Abstract
Heme, a major iron source, is transported through the outer membrane of Gram-negative bacteria by specific heme/hemoprotein receptors and through the inner membrane by heme-specific, periplasmic, binding protein-dependent, ATP-binding cassette permeases. Escherichia coli K12 does not use exogenous heme, and no heme uptake genes have been identified. Nevertheless, a recombinant E. coli strain expressing just one foreign heme outer membrane receptor can use exogenous heme as an iron source. This result suggests either that heme might be able to cross the cytoplasmic membrane in the absence of specific carrier or that there is a functional inner membrane heme transporter. Here, we show that to use heme iron E. coli requires the dipeptide inner membrane ATP-binding cassette transporter (DppBCDF) and either of two periplasmic binding proteins: MppA, the l-alanyl-γ-d-glutamyl-meso-diaminopimelate binding protein, or DppA, the dipeptide binding protein. Thus, wild-type E. coli has a peptide/heme permease despite being unable to use exogenous heme. DppA, which shares sequence similarity with the Haemophilus influenzae heme-binding protein HbpA, and MppA are functional heme-binding proteins. Peptides compete with heme for binding both “in vitro” and “in vivo.”
Keywords: heme uptake, peptide and heme competition, peptide uptake, ATP-binding cassette
Heme is abundant in all organisms. In addition to its role in vital reactions, it serves as a major iron source, mostly for pathogenic microorganisms (1). Bacteria internalize intact heme into the cytosol to retrieve the iron for use as a cofactor (2). Heme is transported through the Gram-negative outer membrane by specific receptors. These receptors belong to a family of outer membrane proteins involved in the uptake of siderophores, vitamin B12 and transferrin/lactoferrin iron. They require the proton motive force driven by the TonB-ExbB-ExbD inner membrane protein complex for activity (3). All characterized Gram-negative bacterial heme receptors share sequence similarities (25–90% identity) and have two conserved histidine residues involved in heme binding (4). Most heme receptors present specificity either for free heme or a protein carrying heme such as hemoglobin, hemoglobin-haptoglobin complex, hemopexin, or hemophore. However, the specificity for one type of heme-containing molecule is not absolute, and several hemoprotein receptors also bind heme directly (5).
Once in the periplasm, heme is transported through the inner membrane by permeases. Heme permeases have the overall molecular organization common to all binding protein-dependent, bacterial ATP-binding cassette (ABC) importers, including (i) a soluble periplasmic substrate-binding protein (PBP), (ii) two membrane-associated ABC modules, and (iii) two transmembrane modules. These modules can be either a single polypeptide or composed of homodimeric or heterodimeric subunits. The set of transmembrane- and membrane-associated ABC modules is called the ABC transporter. In association with a PBP, it forms a complete uptake system (permease) (see ref. 6 for a review).
All known heme permeases are similar in sequence and organization to the first heme permease identified, that from Yersinia enterocolitica comprising one heme-binding protein (HemT), one transmembrane protein (HemU), and one ABC protein (HemV) (7). By analogy with other Gram-negative permeases, it is likely (but not demonstrated) that the transporter is made up of a homodimer of each HemV and HemU.
In most species, the heme permeases are encoded by genes clustered in an operon. The operon is repressed by the iron-loaded Fur protein, which binds to a conserved operator sequence in the promoter region, named the Fur box (8).
Many bacteria have several heme outer membrane receptors with, in some cases, partially redundant functions and, thus, can exploit the diversity of potential heme sources. In contrast, most species apparently have only a single heme permease for collecting free heme in the periplasm. The heme permease operon often includes a gene encoding one heme outer membrane receptor (5). Inactivation of one outer membrane heme receptor gene does not generally lead to the loss of heme utilization as an iron source because of the redundancy of these genes (7). Unexpectedly, mutations in the single heme-specific permease genes only reduce, but do not abolish, heme utilization as an iron source in Y. enterocolitica, Yersinia pestis, and Vibrio cholerae (2, 9, 10). These results suggest the existence in these species of another unidentified heme permease.
Two other observations further support this hypothesis. Neisseria and Haemophilus species have outer membrane heme/hemoprotein receptors and use heme as an iron source (11, 12). Nevertheless, BLAST searches of their genomes for homology with hemTUV genes did not reveal any homologous genes (13). Also, expression of a foreign outer membrane heme receptor (the Serratia marcescens HasR receptor, the Y. enterocolitica HemR receptor, or the Shigella dysenteria ShuA receptor) in Escherichia coli K12 is sufficient to allow the use of heme as an iron source, despite the E. coli genome containing no genes homologous to hemTUV (2, 14, 15).
