Abstract
Uropathogenic Escherichia coli (UPEC) strain 536 (O6:K15:H31) is one of the model organisms of extraintestinal pathogenic E. coli (ExPEC). To analyze this strain's genetic basis of urovirulence, we sequenced the entire genome and compared the data with the genome sequence of UPEC strain CFT073 (O6:K2:H1) and to the available genomes of nonpathogenic E. coli strain MG1655 (K-12) and enterohemorrhagic E. coli. The genome of strain 536 is ≈292 kb smaller than that of strain CFT073. Genomic differences between both UPEC are mainly restricted to large pathogenicity islands, parts of which are unique to strain 536 or CFT073. Genome comparison underlines that repeated insertions and deletions in certain parts of the genome contribute to genome evolution. Furthermore, 427 and 432 genes are only present in strain 536 or in both UPEC, respectively. The majority of the latter genes is encoded within smaller horizontally acquired DNA regions scattered all over the genome. Several of these genes are involved in increasing the pathogens' fitness and adaptability. Analysis of virulence-associated traits expressed in the two UPEC O6 strains, together with genome comparison, demonstrate the marked genetic and phenotypic variability among UPEC. The ability to accumulate and express a variety of virulence-associated genes distinguishes ExPEC from many commensals and forms the basis for the individual virulence potential of ExPEC. Accordingly, instead of a common virulence mechanism, different ways exist among ExPEC to cause disease.
Keywords: fitness, genome comparison, uropathogenic Escherichia coli
Uropathogenic Escherichia coli (UPEC) are the most common cause of community-acquired urinary tract infection (UTI) and are responsible for 70–90% of the estimated 150 million UTIs diagnosed annually (1). UPEC also cause ≈40% of all nosocomial UTI, thus representing one of the most frequently isolated nosocomial pathogens (2). These frequencies illustrate the magnitude of the problem but do not reflect disease diversity in the urinary tract. UTI may be acute, symptomatic with a varying severity and localization, but may also be sporadic, recurrent, or chronic. It is essential to understand the molecular basis of disease diversity on the bacterial side that determines the different disease types. UPEC are a geno- and phenotypically heterogeneous group of isolates restricted to a small number of O-serogroups that seem to represent different subclasses of facultative pathogens (3–5).
UPEC virulence factors are frequently encoded on pathogenicity islands (PAIs) (6–9). The two O6 strains 536 (pyelonephritis isolate) and CFT073 (urosepsis isolate) became generally accepted UPEC model organisms, and several PAIs of them have been described in detail (10–16). The complete CFT073 genome sequence shows a mosaic structure in terms of the distribution of backbone genes conserved in E. coli, and “foreign” genes, which presumably have been acquired horizontally (17). Genome comparison of CFT073, O157:H7 strain EDL933, and K-12 strain MG1655 revealed that only 39.2% of their combined set of proteins are common to all three strains (17–20), underlining the astonishing diversity among E. coli. Furthermore, the genome sequence of CFT073 revealed 1,623 strain-specific genes (21.2%). Comparison of both UPEC phenotypes and their genomes with other complete E. coli genome sequences should therefore help to identify sets of “UPEC-specific” and strain-specific proteins, respectively, that may form the basis of their different individual phenotypes and uropathogenic potential.
Results and Discussion
E. coli 536 Genome Sequence Determination and Comparative Analysis.
The genome consists of a single circular chromosome of 4,938,875 bp. No plasmids were found. The 536 genome is 292 kb smaller than that of strain CFT073. Essentially, the additional DNA in CFT073 harbors genes of five cryptic prophages, which are absent from strain 536, as well as genes that are located in islands absent from other E. coli. The E. coli 536 genome contains one cryptic prophage region.
For the 536 genome, 4,747 putative coding sequences were predicted, ≈3,650 of which (77%) have highly similar orthologs in MG1655 (Fig. 1). From the remaining ORFs, 524 are also present in CFT073, which means that 89% of all ORFs of E. coli 536 have highly similar orthologs in the UPEC CFT073 genome (Table 1 and Table 4, which is published as supporting information on the PNAS web site). Further comparison with the genome sequences of enterohemorrhagic E. coli (EHEC) O157:H7 strains Sakai and EDL933 (18, 19) revealed ≈3,560 ORFs (75%) with highly similar orthologs in all published complete E. coli genomes. Of the remaining ORFs present in the genomes of strain 536 and at least one of these other four E. coli strains, 427 are mainly located within a region of the cryptic prophage or within the major PAIs of strain 536 (Table 5, which is published as supporting information on the PNAS web site).
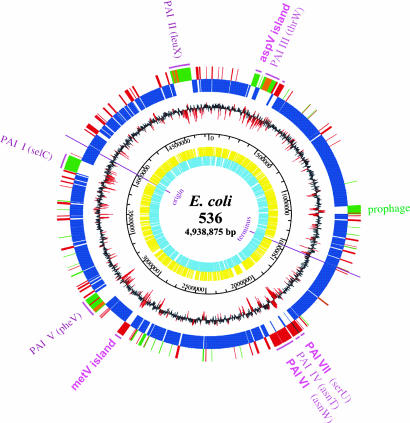
Fig. 1.
