Abstract
Clinical trials are testing oncolytic viruses (OVs) as therapies for cancer. We have shown that animals that have brain tumors and are treated with a herpes simplex virus (HSV)-derived OV live significantly longer when cyclophosphamide (CPA) is preadministered. Here, we explore the mechanisms behind this finding. In a syngeneic rat glioma model, intratumoral HSV administration is associated with rapid increase of natural killer cells, microglia/macrophages (CD68+ and CD163+), and IFN-γ. Pretreatment with CPA enhances HSV replication and oncolysis and reduces an HSV-mediated increase in CD68+ and CD163+ cells and intratumoral IFN-γ. Molecular imaging shows CPA pretreatment to inhibit HSV-induced infiltration of tumor-associated phagocytic cells. Our results reveal molecular and cellular mechanisms that inhibit intratumoral spread of HSV and suggest a therapeutic path for improving the efficacy of virotherapy as a treatment for cancer.
Keywords: gene therapy, innate immunity, oncolytic virus, brain tumor, herpes simplex virus
Advances in virology and tumor biology have enabled development of oncolytic viruses (OVs), which replicate selectively in tumor cells (1–6). OV progeny propagate throughout tumors, lysing tumor cells but not normal cells. Phase I clinical trials have shown OV therapy to be safe (7–13) but with limited efficacy. A brisk host response to OV therapy has been seen. It includes intratumoral immune cells (7) and acute-phase reaction to intravascular virus (13). Innate immune responses may be a common side effect of OV therapy, similar to the radionecrosis of radiotherapy or myelosuppression of chemotherapy.
The role of host immune responses in the efficacy or toxicity of OV therapy is poorly defined. Such responses are thought beneficial because oncolysis stimulates adaptive immunity, setting up an anticancer vaccination effect (14–16). However, initial innate responses to OVs may reduce efficient anticancer effects (17–21). For example, we have shown a herpes simplex virus (HSV)-based OV therapy to be more efficient when cyclophosphamide (CPA) is present (22–25), and this increased efficiency is credited to CPA's immunosuppressive action. However, the specific immune pathways of the observed effects have not been analyzed.
Results
Preadministering CPA Inhibits Clearance of Viral Particles and Increases HSV Replication Within Injected Tumors.
We have reported that CPA allows increased replication of HSV in injected tumors by suppressing immune activity (25). To determine intratumoral persistence of HSV, we compared intratumoral viral-mediated LacZ (Fig. 1) and ICP4 (infected-cell protein 4) (Fig. 6, which is published as supporting information on the PNAS web site) gene expression in immunocompetent rats 6 and 72 h after treatment with HSV, with and without CPA pretreatment. Six hours after intratumoral HSV delivery, ≈50% of tumor cells showed viral-mediated gene expression, regardless of CPA (Fig. 1 A and C). However, by 72 h, without CPA, <10% of tumor cells showed viral-mediated gene expression, despite the replicating ability of the injected virus (Fig. 1 B and C); with CPA, ≈80% of tumor cells exhibited HSV-mediated gene expression (Fig. 1 B and C). These results suggested that CPA's immunosuppressive activity inhibited the rapid natural clearance of intratumoral HSV and increased its ability to replicate, agreeing with previous findings (25).
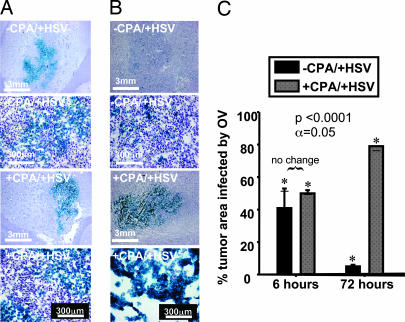
Fig. 1.
Tumor infection and intratumoral proliferation and persistence of HSV (LacZ expression). (A and B) Intratumoral distribution of HSV-mediated gene expression is shown by blue stain of infected cells expressing E. coli β-galactosidase. Two magnifications show tumors extracted from control rats (−CPA/+HSV, two upper rows) and from rats treated with CPA 48 h before injection with virus (+CPA/+HSV, two lower rows). Microphotographs show tumors 6 h after viral injection (48 h after CPA treatment), before viral replication (A) and 72 h after viral injection (5 days after CPA treatment), when several rounds of HSV replication should have occurred (B). (C) Quantitative enumeration of the percentage of tumor area infected by HSV at 6 and 72 h after viral injection in control rats and rats pretreated with CPA (three rats per treatment group). Significant differences were observed for the 6-h versus 72-h time points and for control animals versus those treated with CPA at 72 h.
CPA Reverses HSV-Mediated Increases in Mononuclear Cells After Infection of Brain Tumors.
To identify the immune cells causing CPA-mediated enhancement of HSV replication in vivo, we performed immunohistochemistry (IHC) staining for markers of microglia/macrophages (CD68 and CD163), natural killer (NK) cells (NKR-P1), T and B lymphocytes (CD4, CD8, CD43, and CD45), and neutrophils and granulocytes (CD43 and morphology) at 6 and 72 h after inoculation of HSV into tumors of animals pretreated with either CPA or vehicle.