We show here that heme uptake through the E. coli K12 inner membrane requires a permease made up of DppBCDF (the dipeptide inner membrane transporter) as the ABC transporter and either MppA (the l-alanyl-γ-d-glutamyl-meso-diaminopimelate periplasmic binding protein) or DppA (the dipeptide periplasmic protein) as the PBP. We show that both MppA and DppA are able to bind heme “in vitro.” Possibly, the heme permease in bacteria naturally unable to use exogenous heme is involved in endogenous periplasmic heme recycling. The Dpp permease may also allow pathogens that lack the heme-specific HemTUV type permease to use exogenous heme as an iron source.
Results
Transposon Mutagenesis and Isolation of Mutants Unable to Use Heme as an Iron Source.
We used an E. coli recombinant strain expressing the S. marcescens outer membrane receptor, HasR, allowing use of exogenous heme as an iron source. We searched for mutants of this strain that had lost this ability.
To optimize the utilization of heme as an iron source, the strain FB827 was used because it carries the entF::Tn10 mutation to suppress enterobactin production, making the strain more sensitive than the FB8 entF+ parental strain to iron chelation. FB827 carrying a control plasmid (pAM 238) or a plasmid encoding the S. marcescens HasR receptor (pAM 238-hasR) did not grow on minimal medium not supplemented with iron (M63*) containing 100 μM 2,2′dipyridyl (Dip) to chelate any residual iron (M63* Dip). When hemoglobin was added to the medium to 40 μM [M63*, Dip, bovine hemoglobin (Hb)], strain FB827 (pAM 238-hasR) grew, but FB827(pAM 238) did not. Growth was dependent on the hemoglobin concentration and could be detected down to 4 μM (data not shown). Bovine hemoglobin is more soluble and easier to handle than heme and was used in most experiments: Similar results were obtained with heme and hemoglobin (data not shown). Strain FB827 (pAM 238-hasR) was mutagenized with TnSC189 and plated on LB plates with appropriate antibiotics, and 10,000 colonies were replicated onto M63 media and then onto M63*, Dip, Hb plates with appropriate antibiotics. Mutants that grew on the first, but not on the latter, were studied. One expected class carrying transposon insertions in hasR on the plasmid was found and discarded. All chromosomal transposon insertions were P1 transduced into FB827 (pAM 238-hasR) with selection for the transposon-linked antibiotic resistance to test whether the inability to grow on M63* Glu, Dip, Hb plates was genetically linked to the transposon. Only one isolate was found for which such linkage was the case, indicating that our transposon mutagenesis had not saturated the genome. In addition, there may be a selection against clones with poor growth on microplates. The precise site of the insertion was determined by DNA sequencing with transposon-specific primers: It was within the dppF gene. dppF encodes one ABC protein subunit of the dipeptide permease (16). We therefore tested whether the dipeptide permease is involved in iron heme utilization.
Insertion in Each dppB, dppC, dppD, and dppF, but Not in dppA, Abolishes Use of Heme as an Iron Source.
Mutants carrying kanamycin (Km)-cassette insertions into dppA, dppB, dppC, or dppD were transduced into FB827 (pAM 238-hasR), and the resulting strains were tested for growth on M63*, Dip, Hb plates. Mutations in each dppB, dppC, dppD, and dppF abolished heme utilization (Table 1). The dpp genes are in a single operon, and, therefore, mutations in dppB, dppC, and dppD could have a polar effect on downstream genes. FB827 (pAM 238-hasR) carrying either dppB::Km or dppC::Km or dppD::Km were transformed with plasmids carrying the downstream genes dppC-dppD-dppF or dppD-dppF or dppF, respectively, expressed under the control of the lac promoter. None of these constructs restored growth, whereas a plasmid encoding the whole operon (pTRC 99 dppA-dppB-dppC-dppD-dppF) complemented all of the mutants for iron heme utilization (Table 1). These results show that DppB, DppC, and DppD and DppF are each individually required for iron heme utilization and are constituents of the E. coli heme ABC transporter. The complete loss of growth of the dpp mutants on M63*, Dip, Hb demonstrates that the Dpp transporter is the only route for E. coli to take up heme through the inner membrane; nonspecific diffusion does not contribute.