Genetic map of the UPEC strain 536 chromosome. The two inner circles represent all putative genes, depending on ORF orientation. The third circle from the center gives the scale. The fourth circle from the center shows the G + C distribution. Regions with a highly aberrant G + C content (greater than or less than 2-fold standard deviation) are highlighted in red. The outermost circles show the result of a three-way genome comparison with the UPEC strain CFT073 and E. coli MG1655 (K-12) genomes: blue, backbone genes found in all three strains; red, genes present in 536 and CFT073 but absent from MG1655; green, genes found in 536 only; orange, genes of 536, which are present in CFT073 but located in a different genomic region. Pathogenicity and genomic islands (GEIs) are highlighted; flanking tRNAs are given in brackets.
Table 1.
General features of the E. coli 536 genome compared with those of other sequenced E. coli strains
| Strains | Chromosome size, bp | Plasmids (bp) | No. of prophage-like elements | DNA coding sequence, % | No. of ORFs | C + G content, % | No. of rRNA genes | No. of tRNA genes | No. of predicted miscellaneous RNAs† | Backbone, %* | No. of strain-specific ORFs (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 536 (UPEC) (O6:K15:H31) | 4,938,875 | – | 1 | 88 | 4,747* | 50.5 | 22 | 81 | 45 | 77 | 374 (8) |
| CFT073 (UPEC) (O6:K2:H1) | 5,231,428 | – | 5 | 89 | 5,533 (5,379*) | 50.5 | 22 | 88 | 51 | 71 | 867 (16) |
| MG1655 (K-12) | 4,639,221 | – | 10 | 89 | 4,411 | 50.8 | 22 | 87 | 47 | 81 | 406 (9) |
| EDL933 (EHEC) (O157:H7) | 5,528,445 | pO157 (92,721) | 16 | 88 | 5,361 | 50.5 | 22 | 100 | 53 | 67 | 1,270 (24) |
| Sakai (EHEC) (O157:H7) | 5,498,450 | pO157 (92,721), pOSAKI (3,308) | 24 | 88 | 5,981 | 50.5 | 22 | 103 | 13 | ND | ND |
ND, not determined.
*Data were found by using the ORF prediction program YACOP.
†Data were found by using the Rfam database (www.sanger.ac.uk/Software/Rfam).
Key features of the completely sequenced E. coli genomes are summarized in Table 1. To visualize chromosomal regions of strain 536, which may contribute to urovirulence, a three-way comparison between the two UPEC genomes and the K-12 genome was done (Fig. 1), revealing a highly mosaic genome structure. Divergences from the mean G + C content are often found in genomic regions absent in strain MG1655. Genomic differences are not exclusively linked to the presence or absence of large PAIs. Instead, the presence of smaller gene clusters, often flanked by mobile elements (Fig. 1), seems to confer strain-specific traits.
Detection of UPEC-Specific DNA Regions in Pathogenic and Nonpathogenic E. coli.
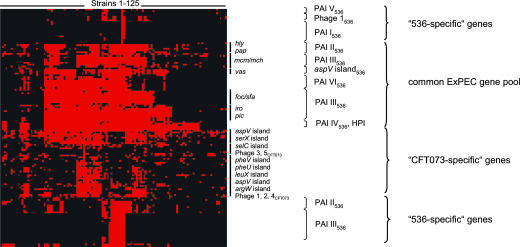
Distributional analyses of the chromosomal regions present in UPEC strain 536 and/or strain CFT073 but not in K-12 or both EHEC O157:H7 strains demonstrate that these DNA stretches are usually more frequently present in extraintestinal pathogenic E. coli (ExPEC) than in commensals and intestinal pathogenic E. coli (IPEC). DNA regions absent from IPEC, many of which represent phage or transposase associated DNA, are either specific for the respective UPEC strain or are absent from the other UPEC and the E. coli K-12 strain. According to hierarchical cluster analysis, a common UPEC gene pool can be detected that is widely distributed among other ExPEC and many fecal E. coli isolates of ECOR group B2 and D (Fig. 2 and Fig. 4, which is published as supporting information on the PNAS web site). These genes include the determinants coding for ExPEC virulence factors, such as α-hemolysin, microcins, P- and S-/F1C fimbriae, salmochelin, and autotransporter serine proteases. Two gene clusters encoding a putative polyketide and the yersiniabactin biosynthesis pathway, respectively, also belong to this gene pool. Additionally, a gene set present in strain 536 but not in CFT073 (and vice versa) can be distinguished. Many of these 536-specific genes, which are only present in a small subset of ExPEC, are localized on so-called PAI I, II, III and V, thus supporting the results of the genome comparison (Fig. 1). In contrast, the genes selected from various islands and phages of the CFT073 genome, which are absent from strain 536, can be detected among many ExPEC and fecal E. coli strains.
Fig. 2.
Distribution of UPEC strain 536 and CFT073-specific genes among 125 nonpathogenic and pathogenic E. coli isolates. Genes were grouped with the CLUSTER software based on the presence or absence of genes. Red and black denote the presence or absence of ORFs, respectively. Groups of genes belonging to distinct islands or phages are indicated. The genomic relationships of the 125 E. coli strains are indicated in Fig. 4.
PAIs Contribute to Virulence of Strain 536.
Four hundred and thirty-two genes of strain 536 are found in CFT073 but are absent from the two EHEC O157:H7 genomes and from E. coli K-12 (Table 5). These genes could contribute to urovirulence. Furthermore, 427 genes of E. coli 536 are absent from all published E. coli genomes completely sequenced so far, indicating their implication in an E. coli 536-specific phenotype (Table 6, which is published as supporting information on the PNAS web site). Many of these genes are organized in PAIs, which have been described in refs. 13 and 14.
