review). Recently, imatinib was shown to be a substrate for the breast cancer resistance protein BCRP/ABCG2 (20), and overexpression of the MDR1/ABCB1 gene decreased imatinib uptake and conferred imatinib resistance in cancer cell lines (21–23). Because ABC transporters are regulated at the level of transcription (19), we sought to determine whether RNA expression of any of these transporters was modified upon imatinib treatment in GIST. We analyzed the expression level of 10 transporters implicated in resistance to chemotherapeutics (19). Although none of these transporters were identified in our previous ANOVA and t test analysis, we reexamined the effect of imatinib treatment with the less-stringent t test analysis (Tables 6 and 7, which are published as supporting information on the PNAS web site). Only ABCC5 was found to have a reasonably significant P value of 0.0095 after 6 h of treatment, but MDR1/ABCB1 and BCRP/ABCG2 were also found to be up-regulated, with P values of 0.0016 and 0.0040, respectively, after 24 h of treatment. These results were validated by real-time PCR (Table 5) and, whereas the three genes were found to be up-regulated after 6 and 24 h of imatinib treatment, the high standard deviation indicated variability between tumors for the regulation of these genes. These results may suggest that imatinib treatment can induce an up-regulation of ABC transporters in GIST, and this could contribute to the development of imatinib resistance.
Human GISTs have been shown to have rather distinct gene expression profiles. To further validate whether the GIST mouse model replicates the human disease, we compared the expression profile of murine GIST with the human GIST signature derived from the comparison of 181 different sarcomas (24). The human GIST signature represents a list of 295 weighted genes and among them, 173 are present on the MOE430A chip and thus could be compared with the mouse GIST expression profile. Importantly, 144 genes from the human GIST signature were found to be expressed in the mouse GIST, and 29 were absent (Table 8, which is published as supporting information on the PNAS web site). The top discriminators of the human signature, including KIT, HRASLS3, PLAT, GHR, and IGF2 (24), were also present in mouse GIST. This indicates a substantial degree of similarity between human and mouse GIST expression profiles. Interestingly, none of the human signature genes found in the mouse GIST expression profiles are affected by imatinib treatment.
Effects of mTOR Inhibition in GIST.
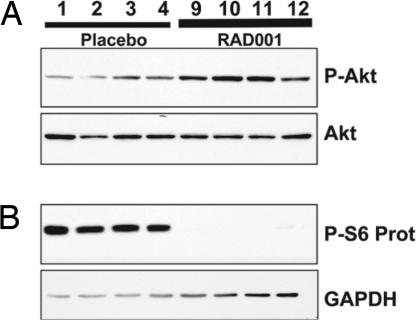
Our previous results indicated that Akt signaling by mTOR may be involved in oncogenic Kit signaling in GIST. To determine whether inhibition of mTOR is sufficient to reproduce the effects of imatinib on murine GIST, we treated KitV558Δ/+ mice with the mTOR inhibitor RAD001. Mice were treated by gavage with 5 mg/kg RAD001 daily. Tumor extracts were prepared after 24 h and characterized by Western blot analysis. We observed that ribosomal protein S6 phosphorylation was completely abolished after treatment, indicating that the dose administered was sufficient to block mTOR signaling in GIST (Fig. 4). We confirmed that phosphorylation of upstream components of mTOR signaling, including Y719 phosphorylation of Kit and phosphorylation of Akt, were not affected by RAD001 treatment; only a variable slight increase in Akt phosphorylation was observed (Fig. 4 and results not shown). These results indicate rapid and effective inhibition of mTOR signaling in GIST by RAD001.
Fig. 4.
RAD001 treatment inhibits mTOR signaling in GIST of KitV558Δ/+ mice. Tumor extracts from four placebo and four RAD001-treated mice were fractionated by SDS/PAGE and subjected to immunoblot analysis with antibodies for phospho-Akt T308 and Akt proteins (A) and phospho-ribosomal protein S6 S235/236 and GAPDH (B).