Table 1.
Effect of dppABCDF and mppA mutations on the use of hemoglobin as an iron source and of δ ALA for aerobic growth
| Relevant mutations | Complementing genes carried on pTRC99 | Growth on M63* Dip + 40 μM Hb | Growth on M63 + 0.5 μg/ml δ ALA |
|---|---|---|---|
| No relevant mutation | None | + | + |
| dppA::Km | None | + | + |
| dppB::Km | None | − | − |
| dppC::Km | None | − | − |
| dppD::Km | None | − | − |
| dppF::Km | None | − | − |
| mppA::Cm | None | + | + |
| dppA::Km−mppA::Cm | None | − | − |
| dppB::Km | dppCDF | − | − |
| dppB::Km | dppABCDF | + | + |
| dppC::Km | dppDF | − | − |
| dppC::Km | dppABCDF | + | + |
| dppD::Km | dppF | − | − |
| dppD::Km | dppABCDF | + | + |
| dppF::Km | dppF | + | + |
| dppF::Km | dppABCDF | + | + |
The relevant chromosomal mutations carried either by strain FB827 (pAM 238-hasR) or by strain POP3 hemA are indicated in the first column. The plasmid-borne complementing genes carried on pTRC 99 are indicated in the second column. When pTRC 99 derivatives were present, 1 mM isopropyl β-d-thiogalactoside was added to induce the complementing genes expressed under the control of a lacpromoter. The third column indicates the growth of strain FB827 (pAM 238-hasR) mutants in iron-chelated medium M63* supplemented with hemoglobin (Hb). The various strains were streaked on the media, and plates were incubated for 72 h at 37°C. All of the experiments were repeated three times. +, all strains grew on M63 medium; −, no strains grew on M63* Dip without Hb. The fourth column indicates the growth of POP3 hemA mutants in M63 supplemented with 0.5 μg/ml ALA. The various strains were streaked on the media indicated and incubated for 48 h at 37°C. All of the experiments were repeated three times. +, all strains grew on M63 medium supplemented with 5 μg/ml δ ALA; −, no strains grew on M63 without δ ALA. None, an empty vector with no complementing gene.
Transposon insertion into dppA had no effect on iron-heme utilization (Table 1) suggesting that another binding protein is involved. The growth of the dppA::Km mutant also indicated that despite the gene being in the dpp operon, and despite the previously described polar effect of a dppA::Km mutation on the dpp operon for dipeptide transport (16), the transposon insertion does not exert a polar effect strong enough to affect the use of heme iron.
Double mppA dppA Mutant Is Unable to Use Heme as Iron Source.
Substrate recognition and import by ABC permeases always involve periplasmic binding proteins, and we therefore tested whether other periplasmic binding proteins were required for heme uptake. Transposon insertions into genes encoding periplasmic proteins similar to dppA (sapA, nikA, oppA, and mppA) were transduced into FB827 (pAM 238-hasR), and heme iron utilization was measured. None of the mutations affected heme iron utilization (data not shown). PBPs have some overlapping specificity and, in some cases, two periplasmic binding proteins interact with one ABC transporter (17). We therefore tested whether any combination of PBP mutations and inactivation of dppA led to the loss of iron-heme utilization. FB827 dppA::Km mppA::chloramphenicol (Cm) (pAM 238-hasR) did not grow on M63* Dip, Hb plates (Table 1). Thus, besides DppA and MppA, there is no other periplasmic heme-binding protein able to promote heme uptake. DppB-DppC-DppD-DppF is the sole E. coli heme ABC transporter, and MppA replaces DppA as binding protein, and, consequently, MppA probably delivers heme to the Dpp transporter. MppA is encoded by an isolated gene unlinked to genes encoding ABC transporter. It binds the murein tripeptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate (18).
Transport of δ Aminolevulinic Acid (ALA).