The contribution of PAI I536–PAI V536 to this strain's virulence potential was analyzed in a murine model of ascending UTI (Table 2). The individual loss of PAI I, II, or III resulted in a 2–3 log increase of LD50. The impact of PAI loss seemed to be cumulative, because parallel deletion of PAI I and II further attenuated virulence. This phenomenon cannot entirely be explained with the loss of α-hemolysin (hly) determinants, because a less pronounced increase in LD50 was reported in case of isogenic hlyI, hlyII, and double hly mutants in the same experimental setup (21). These results confirm that other PAI-encoded factors affect in vivo virulence and support the establishment of UTI.
Table 2.
Impact of PAIs on virulence of strain 536 in infant murine model of ascending UTI
| Strain used for infection | LD50 |
|
|---|---|---|
| cfu/ml* | Change† | |
| 536 wild type | 4.89 × 103 | – |
| 536ΔPAI I (536-114) | 3.46 × 106 | 709 |
| 536ΔPAI II (536-225) | 4.57 × 106 | 935 |
| 536ΔPAI III | 2.23 × 106 | 456 |
| 536ΔPAI IV | 1.66 × 103 | 0.34 |
| 536ΔPAI V | 8.25 × 103 | 1.69 |
| 536ΔPAI IΔPAI II (536-2I) | >109 | >2 × 105 |
*Approximately 25 μl of inoculum was injected into the bladder of mice.
†Relative fold increase compared with the wild-type strain.
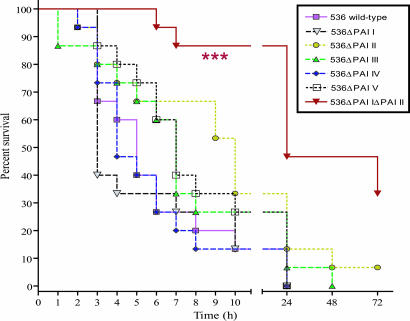
The contribution of PAIs to urosepsis was tested in a murine sepsis model (Fig. 3). Deletion of any single PAI did not significantly affect the survival curves. Simultaneous loss of PAI I and II, however, elicited significantly different survival (P < 0.001). A similar attenuation could be detected in case of isogenic hly mutants (G.N., unpublished work). Accordingly, most of these PAIs may be important for provoking an ascending UTI. However, the final fate of infection (urosepsis) is largely determined by a single PAI-encoded virulence trait, α-hemolysin. Acquisition of two hly determinants by strain 536 may be responsible for the higher i.v. virulence relative to CFT073 (Table 3).
Fig. 3.
Contribution of PAIs I536–V536 to i.v. virulence of UPEC strain 536. In two independent experiments, groups of 15 mice were infected with 5 × 108 cfu of strain 536 or its PAI deletion mutants. Survival curves were statistically analyzed by the log-rank test. ∗∗∗ indicates significant difference (P < 0.001).
Table 3.
Comparison of virulence-associated traits of UPEC O6 strains 536 and CFT073
| Characteristics | Strains |
|
|---|---|---|
| 536 (B2, ST 92; O6:K15:H31) | CFT073 (B2, ST 73; O6:K2:H1) | |
| Hemolysis | ++ | + |
| Smooth phenotype (long-chain LPS) | + | + |
| Capsule | + | + |
| Serum resistance | + | + |
| Motility | + | + |
| Adhesion to T24 cells | 139 ± 57 | 123 ± 51 |
| Adhesion to RT112 cells | 133 ± 56 | 76 ± 68 |
| Invasion rate into T24 cells, % | 3.2 ± 2.8 | 0.7 ± 0.6 |
| Invasion rate into RT112 cells, % | 1.7 ± 0.7 | 0.3 ± 0.3 |
| Enterochelin (Ent) | + | + |
| Salmochelin (Iro) | + | + |
| Salmonella iron transport (Sit) | – | + |
| Aerobactin (Iuc) | – | + |
| Yersiniabactin (Ybt) | + | −* |
| Hemin uptake system (Chu) | + | + |
| No. of putative fimbrial operons | 14 | 13 |
| No. of autotransporters | 9 | 11 |
| Biofilm formation (polyethylene) | + | + |
| Intravenous infection, cfu (LD50 values) | 2.6 × 108 | 6 × 108 |
Strains 536 and CFT073 belong to ECOR group B2. Sequence type (ST) was determined according to multi locus sequence typing (MLST) as described on the following web site: http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli/documents/primersColi_html. 06:K15:H31 and O6:K2:H1 are the serotypes of 536 and CFT073, respectively. For adhesion to both T24 and RT112 cells, bacteria were quantified in microtiter plate adhesion assays as adherent bacteria per well × 100. For the invasion rate into T24 and RT112 cells, bacteria were quantified as the percentage of the inoculum used for infection. LD50 values were determined as described in the text.
*The yersiniabactin determinant is nonfunctional because of frameshift mutations in several ORFs.
Competitiveness of strain 536 relative to strain MG1655 during intestinal growth was studied (Fig. 5A, which is published as supporting information on the PNAS web site). UPEC 536 outcompeted E. coli K-12 in the mouse intestine indicating that traits of strain 536 increase its competitiveness during intestinal growth relative to E. coli K-12. Outcompetition was not due to secreted factors of strain 536 or markedly different growth rates in vitro, because the ratio of UPEC versus K-12 was not significantly affected upon competitive growth in LB medium (Fig. 5B).
PAI I–III and V contain genes that either are absent in strain CFT073 [e.g., several adhesin gene clusters and the cdiAB genes required for contact-dependent growth inhibition of other bacteria (22)] or which are located in similar islands located elsewhere in the CFT073 genome. PAI IV is very similar in both UPEC strains.