Six hours after HSV injection, CD68+ and NKRP1+ markers were up-regulated (Fig. 2). With CPA, intratumoral up-regulation of CD68+ cells was inhibited (Fig. 2 A–C), but the number of NKR-P1+ cells was not affected (Fig. 2 D–F).
Fig. 2.
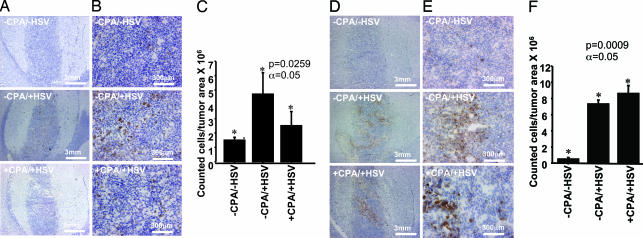
Intratumoral infiltration of CD68+ and NKRP1+ cells 6 h after injection with HSV as a function of HSV infection and treatment with CPA. Rodent brains (three per treatment group) were harvested 6 h after inoculation with HSV or mock treatment (48 h after pretreatment with CPA or mock treatment). Density of CD68+ and NKRP1+ cells was determined for each animal in three different randomly selected tumor regions in which distribution of HSV (assayed by LacZ gene expression) was greatest. (A and B) IHC staining for CD68+ cells at two magnifications in tumors from rats receiving no treatment (−CPA/−HSV) (Top), treatment with HSV alone (−CPA/+HSV) (Middle), and treatment with both CPA and HSV (+CPA/+HSV) (Bottom). (C) Quantitative representation of CD68+ cells in tumors of rats treated as described. (D and E) IHC staining for NKRP1+ cells in the same tumors. (F) Quantitative representation of tumor infiltration of NKRP1+ cells.
At 72 h, HSV induced an increase of CD68+ microglia/macrophages as well as CD163+ peripheral macrophages (Fig. 3), but NK cells remained sparse or absent (data not shown). CPA inhibited increases in CD68+ and CD163+ cells (Fig. 3). Induction of CD4 and CD8 lymphocytes was weak at 72 h but became more significant later (data not shown). Neutrophils and B lymphocytes were absent or sparse in all treatment groups. Thus, NK and monocyte-derived cells characterized a very early innate immune response to HSV infection of tumors, and CPA pretreatment inhibited early infiltration of monocytic cells.
Fig. 3.
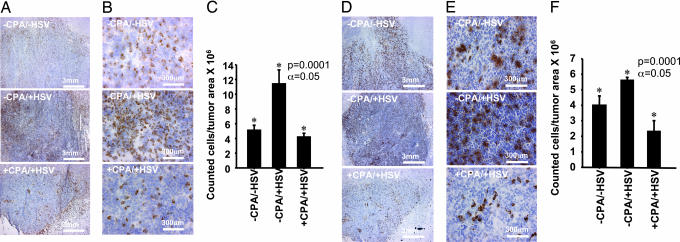
Intratumoral infiltration of CD68+ and CD163+ cells as a function of infection with HSV and treatment with CPA 72 h after HSV injection. Rodent brains (three per treatment group) were harvested 72 h after inoculation with HSV or mock treatment (5 days after pretreatment with CPA or mock treatment). Density of CD68+ and CD163+ cells was determined for each animal in three different randomly selected tumor regions in which distribution of HSV (assayed by expression of LacZ gene) was greatest. (A and B) IHC staining for CD68+ cells at two magnifications in tumors from rats receiving no treatment (−CPA/−HSV) (Top), treatment with HSV alone (−CPA/+HSV) (Middle), and treatment with both CPA and HSV (+CPA/+HSV) (Bottom). (C) Quantitative representation of CD68+ cells in tumors of rats treated as described. (D and E) IHC staining for CD163+ cells in the same tumors described above. (F) Quantitative representation of tumor infiltration of CD163+ peripheral macrophages.
Molecular Imaging Suggests That Tumor Infiltration of Phagocytic Cells Changes as a Function of HSV and CPA.
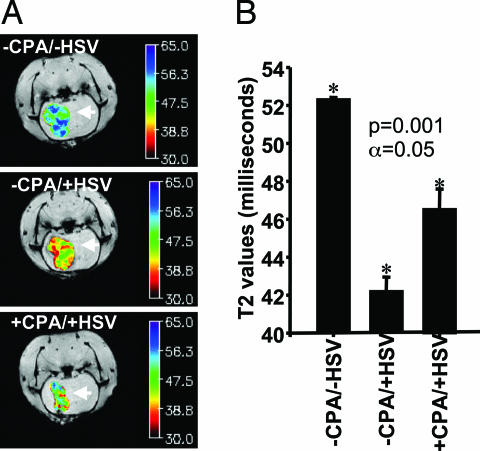
To determine the involvement of tumor-associated phagocytic cells in immune response to HSV administration, we administered i.v. (72 h after HSV infection of rat gliomas) dextran-coated monocrystalline iron oxide particles (MIONs), which were phagocytosed by circulating macrophages. Ingested MIONs give macrophages superparamagnetic properties that enable MRI without affecting their function (26). Brain tumors of rats were imaged as a function of HSV injection and/or CPA pretreatment. HSV markedly decreased the T2-weighted signal (greater magnetic susceptibility) of tumors (Fig. 4A), indicating an increase in intraneoplastic MION-loaded macrophages. CPA pretreatment blocked this phenomenon. The lower T2 signal in tumors treated with HSV averaged 42 ms, compared with >50 ms in control tumors and 46 ms in tumors pretreated with CPA (Fig. 4B). Thus, the increase in CD68+ and CD163+ markers appears to correspond to intratumoral infiltration of phagocytic cells that may be responsible for observed early viral clearance.