Dpp permease is also required for the uptake of the heme precursor, δ ALA (19). To clarify which of the Dpp components are necessary for δ ALA uptake and to test whether the functional redundancy of MppA and DppA for heme utilization also extended to δ ALA, each dpp::Km mutation and the mppA::Cm mutation were P1 transduced into the POP3 hemA strain defective in δ ALA biosynthesis. Mutant strains were tested for fast aerobic growth on M63 plates in the presence of various δ ALA concentrations. POP3 hemA grew well on M63 plates supplemented with 0.5 μg/ml of δ ALA, but strains with a Km insertion in one of the genes encoding the Dpp ABC transporter did not. Transformation with plasmids carrying the genes downstream from the transposon did not restore growth, whereas transformation with a plasmid expressing dppA-dppB-dppC-dppD-dppF complemented the growth defect of each mutant. These results indicate that each of the dppB, dppC, dppD, and dppF genes is individually required for δ ALA uptake as is the case for iron heme utilization (Table 1). Single dppA::Km or mppA::Cm mutants grew with 0.5 μg/ml δ ALA, but the double dppA::Km, mppA::Cm mutant did not grow, indicating that MppA and DppA have partially redundant functions and that both contribute to δ ALA uptake. All mutants formed normal-sized colonies at ten times the δ ALA concentration (5 μg/ml), indicating that other permeases may contribute, albeit less efficiently, to δ ALA uptake.
In Vivo Competition Between Heme and Peptides for Heme Utilization.
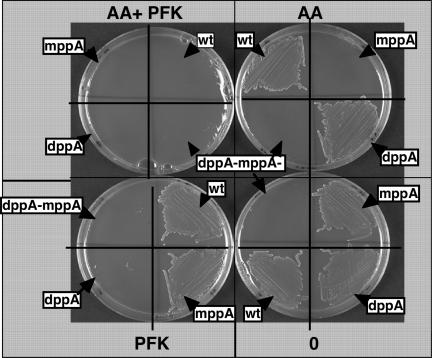
Strain FB827 (pAM 238-hasR) fails to grow on LB Dip, Hb plates suggesting that heme iron acquisition is impaired in medium containing peptides (data not shown). To test this hypothesis and to determine whether the inhibition involved the peptide-binding proteins (DppA and MppA) or the ABC transporter (DppBCDF), heme acquisition was tested in minimal medium supplemented with peptides. We used the dipeptide Ala–Ala (AA), which binds to DppA with a high affinity and the tripeptide Pro–phe–Lys (PFK), which has a low affinity for DppA and is transported via MppA (18, 20). FB827 (pAM 238-hasR) strains carrying either a dppA::Km or a mppA::Cm single mutation or the double dppA::Km, mppA::Cm mutations were grown on M63* Dip, Hb medium (with the appropriate antibiotic) supplemented with AA or PFK or a mixture of both at 100 μM. The addition of dipeptide AA mimicked a dppA mutation and had no effect on the wild-type strain or on the dppA mutant but abolished heme utilization by strain FB827 mppA::Cm (pAM 238-hasR), leading to a DppA− MppA− phenotype. Similarily, addition of the tripeptide PFK had no effect on the wild-type strain or on the mppA mutant but abolished heme utilization by FB827 dppA::Km (pAM 238-hasR) leading to a DppA− MppA− phenotype. The presence of both peptides abolished the growth of both the single mutants and the parental wild-type strain FB827 (pAM 238-hasR) (Fig. 1). However, the presence of both peptides had no effect on the growth of these strains on iron-rich M63 Glu media (data not shown). The different effects of these two peptides on heme acquisition suggest that the inhibition does not involve the single heme ABC transporter (DppBCDF) but the peptide binding proteins: Presumably the dipeptide AA competes with heme for binding to DppA, whereas the tripeptide PFK competes with heme for binding to MppA.
Fig. 1.
Effect of dipeptide (AA) and tripeptide (PFK) alone or mixed (AA+PFK) on iron heme utilization. The strains FB8 entF::Tn10 (pAM 238-hasR) carrying the relevant chromosomal mutations indicated around the Petri dishes were streaked on media containing M63* Glu, Dip, Hb either with or without the dipeptide AA, the tripeptide PFK, or a mixture of both peptides (AA+ PFK), all at a final concentration of 10−4 M. Added peptides are indicated below the plates. Plates were incubated for 72 h at 37°C and photographed. All of the experiments were repeated three times.
DppA and MppA Proteins Bind Heme.