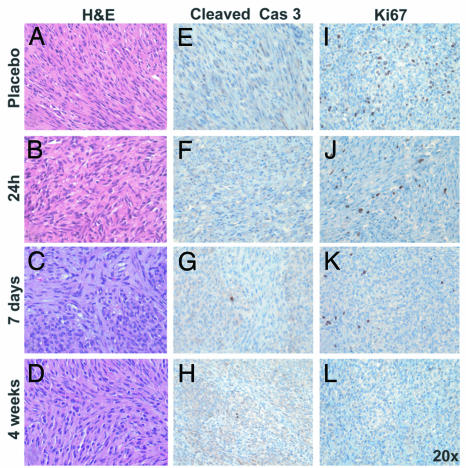
Despite the effect of RAD001 blocking mTOR activity, KitV558Δ/+ mice treated with 5 mg/kg RAD001 for up to 4 weeks did not show a histologic response in tumor sections prepared from treated mice, and no significant decrease in tumor cellularity or increase in myxoid stroma was observed compared to control mice (Fig. 5A–D). Similarly, RAD001 treatment did not produce apoptosis, as demonstrated by the absence of cleaved caspase 3 staining in treated tumor sections (Fig. 5 E–H). However, RAD001 did affect cell proliferation in GIST. Ki67 staining was decreased by 24 h of treatment, which became more pronounced after 7 days and led to an almost complete absence of Ki67 staining by 4 weeks (Fig. 5 I–L and Fig. 9C, which is published as supporting information on the PNAS web site). Treatment of mice with a higher dose of 10 mg/kg RAD001 still produced no effect on histology or apoptosis in a group of five treated animals (Fig. 9A and results not shown), but near-complete arrest of cell cycle progression was evident after only 7 days of treatment (Figs. 9 B and C). Of note is that the onset of cell cycle arrest upon treatment with RAD001 is delayed compared to cell cycle arrest induced by imatinib.
Fig. 5.
RAD001 treatment induces cell cycle arrest in GIST lesions of KitV558Δ/+ mice. (A–D) H&E staining of tumor sections of placebo and RAD001-treated mice (5 mg/kg). (E–L) Immunostaining of tumor sections of placebo and RAD001-treated mice (5 mg/kg) with antibody for cleaved caspase 3 (E–H) and Ki67 (I–L). The quantification of proliferating cells is presented in Fig. 9.
Combination Treatment of GIST with Imatinib and RAD001.
The preceding results showed that mTOR inhibition by RAD001 down-regulated S6 phosphorylation and impaired cell cycle progression in the mouse GIST model. They also showed that signaling through mTOR is strongly affected by imatinib treatment. To determine whether imatinib treatment synergized with RAD001 in GIST, mice were treated with 45 mg/kg imatinib and 10 mg/kg RAD001 for 7 days. The histology of tumor sections showed a response to the treatment with a decrease in cellularity and increase in myxoid stroma (Fig. 10, which is published as supporting information on the PNAS web site). However, the response observed in the eight mice treated with both imatinib and RAD001 varied from mild to moderate and was similar to that obtained by imatinib treatment alone (Fig. 1 E and F). Similarly, cleaved caspase 3 staining revealed that the number of apoptotic cells in the combination treatment was similar to that in imatinib treatment alone (not shown). We conclude that the two drugs do not synergize in mouse GIST.
Discussion
The protein tyrosine kinase inhibitors imatinib, erlotinib, and gefitinib, inhibitors of the Kit, PDGFR, Bcr-Abl, and EGFR tyrosine kinases are being used successfully to treat patients with GIST, chronic myelogenous leukemia, and lung adenocarcinomas, respectively. However, the development of drug resistance is a limiting factor in targeted single-agent therapy. Resistance often involves the acquisition of second-site receptor tyrosine kinase mutations, which interfere with tyrosine kinase inhibition. A detailed understanding of the signaling pathways involved in the development and maintenance of GIST may help to identify effector molecules that could be targeted with other specific inhibitors and to uncover combinational therapies to more effectively block critical signaling pathways involved in tumor growth and maintenance. In the present study, we used a mouse model of GIST to investigate Kit-mediated oncogenic signaling pathways and evaluate the consequences of imatinib inhibition on posttranscriptional modifications and gene expression in vivo. In our GIST mouse model, the juxtamembrane domain KitV558Δ mutation found in a human familial GIST case was introduced into the mouse germ line by using a knock-in strategy. The KitV558Δ/+ mice develop GIST with complete penetrance and indistinguishably from the human disease. We show that the response to imatinib in our murine GIST model is similar to the histologic response in human GIST patients, with replacement of cellular areas by myxoid stroma and focal necrosis. These features provided a rationale to investigate the consequences of imatinib treatment in the GIST mice.