Fig. 4.
MRI quantification of tumor-associated phagocytic cells. Magnetic resonance-based macrophage imaging of rodent brain tumors as a function of treatment with CPA and HSV is shown. (A) Tumors of animals receiving mock treatment (−CPA/−HSV, Top), intratumoral HSV (−CPA/+HSV, Middle), or preadministration of CPA and intratumoral HSV (+CPA/+HSV, Bottom) were imaged for tumor-associated phagocytic cells infiltrating the tumor. Imaging was done 72 h after HSV injection (5 days after CPA pretreatment). Magnetic resonance signals were color-coded. Shift toward red indicates a darkening T2-weighted signal; shift toward blue indicates a paled signal. The darker the T2-weighted signal, the more superparamagnetic iron oxide particles are incorporated in the tumor by infiltrating peripheral phagocytic cells. (B) Each bar represents the mean T2 value from 16 images of three rats per treatment group (±SD). Higher T2 values correspond to decreased intratumoral infiltration of peripheral macrophages. Macrophage imaging showed significant difference among groups.
Relevance of IFN-γ in Glioma Virotherapy.
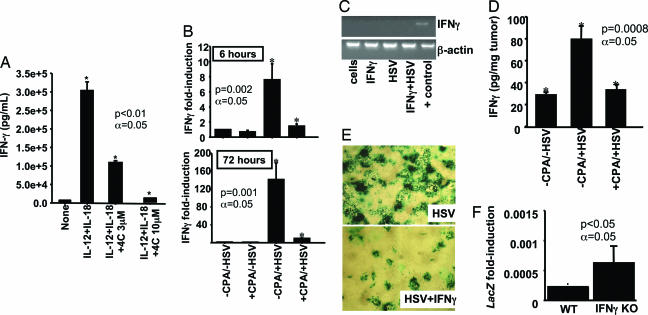
NK cells mediate innate immune responses partly by generating IFN-γ, a chemoattractor of macrophages (27). To determine whether CPA impairs the function of NK cells without affecting their viability, we analyzed their production of IFN-γ in vitro (27) after treatment with 4-hydroxy-CPA (4HC), the intermediate active metabolite of CPA. Low doses of 4HC inhibited IFN-γ release from isolated NK cells grown in vitro (Fig. 5A). IFN-γ release was reduced by 60% with 3 μM 4HC and 100% at higher concentrations. In 48 h of in vitro treatment, at a concentration (100 μM) that is normally high enough to kill tumor cells in 24 h (data not shown), 4HC did not kill NK cells.
Fig. 5.
IFN-γ elevation in response to HSV neutralizes viral replication and is inhibited by CPA. (A) NK cells were isolated from the blood of human donors. Release of IFN-γ from these cells was stimulated by treatment with IL-12/IL-18 and analyzed by ELISA. Addition of 3 and 10 μM 4HC led to 60% and 100% inhibition of release of IFN-γ, respectively. (B) Quantitative RT-PCR for IFN-γ mRNA in rat gliomas receiving mock treatment or treatment with CPA, HSV, or HSV and CPA. CPA treatment occurred 2 days before HSV administration, and brain tumor RNA was isolated 6 h and 72 h after HSV administration. RNA was extracted from three rats per treatment group. The bars compare the mean (±SD) fold induction of gene expression in tumors receiving treatment and in the control group (−CPA/−HSV). (C) Rat D74HveC glioma cells were treated in vitro with HSV, exogenous IFN-γ, or both. Endogenous IFN-γ levels were analyzed by RT-PCR. A brain tumor infected with HSV was used as positive control for IFN-γ; β-actin was used as an internal control. (D) Average values (±SD) of IFN-γ protein (in picograms per milligram of tumor) in tumors extracted from three treatment groups: −CPA/−HSV, −CPA/+HSV, and +CPA/+HSV. Analysis was performed at 72 h. (E) D74 glioma cells were pretreated with 10 ng/ml exogenous IFN-γ 12 h before virus infection, and HSV replication was evaluated by staining with β-galactosidase. The experiment was performed in triplicate and repeated several times; two representative wells are shown. (F) HSV-mediated LacZ expression from tumors of WT and IFN-γ−/− [IFN-γ knockout (KO)] mice was evaluated by quantitative RT-PCR 3 days after viral delivery. The bars show the average of LacZ mRNA induction levels in three animals for each mouse genotype.