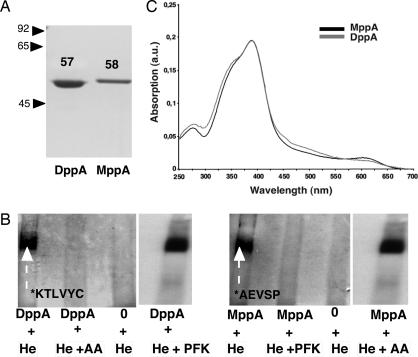
DppA and MppA proteins were overproduced from pTRC 99-dppA and pTRC 99-mppA. The three-step purification procedure described in Materials and Methods resulted in, for each case, a single band on SDS/PAGE with apparent molecular weights of 57 kDa for DppA and 58 kDa for MppA (Fig. 2A). Proteins purified from cultures grown in minimal medium did not contain significant amounts of heme as measured by visible absorption spectroscopy. Reconstitution with heme (10−4 M) was tested on a nondenaturing polyacrylamide gel system allowing separation of free heme, apo-proteins and heme-loaded proteins without dissociating heme from the proteins. Proteins were transferred to nitrocellulose filters, and heme peroxidase activity was detected by chemiluminescence. This method is the most sensitive test available for detecting heme bound to proteins (21). The N-terminal sequences of the bands on the nitrocellulose loaded with heme (Fig. 2B) were determined. The N-terminal sequences (KTLVYC and AEVPS) were identical to those of DppA (20) and MppA (18). Thus, the two protein bands loaded with heme were indeed DppA and MppA. Absorption spectroscopy analysis of the two protein bands cut out from the nondenaturing gel revealed a Soret peak at 392 nm (Fig. 2C) absent from protein bands from samples that were not loaded with heme.
Fig. 2.
Purification, heme-binding properties, and adsorption spectra of DppA and MppA. (A) SDS/PAGE analysis of the purified DppA and MppA proteins. Molecular weights in kilodaltons are indicated on the left. Apparent molecular weights of DppA and MppA are indicated on the SDS/PAGE. (B) ECL detection after nondenaturing PAGE and transfer onto a nitrocellulose membrane. Proteins were incubated either with heme or with a mixture of heme and peptides as indicated below the lanes. 0+He corresponds to heme alone. Heme and peptides were each at a final concentration of 10−4 M. After nondenaturing PAGE, the protein bands loaded with heme were also transferred onto nitrocellulose membrane for N-terminal amino acid sequencing. The sequences are indicated below each protein. (C) Absorption spectra of purified holo-DppA and holo-MppA. Protein bands were sliced out of the gel after nondenaturing PAGE, and absorption was measured in a Beckman DU spectrophotometer.
MppA and DppA were incubated with various concentrations of heme; the samples were analyzed as above, and heme bound to proteins was determined by ECL. Heme binding to each protein was saturable (data not shown), and the affinities of DppA and MppA for heme were ≈10−5 M and 5 × 10−5 M, respectively.
Heme Binding to DppA and MppA Is Inhibited by Dipeptide and Tripeptide, Respectively.
We tested whether peptides competed with heme for binding to purified DppA and MppA, by adding heme (10−4 M) in the presence of the tripeptide PFK or the dipeptide AA (each at 10−4 M). DppA loading with heme was abolished by AA and that of MppA by PFK. DppA incubation with PFK and MppA incubation with AA had no effect on heme loading (Fig. 2B). Thus, the dipeptide is a specific competitor for heme binding in vitro to DppA, and the tripeptide is a specific competitor for heme binding in vitro to MppA.
These results confirm the conclusions drawn from in vivo competition experiments, demonstrating that peptides compete with iron heme utilization by their corresponding binding proteins.
Discussion
Heme can be used as an iron source by E. coli K12, provided it expresses an outer membrane heme receptor. We isolated and characterized mutants of an E. coli strain expressing the S. marcescens HasR receptor having lost the ability to use heme as an iron source. Analysis of these mutants showed that heme uptake through the inner membrane requires the entire inner membrane dipeptide ABC transporter: DppB, DppC, DppD, and DppF. ABC importers collaborate with PBPs. We found that either DppA or MppA is required for heme delivery: Each single mutant was able to use heme, but the double mutant was not. We provide evidence that these two peptide-binding proteins are the functional heme-binding proteins in the E. coli periplasm because (i) both purified DppA and purified MppA bind heme with a Soret maximum at 392 nm and with an affinity of ≈5 × 10−5 to 10−5 M and (ii) peptides compete for heme binding to DppA and MppA in vitro and in vivo. The dipeptide AA, which has a higher affinity for DppA, competes with heme binding to DppA but not to MppA, whereas the tripeptide PFK, which has a higher affinity for MppA, competes with heme binding to MppA but not to DppA. Heme iron acquisition was not possible in rich medium because of its high peptide content. However, previous studies have reported different results with E. coli expressing either the S. marcescens HasR (22) or the Sh. dysenteria ShuA (15) receptors. These heme iron acquisition tests were done by measuring growth around spots of heme, and it is likely that the heme concentration in spots (0.1 to 1 mM) was high enough to compete with the pepdides present in the LB medium.