Additional newly identified island-like regions >20 kb are described in the following paragraphs. Upstream of PAI III, an island-like region is associated with the aspV tRNA gene. This island is absent from CFT073 but present in the EHEC O157:H7 genomes (17–19) and in many ExPEC and fecal isolates (see also AY395687 in Fig. 2). It encodes 28 proteins, the majority of which (ECP0224–ECP0238) show similarity to components of the recently predicted virulence-associated secretion system of Vibrio cholerae required for this pathogen's cytotoxicity to macrophages (23). Some of the encoded proteins are similar to IcmF and IcmH, which are components of a type IV secretion system responsible for macrophage killing and intracellular survival of Legionella pneumophila (24). Other proteins are similar to Hcp, a secreted protein of V. cholerae, which is coordinately regulated with the hemolysin HlyA (25) or VgrG, both of which are effectors secreted by the virulence-associated secretion system pathway in V. cholerae (23). This partially conserved gene cluster was identified in several genomes, including Yersinia, Vibrio, and Salmonella species, designated IcmF-associated homologous proteins (IAHP) cluster (26). Many aspV island-encoded proteins have an equivalent on the functional level within the genome; the corresponding genes, although less conserved, are organized in an island-like region associated with the metV tRNA gene. However, its genetic structure differs from that of the aspV island, indicating that both islands have been acquired independently from different sources. In contrast to the aspV island, the metV-associated region is present in CFT073 (17). Whether the two IAHP clusters complement each other or represent independent secretion systems that may contribute to urovirulence must still be investigated.
Downstream of the asnW tRNA gene, the so-called PAI VI encodes several large proteins, eight of which represent nonribosomal hybrid peptide synthetases/polyketide synthases (NRPS/PKS), which are enzymatic systems responsible for the formation of various natural products (27). Polyketides and polypeptides have been shown to be involved in, e.g., intercellular communication, iron acquisition, competition, or self-defense (28–31) and may represent bacterial virulence factors (32). Their role during UTI is still unknown.
PAI VII, a small island directly downstream of the tRNA locus serU, mainly contains genes of unknown function, except for an ORF with similarity to a shufflon encoded on the plasmid R64, which is involved in the biological switch for generating alternative type IV pili (33), and an ORF that codes for a histone-like protein (34).
Comparison of Virulence-Related Traits of UPEC Strains 536 and CFT073.
Although both strains belong to the same serogroup, they differ in several virulence-associated traits that may also correlate with their pathogenic potential. The majority of classical UPEC virulence determinants is located on only five PAIs, and all major classes of virulence-associated factors (toxins, adhesins, siderophore systems, proteases, capsules, LPS) are expressed in both strains (Fig. 1 and Fig. 6, which is published as supporting information on the PNAS web site). Both isolates behave similarly with respect to motility, serum resistance, capsule and smooth LPS expression, biofilm formation, and adhesion and invasion into uroepithelial cell lines in vitro. They differ, however, in the presence or expression of α-hemolysin, adhesins, and siderophore systems (Table 3). The stronger hemolytic phenotype of strain 536 is due to the presence of two hly determinants, whereas only one allele exists in CFT073. The assortment of putative fimbrial determinants absent in E. coli K-12 differs markedly in both strains (Table 6). Strain 536, but not CFT073, contains several putative operons encoding Pix fimbriae and ETEC-like adhesins (F17-, CS12-, CS1-like), whereas CFT073 carries more copies of typical ExPEC adhesins (e.g., pap, type 1-like) in its genome. The functionality, receptor specificity and role during UTI of most of these adhesins are unknown. The amino acid sequences of FimH, the adhesin of type 1-fimbriae, are almost identical in both strains and exhibit several mutations considered as potentially pathoadaptive for UPEC (35, 36).
The availability of siderophore systems affects the virulence potential. Strain 536 expresses four different iron uptake systems whereas CFT073 expresses five (Table 3). The aerobactin genes and the “Salmonella iron transport locus” are only present in the CFT073 genome. PAI IV536, similar to the core region of the “high pathogenicity island” (HPI) of pathogenic Yersinia sp., encodes the yersiniabactin siderophore system (37, 38). Two of its proteins, HMWP1 (ECP1943) and HMWP2 (ECP1942), are nonribosomal peptide synthetases/polyketide synthases (NRPS/PKS) (30). Unlike in strain 536, both highly homologous ORFs have in-frame stop codons in CFT073. This finding corresponds to the absence of detectable yersiniabactin in this strain in contrast to strain 536 (data not shown).
Small Gene Clusters/Single Genes That May Contribute to Urovirulence.
Apart from large PAIs, other regions within the E. coli 536 genome were identified, that are absent from E. coli K-12. Tables 5 and 6 summarize the location and functional category of some gene(cluster)s found in such regions, some of which will be discussed below. The majority of the regions encode proteins, which have either an implication for the provision or modification of cell membrane/surface components or are involved in specialized metabolic activities.
Nine autotransporter genes exist in the E. coli 536 genome encoding antigen 43 variants and several other autotransporters with conserved pertactin domains and a serine protease. A similar set of 11 putative autotransporter genes can be predicted in the CFT073 genome, which includes two additional serine protease genes absent in strain 536. Two autotransporter genes of strain 536 (ECP0433, ECP1410) are disrupted by transposase genes in strain MG1655. ORF ECP1410 encodes a putative 250-kDa protein, which contains several cell–cell adhesion domains. Several other ORFs are disrupted in MG1655 but are intact in strain 536, e.g., ORF ECP1994, a TonB-dependent receptor with similarity to outer membrane receptors for ferrienterochelin and colicins, and ECP2376, an outer membrane usher protein located within a region encoding fimbrial genes.