We then performed RT-PCR for IFN-γ mRNA in rats undergoing virotherapy and in glioma cells infected with HSV in vitro. Six hours after rat gliomas were treated with HSV, IFN-γ mRNA was increased 10 times in tumors without CPA pretreatment but not in those with CPA (Fig. 5B). At 72 h after injection, intratumoral levels of IFN-γ mRNA were increased >120 times by HSV, a response that was inhibited by CPA (Fig. 5B). D74 glioma cells grown in vitro in the presence of exogenous IFN-γ and/or HSV expressed no IFN-γ mRNA, suggesting stromal derivation of intratumoral IFN-γ (Fig. 5C). CPA pretreatment also inhibited HSV-mediated increase of IFN-γ protein concentration in tumors 72 h after HSV injection (Fig. 5D).
To further clarify IFN-γ's effect during virotherapy, we analyzed the replicative capacity of HSV in tumor cells grown in vitro in the presence of exogenous IFN-γ and in a mouse glioma established in IFN-γ knockout (KO) transgenic mice. IFN-γ pretreatment inhibited viral replication in vitro (Fig. 5E), and HSV-mediated gene expression in tumors established in IFN-γ KO mice was 3-fold that in those with WT strain (Fig. 5F). Thus, rapid up-regulation of IFN-γ in glioma in response to HSV derives from stromal cells, not glioma cells, and inhibits HSV's ability to replicate in tumor cells. CPA inhibits HSV-mediated up-regulation of IFN-γ.
Discussion
Testing of OV therapies continues in clinical trials for a variety of malignancies (7–13, 28), and results reveal the importance of understanding why OV replication is not as effective in vivo as in vitro. We have questioned here whether host innate immune responses to active viral tumor infection are critical to the overall efficacy and toxicity of therapy. We report that within 72 h of viral injection, the tumor volume that exhibits HSV-mediated gene expression decreases significantly. This phenomenon is associated with a rapid increase (within 6 h of infection) of intratumoral NK cells, CD68+ microglia/macrophages, and IFN-γ mRNA and protein deriving from stromal cells in tumors. Further increases in CD68+ microglia/macrophages, CD163+ peripheral macrophages, and IFN-γ are observed 72 h after HSV infection. CPA inhibits intratumoral infiltration of mononuclear cells, increase of IFN-γ, and in vitro production of IFN-γ by NK cells. Finally, we show that administering exogenous IFN-γ is sufficient to inhibit viral replication in glioma cells grown in vitro and that HSV-mediated gene expression is greater in tumors established in IFN-γ knockout mice than in the correspondent WT strain. Thus, the innate immune responses of the host shortly after intratumoral viral infection, including IFN-γ production and activation of microglia/macrophages, suggest a critical role in limiting viral oncolysis.
The changes in levels of NK cells, CD68+ microglia/macrophages, and IFN-γ within 6 h of HSV infection suggest these responses as critical participants of a cascade of innate immune responses to the virus. Described as circulating cells attracted by viral infection of a host, NK cells start the immune process at least partly by releasing IFN-γ before exiting the site of inflammation, giving way to secondary responses (29, 30). The temporal kinetics of NK cell behavior in our model agrees with the published model of NK cell behavior, and specific interactions between NK and host cells infected with WT HSV have been described (31, 32). NK cell-derived IFN-γ is the prototypic monocyte/macrophage activator, without which monocytes/macrophages cannot clear intracellular organisms (33). Our in vitro data show that pretreatment with CPA inhibits production of IFN-γ by NK cells but does not affect their viability within 48 h, probably because of the slow replication cycle of these cells. Thus, the in vivo mechanism of CPA modulation of innate immunity against HSV may be related to the inhibition of IFN-γ production by NK and monocytic cells in response to the virus and to the decrease of intratumoral density of HSV-stimulated macrophages. When making IFN-γ, NK cells make a plethora of cytokines and chemokines (34), many of which may serve as chemoattractants, decreasing viral load (35), and are likely to be similarly affected by CPA.
Several types of macrophages have been described in the brain (36). One, found in perivascular spaces, derives from bone marrow and can move rapidly from the vasculature into the brain parenchyma in response to infection with a pathogen. Other cells with potential phagocytic activity in the brain are microglia, which are believed to be derived from monocytes and thus are able to replicate the function of peripheral macrophages (36). CD68 antigen, a marker of peripheral macrophages and brain microglia, is induced very early after viral delivery (6 h). HSV-mediated infiltration of CD163, a specific marker of peripheral macrophages, occurs only 72 h after viral infection. The first innate immune response is probably mediated by local microglia cooperating with NK cells. Both CD68+ and CD163+ cells were found in perivascular spaces. Moreover, CD68+ cells are often found in gliomas as monocytes with no phagocytic activity, but their function in these tumors is unknown (37). CPA significantly inhibited expression of both CD68+ and CD163+ cells in tumors, but it is unclear whether the depletion of these cells is direct or mediated by the inhibition of NK cell production of IFN-γ. Current experiments aimed at individually depleting NK cells, microglia, or macrophages will clarify the relative contribution of each of these cell types in inhibiting viral replication and persistence in vivo. Because MRI capturing a change in T2 signal after administering superparamagnetic MIONs confirms the presence of phagocytosed MIONs in brain tumors, a correlative imaging technique for monitoring viral oncolysis in clinical trials may also arise from this report. Molecular imaging of CPA-mediated macrophage depletion indicates the persistence of a small percentage of HSV-induced phagocytic cells that are not detectable by IHC for CD68+ and CD163+ cells, perhaps because of the existence of other microglia/macrophages, such as ED3 cells, that are not strongly repressed by CPA and are detectable by MRI (G.F. and E.A.C., unpublished data). The intratumoral presence of these cells may explain why CPA pretreatment increases animal survival but does not cure the animal (25).