Normal Kit ligand-induced Kit receptor signaling is known to activate several signaling molecules and cascades in vitro, including the PI3-kinase signaling network, the Ras/-MAP kinase cascade, Src kinase family signaling, SHP1/2 signaling, and the E3 ubiquitin ligase c-cbl. The characterization of posttranslational modifications of signaling molecules suspected to have a role in Kit signaling in GIST indicates strong activation of the PI3-kinase signaling cascades, including the translational response and down-regulation of the activating modifications upon drug treatment. Furthermore, phosphorylation of the STAT transcription factors, STAT3 and STAT5, was inhibited by imatinib as well. Surprisingly, the Ras-MAP kinase pathway, although activated in GIST samples, was not affected by imatinib treatment. Because all tumor samples showed histologic and biochemical evidence of response to imatinib, the lack of an effect on ERK1/2 activation suggests this pathway is insufficient for oncogenic Kit signaling. However, it is possible that qualitatively superior responses could be seen if ERK1/2 activation were inhibited in imatinib-treated mice. The availability of such inhibitors will help to clarify the role of MAP kinase signaling in GIST.
The analysis of gene expression profiles revealed that the genes affected by imatinib fall into several categories. First, imatinib treatment affects the expression of cell cycle regulators. The cyclins E and D and their associated kinases are positive regulators of cell division. The cyclin D family members (D1, D2, and D3) are expressed in various combinations in different cell types. They bind and activate the cyclin-dependent kinases Cdk4 and –6, which lead to the phosphorylation and inactivation of pRb and subsequent transcription of E2Fs-dependent genes required for S phase entry. Two families of inhibitors restrain cyclin/cdk activity: the ink4 and Cip/Kip families (for reviews, see refs. 25–27). Our gene expression profiling experiments revealed that the three members of the cyclin D family were down-regulated upon imatinib treatment, whereas the inhibitor p18ink4c was up-regulated. This is in agreement with a decrease in cell proliferation, and it underlines the importance of cyclin D in oncogenic Kit signaling in GIST. Cyclin D expression is induced by growth factors and mitogenic signals to mediate progression of the cell cycle, and their overexpression is observed in several cancer types (25). In agreement with our biochemical results showing diminished STAT5 phosphorylation upon imatinib treatment, STAT5 has been shown to bind and activate the cyclin D1 promoter (28). Small molecule inhibitors of cyclin/cdk activity have been developed recently as possible therapeutics (29, 30). It will be interesting to know whether GIST patients resistant to imatinib therapy could benefit from these inhibitors.
Both Imatinib and RAD001 similarly down-regulate the translational response in GIST, as demonstrated by the greatly diminished ribosomal protein S6 phosphorylation, and both drugs induced cell cycle arrest. But, in contrast to imatinib, RAD001 treatment did not induce apoptosis. This difference could be explained by the fact that mTOR forms two different complexes, the raptor–mTOR and the rictor–mTOR complex. The raptor–mTOR complex has been shown to directly phosphorylate the hydrophobic motif site of S6K1 and to regulate cell growth (31) by ribosomal protein S6 phosphorylation. This complex is sensitive to rapamycin and RAD001 inhibition. In contrast, rapamycin does not associate with the rictor–mTOR complex (32), which has been shown to be a kinase for Akt in Drosophila and human cells (33) and therefore may play an important role in Akt activation also in GIST. One can hypothesize that in GIST, RAD001 inhibits the raptor–mTOR complex, which is consistent with a decrease in cell proliferation observed in mice treated for 7 days but leaves the rictor–mTOR complex free to activate Akt.