Specialized metabolic activities may also support virulence. d-serine catabolism, carried out by a d-serine deaminase system, composed of the deaminase, a transcriptional activator, and a putative d-serine permease, provides a growth advantage in the murine urinary tract (39). This locus is intact in UPEC strain CFT073 but disrupted in strain MG1655, suggesting that UPEC not only survive in the presence of d-serine but also use it as a N and C source in the urinary tract. Examining the CFT073-homologous genomic region in strain 536, only a truncated cluster could be found, replaced partly by a sucrose utilization gene cluster. Unlike in CFT073, a full-length d-serine deaminase cluster is present within PAI II536, which may indicate that the ability to catabolize d-serine is indeed of essential benefit in the course of a UTI.
Two sucrose utilization systems absent from CFT073 and MG1655 can be found in strain 536. The genes of a phosphotransferase system (PTS)-independent cluster (ECP2386–ECP2389) replace the permease and regulator genes of the above-mentioned d-serine deaminase cluster. The PTS-independent system is composed of a sucrose permease, a fructokinase, a sucrase, and a transcriptional regulator. Their genes are chromosomally inserted at the tRNA gene argW. This operon is located at the same chromosomal locus in both EHEC O157:H7 genomes. The second sucrose utilization operon encodes a PTS-dependent system (ECP2750–ECP2754), which is composed of a fructokinase, a sucrose porin, a sucrase, the sucrose-specific IIBC component of PTS, and a repressor. This system is absent from all, thus far, completely sequenced E. coli strains and has been reported to be plasmid-encoded in Salmonella sp. and some E. coli isolates (40, 41). A similar PTS-dependent utilization system specific for l-sorbose (ECP4233–ECP4239) is present in 536 and other pathogenic E. coli. It is composed of seven genes that catalyze the uptake and conversion of l-sorbose to d-fructose-6-phosphate (42).
Four additional regions encoding sugar utilization systems have been detected that occur only in pathogenic E. coli but that are absent from MG1655: the aga (ECP3221–ECP3233) and deo (ECP2982–ECP2985) operons required for N-acetylgalactosamine and deoxyribose utilization, respectively (43, 44). Two other systems (ECP3754–ECP3761 and ECP4086–ECP3793) still have unclear specificities. The first system may be involved in PTS-dependent fructose utilization.
Several genes present in UPEC, but not in K-12, may be involved in pH homeostasis. The pH of urine usually tends to be rather acidic (pH 4.6–8.0). To respond to acidic conditions, there are several systems available in E. coli 536. Two additional Na+/H+ antiporters, ECP4687 and ECP4615, could be involved in regulating the intracellular H+ concentration. The latter gene is located within PAI II536, however, it is absent from other E. coli strains. Lysine decarboxylase can be involved in acid tolerance by consuming protons and neutralizing the acidic byproducts of carbohydrate fermentation (45). Three copies, each composed of the decarboxylase gene and a lysine/cadaverine antiporter, exist in the E. coli 536 genome, one of which is located within PAI III (ECP327–ECP328). An arginine catabolism system (ECP4496–ECP4500), also found in CFT073, could have a similar role in pH homeostasis, because it has been shown for the oral pathogen Streptococcus gordonii (46). The cluster contains five genes whose products catalyze the conversion of arginine to ornithine by using arginine deiminase and ornithine carbamoyltransferase. An arginine/ornithine antiporter guarantees the interchangeability of substrate and product. In addition to its role in pH homeostasis, the system can also be important for substrate-level ATP synthesis during anaerobic arginine fermentation. Carbamoyl phosphate, the byproduct of the above-mentioned ornithine carbamoyltransferase reaction, is converted to NH3 and CO2 catalyzed by carbamate kinase, also encoded within the cluster. An additional system with implications in substrate-level ATP synthesis preferable under anaerobic conditions could be the 2-oxoglutarate degradation cluster (ECP4274–ECP4282). ATP is generated in the succinyl-CoA synthetase reaction. The cluster also contains the genes of a two-component system putatively responding to C4-dicarboxylates and a dicarboxylate transporter. Another C4-dicarboxylate transport system (ECP4684–ECP4686), absent in strain MG1655, could be found elsewhere in the E. coli 536 genome. The relationship between virulence and functionality of these regions still needs to be elucidated.
Genome Plasticity Is Responsible for Phenotypic Diversity and Evolution of E. coli.
E. coli is a variable species because its genome is highly dynamic (47), and thus, the pathogenic strains associated with human diseases are remarkably diverse (48). Comparative genome analysis with both UPEC O6 strains revealed that horizontal gene transfer, gene loss, and insertion sequence element-mediated chromosomal rearrangements play important roles for their evolution and demonstrated that PAIs are seldom fixed but rather bear the potential for ongoing rearrangements, deletions, and insertions (Fig. 1). Accordingly, genome evolution in these bacteria cannot be simply described by a “backbone and flexible gene pool” model, but must also be described by repeated insertions and deletions occurring in certain parts of the genome reminiscent of palimpsests, i.e., antique writings in which text parts were repeatedly deleted and overwritten. An interesting example represents the transposon-like 15.5-kb region (ECP4593–ECP4612) within PAI II which is flanked by IS629-related sequences and comprises several genes with homology to the pdu operon of Salmonella enterica sv. typhimurium involved in coenzyme B12-dependent 1,2-propanediol catabolism. In Salmonella, the pdu operon is contiguous and coregulated with the cobalamin (B12) biosynthesis cob operon (49). The pdu/cob genes were lost by a common ancestor of E. coli and S. enterica and reintroduced into Salmonellae by horizontal gene transfer (50, 51). PAI II contains eight ORFs with homology to different genes of the pdu gene cluster. Propanediol degradation is an important C source for Salmonella in anaerobic environments, e.g., the large intestine, because propanediol is produced by fermentation of the plant sugars rhamnose and fucose. In murine models, pdu gene cluster expression affects growth in host tissues and contributes to virulence (52, 53). It remains to be shown whether strain 536 has become able to metabolize 1,2-propanediol upon PAI II acquisition.