CD68+ and CD163+ cells are evident in human gliomas treated with ONYX-015 (1, 7), a different OV from that used in our experiments (data not shown). Thus, host responses to HSV infection seem to be independent of OV type or species, implying a shared response to pathogens in the brain. Data from a collaborating group also show that CPA enhances oncolytic effects of adenovirus in gliomas by depleting innate immune cells (M. L. Lamfers and E.A.C., unpublished work). The presence of CD68+ and CD163+ cells in human brains infected with WT HSV suggests confirmation of this hypothesis (data not shown). Further analysis on human gliomas treated with an HSV-derived OV would confirm the relevance of our findings to human patients.
We have reported CPA's enhancement of viral oncolysis, showing prolonged survival of animals with brain tumors (23, 25), increased titers of tumor-associated HSV (25), and a reduction in the dose of HSV (38) required to achieve significant anticancer effects. Here, we provide mechanistic evidence that CPA enhances the oncolytic effects of viruses by inhibiting innate immune cells (NK cells and CD68+/CD163+ microglia/macrophages) and levels of IFN-γ within tumors and that the enhancement may be linked as well to decreased production of IFN-γ by NK cells in response to viral infection. Global gene expression analyses of tumors treated with CPA and with HSV alone reveal CPA's rapid inhibition of pathways linked to innate immune responses against HSV (G.F. and E.A.C., unpublished data), agreeing with present findings. So, we propose a paradigm to improve OV-based tumor therapy based on the need for transient initial suppression of the immune system for successful intratumoral viral replication.
We have evidence that administering virus into tumors can trigger changes in gene expression that reflect activation of immune responses in peripheral blood mononuclear cells 12 h after delivery (G.F. and E.A.C., unpublished data). Combined with increased perfusion of tumor tissue compared with normal brain and with presence of lymphatic and dendritic cells in the meninges and the ventricular zone, these data indicate the virus's capacity for adaptive immune response. Nevertheless, the later infiltration of T lymphocytes compared with NK cells and macrophages suggests the possibility of transiently targeting innate immune responses in early viral replication without hampering the adaptive immune response. Indeed, later, when CPA is no longer active, the viral load of CPA-treated rats again induces infiltration of peripheral CD163+ macrophages, and CD8 and CD4 T cells are increased (G.F. and E.A.C., unpublished data). Thus, the stimulation of antitumoral, OV-induced immune responses in a second phase of therapy appears unhampered by this transient immune suppression.
Materials and Methods
Viruses and Cells.
The OV hrR3, with an Escherichia coli LacZ gene inserted in the ICP6 locus, was derived from HSV-1 (39). D74/HveC rat glioma cells (25) were grown in complete DMEM supplemented with 7.5 μg/ml blasticidin S (Calbiochem/EMD Biosciences, La Jolla, CA).
Animal Studies.
Male Fischer 344 rats aged 6–8 weeks (Taconic Farms, Germantown, NY) were kept according to the guidelines of the Subcommittee on Research Animal Care of Massachusetts General Hospital. Tumors were implanted by using stereotaxy as described in ref. 25 and grown for 5 days before i.p. treatment with 80 mg/kg CPA (Bristol–Myers Squibb, Princeton, NJ) or 1 ml of PBS. Seven days after implantation, 2 × 107 pfu of hrR3 were injected into tumors by using the same procedure and stereotactic coordinates as for the tumor cells. Animals were euthanized at 6 and 72 h after viral injection or at partial or complete paralysis. Male C57BL/6J and IFN-γ−/− mice aged 6–8 weeks were obtained from The Jackson Laboratory (Bar Harbor, ME). Genomic DNA from tail clips was prepared by using a GenomicPrep Cells and Tissue DNA Isolation Kit (Amersham Pharmacia Biosciences, Piscataway, NJ) per the manufacturer's instructions. All animals were genotyped by using PCR primers oIMR126, oIMR127, oIMR128, and oIMR129 (The Jackson Laboratory) and injected stereotactically with 5 × 105 KR158ΔEGFR cells (2 mm lateral to bregma at a depth of 3 mm). Ten days after implantation, tumors were injected with 106 pfu of hrR3 or PBS. Animals were killed 72 h later, and tumor-bearing right hemispheres were excised for analysis.
Extracting and Storing Tissues.
Rodent brains were extracted and frozen in an isopentane dry-ice bath and then sectioned by cryostat to a thickness of 6 μm through the entire tumor volume. Every fifth section was analyzed. Tissue slides were dried overnight at room temperature, fixed in ice-cold acetone, and stored at −20°C for IHC analysis.