Whereas mTOR inhibition by RAD001 induced cell cycle arrest, no concomitant histological or apoptotic response was observed in tumor lesions. In contrast, imatinib inhibited cell cycle progression and induced an increase in apoptosis as well as a histological response in GIST. This indicates that mTOR inhibition induces cell cycle arrest but, to achieve better therapeutic efficacy, other components of oncogenic Kit signaling have to be targeted. These components may lie upstream of mTOR in the PI-3 kinase or STAT pathways. In addition to activating the translational response, the PI3-kinase effector Akt also phosphorylates and inhibits the proapoptotic BAD protein and the forkhead transcription factor, which may account for the lack of induction of apoptosis.
Rapamycin has been reported to have a weak antitumor activity in vivo. In mouse models of lymphoma and chronic myelogenous leukemia, rapamycin alone did not improve survival of transplanted mice significantly. However, when rapamycin was used in combination with cytotoxic agents or with imatinib, synergistic antitumor activity was observed (34, 35). Our results show no benefit of using the rapamycin derivative RAD001 in combination with imatinib to improve the histological response in GIST, indicating that RAD001 does not improve the therapeutic efficacy of imatinib inhibition in GIST carrying the KitV558Δ mutation. Targeting of other pathways in addition to mTOR may improve the efficacy of GIST treatment. High mitotic index values are associated with malignant behavior of human GISTs and poor prognosis. By virtue of abolishing tumor cell proliferation, RAD001 may be of useful in the treatment of patients with imatinib-resistant GIST. Our previous description and the current study convincingly show that KitV558Δ/+ mice represent a unique faithful mouse model of human familial GIST. The current study demonstrates the utility of these mice for preclinical investigations and for elucidating oncogenic signaling mechanisms by using genetic approaches and targeted pharmacological intervention.
Materials and Methods
Mice.
Heterozygous KitV558Δ/+ mice were described in ref. 12. The KitV558Δ/+ mice used in these experiments were backcrossed with C57BL/6J for seven to nine generations. Mice selected for treatment had no apparent signs of disease and were 3–5 months old.
Drug Treatment of Heterozygous KitV558Δ/+ Mice.
Imatinib (Gleevec, STI571) was kindly provided by Novartis. Imatinib was dissolved in water as a 10-mM solution and stored at −20°C. Heterozygous KitV558Δ/+ mice were treated by i.p. injection with 45 mg/kg imatinib, once per day for 6- and 12-h treatments or twice per day for 24 h, 7 days, and 3 weeks. RAD001 (everolimus) [40-O-(2-hydroxyethyl)-rapamycin] was also provided by Novartis as a 20-mg/g emulsion. An emulsion placebo was also provided. The emulsion was diluted in 5% sucrose before administration by gavage, twice for a 24-h treatment and once per day for 7-day and 4-week treatment. After the indicated treatment times, mice were killed 6 h after the last administration of the drug, and the tumors were quickly harvested and fixed in freshly prepared 4% paraformaldehyde for histology and immunohistochemistry or snap-frozen in liquid nitrogen for protein analyses. Five to 10 mice per group of treatment were used.
Microarray Expression Analysis.
Total RNA was prepared from GIST by the TRIzol method (Invitrogen). Biotin-labeled c-RNA prepared from 5 μg of total RNA was fragmented and hybridized to oligonucleotide microarrays MOE430A (Affymetrix). The image files were quantitated with Affymetrix’ MAS 5 software. Initially, the lists of genes were filtered to remove those with <25% present calls in at least one group (placebo, 6 h, 24 h). An ANOVA test was done to find genes whose expression value differed among the groups. Then, to find genes differentially expressed at a given time point, a standard t test was used. To correct for multiple testing, the False Discovery Rate method was used.
Real-Time PCR.
Two micrograms of total RNA was reverse-transcribed at 42°C for 30 min using the iScript cDNA Synthesis kit (Bio-Rad). Forty nanograms of resultant cDNA was used in a quantitative PCR by using an iCycler (Bio-Rad) and predesigned TaqMan gene expression assays (Supporting Text, which is published as supporting information on the PNAS web site). Triplicate cycle threshold values were averaged, and amounts of target were interpolated from the standard curves and normalized to hypoxanthine phosphoribosyltransferase.