Although the CFT073 genome is ≈292 kb larger than that of strain 536, much of the additional genes seem to be of no virulence function, because >50% of them are located within cryptic prophages. In addition, 470 genes of E. coli 536 are absent from CFT073 and MG1655, indicating that individual differences between both UPEC in their potential to cause disease and in the severity of the UTI caused are not due to a simple gene loss or gain, respectively. Rather, they seem to be the result of using common as well as strain-specific gene sets (Fig. 2). Accumulation of three serine protease autotransporters of Enterobacteriaceae (SPATE) and the presence of the aerobactin and two P fimbrial determinants, coding for two well known major UPEC virulence factors (54), in CFT073 may be responsible for this strain's higher virulence potential relative to strain 536. Furthermore, differences between 536 and CFT073 are not exclusively restricted to large islands but also to a considerable number of smaller gene clusters coding for integrative functions such as nutrient utilization or surface modification. Without doubt, large PAIs are of major importance in conferring a virulent phenotype, however, the smaller horizontally acquired clusters are of additional benefit in the course of an infection.
No clear distinction can be drawn between ExPEC and commensal E. coli (55). ExPEC can stably colonize the host intestine and are predominant in ≈20% of healthy people (54). In contrast to IPEC, host acquisition of an ExPEC is not sufficient to cause infection. Instead, bacteria have to reach an extraintestinal site of the host. Because colonizing sites outside the gut are unlikely to provide a selective advantage in terms of transmissibility, so-called “extraintestinal virulence factors” probably evolved to enhance survival in the gut and/or transmission between hosts, and therefore will be shared with at least some commensal strains. The ability of strains to “accumulate” such fitness traits directs their virulence potential. This is corroborated by comprehensive analysis of the contribution of five PAIs to urovirulence of strain 536, indicating that each island increases the strain's adaptability and fitness in the urinary tract (Table 2). ExPEC probably have to possess specific genes to outcompete commensals during intestinal colonization (Fig.5) and to adapt to different specific niches encountered during infection. Sugar catabolism is important for intestinal colonization by and maintenance of E. coli. Cocolonization by different E. coli strains is possible if strains have different preferences for nutrients and/or the ability to switch to an alternative nutrient source (56). After colonization of the gastrointestinal tract, UPEC may be able to cause extraintestinal infection. Here again, metabolic factors might be implicated as suggested by a recent study (39).
Comparison of the available complete genome sequences reveals insights into the genetic diversity underlying the phenotypic diversity of E. coli in general and of E. coli serogroup O6 in particular. The E. coli serogroup O6 is very heterogeneous and includes commensal and IPEC isolates but is also one of the most common serogroups among UPEC (57–59). The presence of several IPEC virulence-associated genes, e.g., the virulence-associated secretion system pathway also found in EHEC as well as putative fimbrial determinants in strain 536 with homology to those known for ETEC, may be reminiscent of the fact that ETEC and verotoxigenic E. coli O6 strains are also frequent causative agents of diarrhea (60). It could thus be hypothesized that UPEC strain 536 may represent an evolutionary state indicative of divergent lineages among E. coli O6, either toward a “true” intestinal pathogen or toward a “true” extraintestinal pathogen.
Materials and Methods
Genome Sequencing.
Details of the sequencing strategy, gene prediction, and annotation may be found in the Supporting Text, which is published as supporting information on the PNAS web site. The genome sequence has been deposited in the GenBank database (accession no. CP000247).
Comparative Genomics.
For comparative analysis, each ORF of E. coli 536 was searched against all ORFs of E. coli CFT073, EDL933, Sakai, and MG1655 by using the blast tool. Orthologous proteins were defined with an amino acid identity of >90% over >90% of query and reference sequence. The presence of 130 PAI- or bacteriophage-associated genes of UPEC strain 536 and/or CFT073 was analyzed by PCR in 125 ExPEC, IPEC, and nonpathogenic fecal E. coli isolates.
Virulence Tests.
Invasion and adherence assays were performed as described in refs. 61 and 62. Biofilm formation was measured after 48 h of growth at 20°C in polyethylene microtiter plates (63). Motility at 37°C was analyzed on LB plates with 0.3% agar. Expression of α-hemolysin was tested at 37°C on LB agar with 5% human blood. Bacterial resistance against human blood serum was tested as described in ref. 64. The infant mouse model of ascending UTI was used as described in ref. 65. In the murine sepsis model, female 8-week-old Naval Medical Research Institute (NMRI) mice were intravenously infected in two independent experiments. For details, see the Supporting Text.