Evaluating HSV Distribution in Tumors.
To evaluate HSV propagation within tumors, every fifth slide of frozen brain tissue was rehydrated in PBS, stained with X-Gal, and counterstained with hematoxylin. MetaVue image analysis software (Universal Imaging, Downingtown, PA) was used to evaluate areas infected with virus on 10 slides, taking in the entire tumor.
IHC and ELISA.
Brain tissue slides were thawed and rehydrated for 5 min in PBS before staining. The central and peripheral areas of the tumor were analyzed for infiltration by macrophages as follows. Endogenous proteins and peroxidases were blocked with serum-free protein block (X0909) and peroxidase-blocking reagent (002428) (DAKO-Cytomation, Glostrup, Denmark), respectively. Sections were then incubated for 1 h at room temperature with the primary antibody: mouse anti-rat CD68 (MCA 341R), CD163 (MCA 342R), CD4 (MCA55R), CD8 (MCA48R), and CD43 (MCA54R) (Serotec, Oxford, U.K.); mouse anti-rat NKR-P1 (CL055AP) and CL083 (clone OX-62, CL083AP) (Cederlane Laboratories, Hornby, ON, Canada); and mouse anti-rat CD45R (14–0460) (eBioscience, San Diego, CA). This procedure was followed by incubation with an HRP-conjugated secondary antibody (ECL anti-mouse IgG, NA931V) (Amersham Pharmacia Biosciences, Buckinghamshire, U.K.). A DAKO Liquid DAB Substrate Chromogen System (catalog no. K3465; DAKO-Cytomation) was used for detection. The sections were counterstained with hematoxylin, dehydrated with increasing concentrations of ethanol, and fixed in xylene. To evaluate intratumoral levels of IFN-γ, the tumors were isolated from the rat brains and homogenized in 8 M urea solution containing 0.2 M NaCl and 0.1 M Tris base for protein extraction, and the proteins were analyzed by ELISA (8837315; eBioscience) per the manufacturer's directions.
Determining Density of NK Cells and Macrophages in Tumors.
We used a CAST image analysis system (Olympus, Albertslund, Denmark) mounted on an upright BX51 Olympus microscope with an integrated motorized stage (Prior Scientific, Rockland, MA) to outline the tumor region and count positively stained infiltrating cells. Cells were counted manually in a 21.8 × 21.8 × 16-μm counting frame by meander sampling, with step length determined to include equal numbers of samples containing at least 100 positive-stained cells for each area of tumor. Density of counted cells was calculated by analyzing the ratio of counted cells per area of tumor.
In Vitro Production of IFN-γ by NK Cells.
Human primary NK cells were enriched from peripheral blood leukopaks of healthy donors (American Red Cross, Columbus, OH) as described in ref. 26. Purified NK cells were first suspended in RPMI medium 1640/10% FCS (Invitrogen, Carlsbad, CA) in 96-well plates at a concentration of 2 × 106 per ml (total volume of 200 μl) with different dosages of freshly dissolved 4HC (Duke University Comprehensive Cancer Center, Durham, NC) and incubated at 37°C with 5% CO2 for 24 h. IL-12 (10 ng/ml; Genetics Institute, Cambridge, MA) and IL-18 (100 ng/ml; BASF Bioresearch, Worcester, MA) were added to each well in the two 96-well plates and incubated another 24 h. Cells from one plate were used to quantify cell survival by using a trypan blue exclusion assay at both 24-h and 48-h time points. Cell-free supernatants from the second plate were collected to detect IFN-γ protein by ELISA using commercially available mAb pairs (Endogen, Woburn, MA) according to the manufacturer's protocol.
Analyzing Gene Expression.
Total RNA from tumors was extracted with TRIzol reagent (15596-026; Invitrogen), following the manufacturer's guidelines. cDNA was synthesized from 2 μg of total RNA by using the Omniscript Reverse Transcriptase kit (205113; Qiagen, Valencia, CA). Quantitative real-time PCR was performed by using the ABI Prism 7900 HT Sequence detection system (Applied Biosystems, Foster City, CA). PCR mix included 25 μl of aqueous solution containing a 0.9 μM concentration of each primer, 0.2 μM 5′2′-chloro-7′-phenyl-1,4-dichloro-6-carboxyfluorescein-3′carboxytetramethylrhodamine-labeled probe, 12.5 μl of 2× Universal Master Mix (4304437; Applied Biosystems), and 5 μl of diluted cDNA. The PCR program included 1 cycle of 2 min at 50°C, 1 cycle of 10 min at 95°C, and 40 cycles consisting of 15 s at 95°C followed by 1 min at 60°C. Each gene was amplified in triplicate. β-Actin was used as an internal control. Relative quantification of gene expression was calculated as 2ΔCt test gene/2ΔCt actin, where Ct is the number of cycles for saturation and ΔCt is the difference between the number of cycles needed for expression of a gene in a tumor from untreated rats (used as baseline) and for expression from rats receiving treatment (HSV alone or HSV plus CPA). Primers and probes, designed with the Primer Express program (Applied Biosystems), were as follows: IFN-γ, forward, catggatgctatggaaggaaaga; reverse, attttcgtgttaccgtccttttg; probe, tcctcttggatatctggaggaactg; lacZ, forward, aatggctttcgctacctgga; reverse, ccatcgcgtgggcgta; probe, cgcccgctgatcctttgcga; β-actin, forward, ctacagatcatgtttgagaccttcaac; reverse, ccagaggcatacagggacaac; probe, cagccatgtacgtagccatccaggct.