Statistical Analysis.
The mitotic index (percentage of proliferating cells) in placebo-treated tumors was obtained by counting total and Ki67-positive cells by using MetaMorph software in 20 pictures of 10 placebo-treated tumors. To determine cell proliferation and apoptosis in tumors, Ki67 and cleaved caspase 3-positive cells were counted under a microscope on 20 fields of at least five different tumors for each time point. Student’s t test assuming unequal variances between the two samples was used to determine the significance of differences of proliferating and apoptotic cells between the placebo and imatinib or RAD001-treated GISTs. Groups were judged to differ significantly at P < 0.05.
Histological and Immunohistochemical Analyses.
For microscopic analysis, 5-μm sections from paraffin-embedded tissue were stained with H&E. Immunohistochemistry antibodies were purchased from Cell Signaling Technology (no. 9661) for cleaved caspase 3, Novacastra (no. NCL-Ki67P) for Ki67, Oncogene (no. PC34) for Kit, and Cell Signaling Technology (no. 2211) for phospho-S6 ribosomal protein.
Immunoprecipitation and Western Blotting.
Western blotting and immunoprecipitation of tumor lysates were performed as described (12). Anti-p85 rabbit polyclonal antibody was purchased from Upstate Biotechnology, and anti-GAPDH were purchased from GeneTex. All other antibodies were rabbit polyclonals obtained from Cell Signaling Technology.
Supplementary Material
Acknowledgments
We thank Sandra Gonzales, Craig Farrell, and Ahmed Fadl of the Molecular Cytology Facility for help with histological analyses and Drs. Yuhong She and Ju Haiying of the Antitumor Assessment Facility for technical assistance with drug administration. We thank Drs. Elisabeth Buchdunger and Heidi Lane of the Novartis Institutes for BioMedical Research and Oncology, Basel, respectively, for providing STI571 and RAD001. We also thank Drs. Eva Besmer and Joe Scandura and Robert Maki for critical reading and comments of this manuscript. This work was supported by the National Cancer Institute and National Institutes of Health Grants CA 102774 and HL/DK55748 (to P.B.).
Abbreviations
- GIST
gastrointestinal stromal tumor
- PI3-kinase
phosphatidylinositol 3-kinase
- ABC
ATP-binding cassette
- mTOR
mammalian target of rapamycin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Huizinga J. D., Thuneberg L., Kluppel M., Malysz J., Mikkelsen H. B., Bernstein A. Nature. 1995;373:347–349. doi: 10.1038/373347a0. [DOI] [PubMed] [Google Scholar]
- 2.Maeda H., Yamagata A., Nishikawa S., Yoshinaga K., Kobayashi S., Nishi K. Development (Cambridge, U.K.) 1992;116:369–375. doi: 10.1242/dev.116.2.369. [DOI] [PubMed] [Google Scholar]
- 3.Torihashi S., Ward S. M., Nishikawa S., Nishi K., Kobayashi S., Sanders K. M. Cell Tissue Res. 1995;280:97–111. doi: 10.1007/BF00304515. [DOI] [PubMed] [Google Scholar]
- 4.Antonescu C. R., Sommer G., Sarran L., Tschernyavsky S. J., Riedel E., Woodruff J. M., Robson M., Maki R., Brennan M. F., Ladanyi M., et al. Clin. Cancer Res. 2003;9:3329–3337. [PubMed] [Google Scholar]
- 5.Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., et al. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 6.Lasota J., Wozniak A., Sarlomo-Rikala M., Rys J., Kordek R., Nassar A., Sobin L. H., Miettinen M. Am. J. Pathol. 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin B. P., Singer S., Tsao C., Duensing A., Lux M. L., Ruiz R., Hibbard M. K., Chen C. J., Xiao S., Tuveson D. A., et al. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- 8.Antonescu C. R., Besmer P., Guo T., Arkun K., Hom G., Koryotowski B., Leversha M. A., Jeffrey P. D., Desantis D., Singer S., et al. Clin. Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 9.Debiec-Rychter M., Cools J., Dumez H., Sciot R., Stul M., Mentens N., Vranckx H., Wasag B., Prenen H., Roesel J., et al. Gastroenterology. 2005;128:270–279. doi: 10.1053/j.gastro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Nishida T., Hirota S., Taniguchi M., Hashimoto K., Isozaki K., Nakamura H., Kanakura Y., Tanaka T., Takabayashi A., Matsuda H., et al. Nat. Genet. 1998;19:323–324. doi: 10.1038/1209. [DOI] [PubMed] [Google Scholar]