Supplementary Material
Acknowledgments
We thank B. Plaschke (Institute for Molecular Biology of Infectious Diseases, Würzburg, Germany), I. Decker (Göttingen Genomics Laboratory, Göttingen, Germany), and M. Grzywa (Göttingen, Genomics Laboratory, Göttingen, Germany) for excellent technical assistance and H. Mobley (University of Michigan Medical School, Ann Arbor, MI) for the gift of UPEC strain CFT073. The Würzburg group was supported by the BFS (AZ 547/02) and the Deutsche Forschungsgemeinschaft (SFB479, TP A1). The Göttingen group was supported by a grant from the Niedersächsisches Ministerium für Wissenschaft und Kultur. L.E. and G.N. were supported by Hungarian Scientific Research Fund (OTKA) Grants T037833 and F048526. Biomax Informatics AG was supported by the BFS (AZ 547/02). We acknowledge the support of W. Rabsch (Robert Koch Institute, Wernigerode, Germany) and S. Schubert (Max von Pettenkofer Institute, München, Germany) with respect to the analysis of yersiniabactin expression of the E. coli strains 536 and CFT073.
Abbreviations
- EHEC
enterohemorrhagic E. coli
- ExPEC
extraintestinal pathogenic E. coli
- IPEC
intestinal pathogenic E. coli
- PAI
pathogenicity islands
- PTS
phosphotransferase system
- UPEC
uropathogenic E. coli
- UTI
urinary tract infection.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The complete genome sequence of E. coli strain 536 has been deposited in the GenBank database (accession no. CP000247).
References
- 1.Stamm W. E., Norrby S. R. J. Infect. Dis. 2001;183:S1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 2.Struelens M. J., Denis O., Rodriguez-Villalobos H. Microbes Infect. 2004;6:1043–1048. doi: 10.1016/j.micinf.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J. R., Delavari P., Kuskowski M., Stell A. L. J. Infect. Dis. 2001;183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 4.Bingen-Bidois M., Clermont O., Bonacorsi S., Terki M., Brahimi N., Loukil C., Barraud D., Bingen E. Infect. Immun. 2002;70:3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahrani-Mougeot F., Gunther N. W., IV, Donnenberg M. S., Mobley H. L. T. In: Escherichia coli: Virulence Mechanisms of a Versatile Pathogen. Donnenberg M. S., editor. San Diego: Academic; 2002. pp. 239–268. [Google Scholar]
- 6.Dobrindt U., Hochhut B., Hentschel U., Hacker J. Nat. Rev. Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 7.Hacker J., Hentschel U., Dobrindt U. Science. 2003;301:790–793. doi: 10.1126/science.1086802. [DOI] [PubMed] [Google Scholar]
- 8.Blum G., Ott M., Lischewski A., Ritter A., Imrich H., Tschäpe H., Hacker J. Infect. Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swenson D. L., Bukanov N. O., Berg D. E., Welch R. A. Infect. Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobley H. L., Green D. M., Trifillis A. L., Johnson D. E., Chippendale G. R., Lockatell C. V., Jones B. D., Warren J. W. Infect. Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao J. S., Stucker D. M., Warren J. W., Mobley H. L. Infect. Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyer D. M., Kao J. S., Mobley H. L. Infect. Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrindt U., Blum-Oehler G., Nagy G., Schneider G., Johann A., Gottschalk G., Hacker J. Infect. Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider G., Dobrindt U., Brüggemann H., Nagy G., Janke B., Blum-Oehler G., Buchrieser C., Gottschalk G., Emődy L., Hacker J. Infect. Immun. 2004;72:5993–6001. doi: 10.1128/IAI.72.10.5993-6001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasko D. A., Phillips J. A., Li X., Mobley H. L. J. Infect. Dis. 2001;184:1041–1049. doi: 10.1086/323602. [DOI] [PubMed] [Google Scholar]
- 16.Hacker J., Knapp S., Goebel W. J. Bacteriol. 1983;154:1145–1152. doi: 10.1128/jb.154.3.1145-1152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch R. A., Burland V., Plunkett G., III, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., et al. Proc. Natl. Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perna N. T., Plunkett G., III, Burland V., Mau B., Glasner J. D., Rose D. J., Mayhew G. F., Evans P. S., Gregor J., Kirkpatrick H. A., et al. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., Han C. G., Ohtsubo E., Nakayama K., Murata T., et al. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 20.Blattner F. R., Plunkett G., III, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 21.Nagy G., Altenhöfer A., Knapp O., Maier E., Dobrindt U., Blum-Oehler G., Benz R., Emődy L., Hacker J. Microbes Infect. 2006 doi: 10.1016/j.micinf.2006.02.029. in press. [DOI] [PubMed] [Google Scholar]
- 22.Aoki S. K., Pamma R., Hernday A. D., Bickham J. E., Braaten B. A., Low D. A. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 23.Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. Proc. Natl. Acad. Sci. USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell M., Shuman H. A. Infect. Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams S. G., Varcoe L. T., Attridge S. R., Manning P. A. Infect. Immun. 1996;64:283–289. doi: 10.1128/iai.64.1.283-289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S., Chaudhuri K. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- 27.Staunton J., Weissman K. J. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 28.Silakowski B., Kunze B., Nordsiek G., Blöcker H., Höfle G., Müller R. Eur. J. Biochem. 2000;267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 29.Paitan Y., Alon G., Orr E., Ron E. Z., Rosenberg E. J. Mol. Biol. 1999;286:465–474. doi: 10.1006/jmbi.1998.2478. [DOI] [PubMed] [Google Scholar]