MRI.
Rats were anesthetized with isoflurane (2.0% at 2 liters/min) and imaged by using a 4.7-T, 16-cm bore MRI system (Bruker Pharmascan, Billerica, MA). Rats were imaged before and after i.v. injection of MION 46 (10 mg/kg of body weight) and 24 h after injection. MRI included T2-weighted gradient echo sequences [repetition time (TR) = 600 ms, echo time (TE) = 6 ms, matrix (MTX) = 128 × 1,286, slice thickness = 0.8 mm, number of excitations (NEX) = 8, and field of view (FOV) = 3.5 cm] and multiple-slice multiple-echo sequences (TR = 2,000 ms, TE = 6.0–104.0 ms, MTX = 128 × 128, slice thickness = 0.8 mm, NEX = 8, and FOV = 3.5 cm). Regions of interest, including the tumor, contralateral brain tissue, and muscle, were selected by using an in-house program, CMIR-Image, to calculate macrophage content, which was scaled to contralateral brain and muscle.
Statistical Analysis.
Statistical analyses were performed with ANOVA tests, which were followed by means comparisons with post hoc Tukey's tests.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants P01 CA69246, NS41571, and R01 CA85139 (to E.A.C.) and CA95426, CA16058, and CA68458 (to M.A.C.).
Abbreviations
- CPA
cyclophosphamide
- 4HC
4-hydroxy-CPA
- MION
monocrystalline iron oxide particle
- OV
oncolytic virus
- NK
natural killer
- HSV
herpes simplex virus
- IHC
immunohistochemistry.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bischoff J. R., Kirn D. H., Williams A., Heise C., Horn S., Muna M., Ng L., Nye J. A., Sampson-Johannes A., Fattaey A., et al. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 2.Chiocca E. A. Nat. Rev. Cancer. 2002;2:938–950. doi: 10.1038/nrc948. [DOI] [PubMed] [Google Scholar]
- 3.Fulci G., Chiocca E. A. Front. Biosci. 2003;8:e346–e360. doi: 10.2741/976. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Manzano C., Balague C., Alemany R., Lemoine M. G., Mitlianga P., Jiang H., Khan A., Alonso M., Lang F. F., Conrad C. A., et al. Oncogene. 2004;23:1821–1828. doi: 10.1038/sj.onc.1207321. [DOI] [PubMed] [Google Scholar]
- 5.Jiang H., Gomez-Manzano C., Alemany R., Medrano D., Alonso M., Bekele B. N., Lin E., Conrad C. C., Yung W. K., Fueyo J. Neoplasia. 2005;7:48–56. doi: 10.1593/neo.04391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martuza R. L., Malick A., Markert J. M., Ruffner K. L., Coen D. M. Science. 1991;252:854–856. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 7.Chiocca E. A., Abbed K. M., Tatter S., Louis D. N., Hochberg F. H., Barker F., Kracher J., Grossman S. A., Fisher J. D., Carson K., et al. Mol. Ther. 2004;10:958–966. doi: 10.1016/j.ymthe.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Csatary L. K., Gosztonyi G., Szeberenyi J., Fabian Z., Liszka V., Bodey B., Csatary C. M. J. Neurooncol. 2004;67:83–93. doi: 10.1023/b:neon.0000021735.85511.05. [DOI] [PubMed] [Google Scholar]
- 9.Khuri F. R., Nemunaitis J., Ganly I., Arseneau J., Tannock I. F., Romel L., Gore M., Ironside J., MacDougall R. H., Heise C., et al. Nat. Med. 2000;6:879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 10.Lorence R. M., Pecora A. L., Major P. P., Hotte S. J., Laurie S. A., Roberts M. S., Groene W. S., Bamat M. K. Curr. Opin. Mol. Ther. 2003;5:618–624. [PubMed] [Google Scholar]
- 11.Markert J. M., Medlock M. D., Rabkin S. D., Gillespie G. Y., Todo T., Hunter W. D., Palmer C. A., Feigenbaum F., Tornatore C., Tufaro F., et al. Gene Ther. 2000;7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 12.Papanastassiou V., Rampling R., Fraser M., Petty R., Hadley D., Nicoll J., Harland J., Mabbs R., Brown M. Gene Ther. 2002;9:398–406. doi: 10.1038/sj.gt.3301664. [DOI] [PubMed] [Google Scholar]
- 13.Pecora A. L., Rizvi N., Cohen G. I., Meropol N. J., Sterman D., Marshall J. L., Goldberg S., Gross P., O'Neil J. D., Groene W. S., et al. J. Clin. Oncol. 2002;20:2251–2266. doi: 10.1200/JCO.2002.08.042. [DOI] [PubMed] [Google Scholar]