- 11.Robson M. E., Glogowski E., Sommer G., Antonescu C. R., Nafa K., Maki R. G., Ellis N., Besmer P., Brennan M., Offit K. Clin. Cancer Res. 2004;10:1250–1254. doi: 10.1158/1078-0432.ccr-03-0110. [DOI] [PubMed] [Google Scholar]
- 12.Sommer G., Agosti V., Ehlers I., Rossi F., Corbacioglu S., Farkas J., Moore M., Manova K., Antonescu C. R., Besmer P. Proc. Natl. Acad. Sci. USA. 2003;100:6706–6711. doi: 10.1073/pnas.1037763100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissel H., Timokhina I., Hardy M. P., Rothschild G., Tajima Y., Soares V., Angeles M., Whitlow S. R., Manova K., Besmer P. EMBO J. 2000;19:1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agosti V., Corbacioglu S., Ehlers I., Waskow C., Sommer G., Berrozpe G., Kissel H., Tucker C. M., Manova K., Moore M. A., et al. J. Exp. Med. 2004;199:867–878. doi: 10.1084/jem.20031983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timokhina I., Kissel H., Stella G., Besmer P. EMBO J. 1998;17:6250–6262. doi: 10.1093/emboj/17.21.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J., Manning B. D., Cantley L. C. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 17.Wendel H. G., Lowe S. W. Cell Cycle. 2004;3:847–849. [PubMed] [Google Scholar]
- 18.Borst P., Elferink R. O. Annu. Rev. Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 19.Scotto K. W. Oncogene. 2003;22:7496–7511. doi: 10.1038/sj.onc.1206950. [DOI] [PubMed] [Google Scholar]
- 20.Burger H., van Tol H., Boersma A. W., Brok M., Wiemer E. A., Stoter G., Nooter K. Blood. 2004;104:2940–2942. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 21.Hamada A., Miyano H., Watanabe H., Saito H. J. Pharmacol. Exp. Ther. 2003;307:824–828. doi: 10.1124/jpet.103.055574. [DOI] [PubMed] [Google Scholar]
- 22.Mahon F. X., Belloc F., Lagarde V., Chollet C., Moreau-Gaudry F., Reiffers J., Goldman J. M., Melo J. V. Blood. 2003;101:2368–2373. doi: 10.1182/blood.V101.6.2368. [DOI] [PubMed] [Google Scholar]
- 23.Thomas J., Wang L., Clark R. E., Pirmohamed M. Blood. 2004;104:3739–3745. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 24.Baird K., Davis S., Antonescu C. R., Harper U. L., Walker R. L., Chen Y., Glatfelter A. A., Duray P. H., Meltzer P. S. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 25.Sherr C. J., Roberts J. M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 26.Sherr C. J., Roberts J. M. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg R. A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 28.Bromberg J. F. BioEssays. 2001;23:161–169. doi: 10.1002/1521-1878(200102)23:2<161::AID-BIES1023>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Malumbres M., Barbacid M. Nat. Rev. Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 30.Swanton C. Lancet Oncol. 2004;5:27–36. doi: 10.1016/s1470-2045(03)01321-4. [DOI] [PubMed] [Google Scholar]
- 31.Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M. Proc. Natl. Acad. Sci. USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 34.Mohi M. G., Boulton C., Gu T. L., Sternberg D. W., Neuberg D., Griffin J. D., Gilliland D. G., Neel B. G. Proc. Natl. Acad. Sci. USA. 2004;101:3130–3135. doi: 10.1073/pnas.0400063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendel H. G., De Stanchina E., Fridman J. S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S. W. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.