- 30.Miller D. A., Luo L., Hillson N., Keating T. A., Walsh C. T. Chem. Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 31.August P. R., Tang L., Yoon Y. J., Ning S., Muller R., Yu T. W., Taylor M., Hoffmann D., Kim C. G., Zhang X., Hutchinson C. R., Floss H. G. Chem. Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 32.George K. M., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., Small P. L. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida T., Furuya N., Ishikura M., Isobe T., Haino-Fukushima K., Ogawa T., Komano T. J. Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson H. S., Free A. Mol. Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 35.Hommais F., Gouriou S., Amorin C., Bui H., Rahimy M. C., Picard B., Denamur E. Infect. Immun. 2003;71:3619–3622. doi: 10.1128/IAI.71.6.3619-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokurenko E. V., Chesnokova V., Dykhuizen D. E., Ofek I., Wu X. R., Krogfelt K. A., Struve C., Schembri M. A., Hasty D. L. Proc. Natl. Acad. Sci. USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carniel E. Microbes Infect. 2001;3:561–569. doi: 10.1016/s1286-4579(01)01412-5. [DOI] [PubMed] [Google Scholar]
- 38.Buchrieser C., Rusniok C., Frangeul L., Couve E., Billault A., Kunst F., Carniel E., Glaser P. Infect. Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roesch P. L., Redford P., Batchelet S., Moritz R. L., Pellett S., Haugen B. J., Blattner F. R., Welch R. A. Mol. Microbiol. 2003;49:55–67. doi: 10.1046/j.1365-2958.2003.03543.x. [DOI] [PubMed] [Google Scholar]
- 40.Titgemeyer F., Jahreis K., Ebner R., Lengeler J. W. Mol. Gen. Genet. 1996;250:197–206. doi: 10.1007/BF02174179. [DOI] [PubMed] [Google Scholar]
- 41.Hardesty C., Ferran C., DiRienzo J. M. J. Bacteriol. 1991;173:449–456. doi: 10.1128/jb.173.2.449-456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wehmeier U. F., Lengeler J. W. Biochim. Biophys. Acta. 1994;1208:348–351. doi: 10.1016/0167-4838(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 43.Brinkkötter A., Klöß H., Alpert C., Lengeler J. W. Mol. Microbiol. 2000;37:125–135. doi: 10.1046/j.1365-2958.2000.01969.x. [DOI] [PubMed] [Google Scholar]
- 44.Bernier-Febreau C., du Merle L., Turlin E., Labas V., Ordonez J., Gilles A. M., Le Bouguenec C. Infect. Immun. 2004;72:6151–6156. doi: 10.1128/IAI.72.10.6151-6156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park Y. K., Bearson B., Bang S. H., Bang I. S., Foster J. W. Mol. Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 46.Dong Y., Chen Y. Y., Snyder J. A., Burne R. A. Appl. Environ. Microbiol. 2002;68:5549–5553. doi: 10.1128/AEM.68.11.5549-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobrindt U., Agerer F., Michaelis K., Janka A., Buchrieser C., Samuelson M., Svanborg C., Gottschalk G., Karch H., Hacker J. J. Bacteriol. 2003;185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaper J. B., Nataro J. P., Mobley H. L. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 49.Bobik T. A., Ailion M., Roth J. R. J. Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bobik T. A., Havemann G. D., Busch R. J., Williams D. S., Aldrich H. C. J. Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence J. G., Roth J. R. Genetics. 1996;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heithoff D. M., Conner C. P., Hentschel U., Govantes F., Hanna P. C., Mahan M. J. J. Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conner C. P., Heithoff D. M., Mahan M. J. Curr. Top. Microbiol. Immunol. 1998;225:1–12. doi: 10.1007/978-3-642-80451-9_1. [DOI] [PubMed] [Google Scholar]
- 54.Johnson J. R. Clin. Microbiol. Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grozdanov L., Raasch C., Schulze J., Sonnenborn U., Gottschalk G., Hacker J., Dobrindt U. J. Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., Anderson A. B., Grissom J. E., Laux D. C., Cohen P. S., Conway T. Proc. Natl. Acad. Sci. USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blum G., Marre R., Hacker J. Infection. 1995;23:234–236. doi: 10.1007/BF01781204. [DOI] [PubMed] [Google Scholar]
- 58.Bettelheim K. A. In: Escherichia coli: Mechanisms of Virulence. Sussman M., editor. Cambridge, U.K.: Cambridge Univ. Press; 1997. pp. 85–109. [Google Scholar]
- 59.Ørskov I., Ørskov F., Jann B., Jann K. Bacteriol. Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sussman M. In: Escherichia coli: Mechanisms of Virulence. Sussman M., editor. Cambridge, U.K.: Cambdrige Univ. Press; 1997. pp. 3–48. [Google Scholar]
- 61.Altenhoefer A., Oswald S., Sonnenborn U., Enders C., Schulze J., Hacker J., Ölschläger T. A. FEMS Immunol. Med. Microbiol. 2004;40:223–229. doi: 10.1016/S0928-8244(03)00368-7. [DOI] [PubMed] [Google Scholar]
- 62.Hess P., Altenhöfer A., Khan A. S., Daryab N., Kim K. S., Hacker J., Ölschläger T. A. Infect. Immun. 2004;72:5298–5307. doi: 10.1128/IAI.72.9.5298-5307.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Toole G. A., Kolter R. Mol. Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 64.Ritter A., Blum G., Emődy L., Kerenyi M., Bock A., Neuhierl B., Rabsch W., Scheutz F., Hacker J. Mol. Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 65.Nagy G., Dobrindt U., Schneider G., Khan A. S., Hacker J., Emődy L. Infect. Immun. 2002;70:4406–4413. doi: 10.1128/IAI.70.8.4406-4413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.