- 14.Andreansky S., He B., van Cott J., McGhee J., Markert J. M., Gillespie G. Y., Roizman B., Whitley R. J. Gene Ther. 1998;5:121–130. doi: 10.1038/sj.gt.3300550. [DOI] [PubMed] [Google Scholar]
- 15.Bennett J. J., Malhotra S., Wong R. J., Delman K., Zager J., St. Louis M., Johnson P., Fong Y. Ann. Surg. 2001;233:819–826. doi: 10.1097/00000658-200106000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todo T., Martuza R. L., Rabkin S. D., Johnson P. A. Proc. Natl. Acad. Sci. USA. 2001;98:6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abordo-Adesida E., Follenzi A., Barcia C., Sciascia S., Castro M. G., Naldini L., Lowenstein P. R. Hum. Gene Ther. 2005;16:741–751. doi: 10.1089/hum.2005.16.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balachandran S., Thomas E., Barber G. N. Nature. 2004;432:401–405. doi: 10.1038/nature03124. [DOI] [PubMed] [Google Scholar]
- 19.Balachandran S., Barber G. N. Cancer Cell. 2004;5:51–65. doi: 10.1016/s1535-6108(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 20.Friedman A., Tian J. P., Fulci G., Chiocca E. A., Wang J. Cancer Res. 2006;66:2314–2319. doi: 10.1158/0008-5472.CAN-05-2661. [DOI] [PubMed] [Google Scholar]
- 21.Lowenstein P. R. Mol. Ther. 2005;12:185–186. doi: 10.1016/j.ymthe.2005.06.439. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda K., Ichikawa T., Wakimoto H., Silver J. S., Deisboeck T. S., Finkelstein D., Harsh G. R., Louis D. N., Bartus R. T., Hochberg F. H., et al. Nat. Med. 1999;5:881–887. doi: 10.1038/11320. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K., Wakimoto H., Ichikawa T., Jhung S., Hochberg F. H., Louis D. N., Chiocca E. A. J. Virol. 2000;74:4765–4775. doi: 10.1128/jvi.74.10.4765-4775.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakimoto H., Johnson P. R., Knipe D. M., Chiocca E. A. Gene Ther. 2003;10:983–990. doi: 10.1038/sj.gt.3302038. [DOI] [PubMed] [Google Scholar]
- 25.Wakimoto H., Fulci G., Tyminski E., Chiocca E. A. Gene Ther. 2004;11:214–223. doi: 10.1038/sj.gt.3302143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore A., Weissleder R., Bogdanov A., Jr. J. Magn. Reson. Imaging. 1997;7:1140–1145. doi: 10.1002/jmri.1880070629. [DOI] [PubMed] [Google Scholar]
- 27.Trotta R., Parihar R., Yu J., Becknell B., Allard J., II, Wen J., Ding W., Mao H., Tridandapani S., Carson W. E., et al. Blood. 2005;105:3011–3018. doi: 10.1182/blood-2004-10-4072. [DOI] [PubMed] [Google Scholar]
- 28.Kirn D., Martuza R. L., Zwiebel J. Nat. Med. 2001;7:781–787. doi: 10.1038/89901. [DOI] [PubMed] [Google Scholar]
- 29.Cooper M. A., Fehniger T. A., Turner S. C., Chen K. S., Ghaheri B. A., Ghayur T., Carson W. E., Caligiuri M. A. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 30.Hickey W. F. Semin. Immunol. 1999;11:125–137. doi: 10.1006/smim.1999.0168. [DOI] [PubMed] [Google Scholar]
- 31.Bishop G. A., Marlin S. D., Schwartz S. A., Glorioso J. C. J. Immunol. 1984;133:2207–2214. [PubMed] [Google Scholar]
- 32.Bishop G. A., Kumel G., Schwartz S. A., Glorioso J. C. J. Virol. 1986;57:294–300. doi: 10.1128/jvi.57.1.294-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper A. M., Fehniger A. T., Caligiuri A. M. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 34.Bluman M. E., Bartynski J. K., Avalos R. B., Caligiuri A. M. J. Clin. Invest. 1996;97:2722–2727. doi: 10.1172/JCI118726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fehniger A. T., Herbein G., Yu H., Para I. M., Bernstein P. Z., O'Brien A. W., Caligiuri A. M. J. Immunol. 1998;161:6433–6438. [PubMed] [Google Scholar]
- 36.Guillemin G. J., Brew B. J. J. Leukoc. Biol. 2004;75:388–397. doi: 10.1189/jlb.0303114. [DOI] [PubMed] [Google Scholar]
- 37.Strik H. M., Stoll M., Meyermann R. Anticancer Res. 2004;24:37–42. [PubMed] [Google Scholar]
- 38.Kambara H., Saeki Y., Chiocca E. A. Cancer Res. 2005;65:11255–11258. doi: 10.1158/0008-5472.CAN-05-2278. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein D. J., Weller S. K. J. Virol. 1988;62:2970–2977. doi: 10.1128/jvi.62.8.2970-2977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.