Thus, the E. coli heme permease, like other Gram-negative permeases, is a PBP-dependent system but uses two optional binding proteins. The Dpp permease is one of the three main E. coli peptide permeases that actively transport peptides of various lengths through the inner membrane. Dpp transports dipeptides (16). The two other permeases are Opp, which transports oligopeptides of 4–6 aa, and Tpp, which transports tripeptides (23). Tpp is an inner membrane protein belonging to the major facilitator superfamily (24), whereas Dpp and Opp permeases are binding protein-dependent bacterial ABC importers, each encoded by five genes organized in an operon. The PBP MppA is, in contrast, encoded by a single gene. It binds the cell-wall murein tripeptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. There is no murein peptide-specific ABC transporter. The loaded binding protein delivers the murein tripeptide to the OppBCDF ABC transporter (18). Peptide ABC transporters are believed to recognize the loaded binding protein but not the peptide itself (25). However, our findings strongly suggest that MppA is able to deliver different substrates to different ABC transporters: murein tripeptide to Opp and heme to Dpp. Thus, the ABC transporter may contribute to substrate selectivity. It is possible that PBP loaded with heme adopts a particular configuration that can be recognized only by the Dpp transporter. Alternatively, the membrane components of the ABC transporter (DppB and DppC) may also be able to bind heme. A two-step recognition (by PBP and by membrane components) has been described for the maltose permease (26).
We show here that the heme precursor δ ALA and heme use the same peptide permease. δ ALA is similarly transported in mammals by peptide transporters (27). The resemblance between δ ALA and GG dipeptide has been used to explain δ ALA uptake by the Dpp permease (19). However, we show that MppA can replace DppA for δ aminolevulinic acid uptake, suggesting that the heme precursor and heme might have other common properties. Indeed, the inhibitory activity of heme on δ ALA synthase (28) also supports this possibility. High-resolution crystal structures of three peptide-binding proteins (DppA, OppA, and AmiA) show an overall two-lobe structure characteristic of PBPs; the peptide-binding site is in a cleft between the two lobes (see ref. 25 for review). The two-lobed peptide-binding structures close upon substrate binding. A related protein, the Haemophilus influenzae HbpA, shares 53% sequence identity with DppA (29) and binds heme. An H. influenzae hbpA insertion mutant is strongly affected in heme acquisition, indicating that the HbpA protein is important but not essential for heme acquistion (30). HbpA has been modeled with heme in the putative binding pocket (31). Heme binding to DppA was not detected in one previous study (32), although, under some purification conditions, DppA samples contain bound peptides competing with heme binding (20). The native dipeptide binding pocket of DppA does not appear to be large enough to accomodate heme, but side chain reorientation might occur upon heme binding.
The Sh. dysenteria heme-binding protein, ShuT, has a five-coordinated heme with a tyrosine as the axial iron ligand (33). On the basis of sequence homologies, ShuT belongs to a class of PBP comprising FhuD and BtuF in which the two lobes are bridged by a single α helix. Note that DppA and MppA, like HbpA, are larger and more flexible PBPs with an additional linker bridging the two lobes (34). Thus, heme-binding PBPs are found in two of the three known PBP classes.
E. coli K12 has no way of importing exogenous heme through the outer membrane, so the physiological function of a heme permease seems obscure. DppA is an abundant protein in the periplasm (35), and its expression is significantly increased in stationary phase (16). The regulation of MppA is unknown. Its function in murein recycling is not the principal pathway for cell-wall peptide reutilization because E. coli has proteins for the transport of muropeptides and their subsequent hydrolysis in the cytoplasm (18). One possible function for such a heme permease system is to retrieve periplasmic heme in conditions of iron shortage and thereby maintain iron homeostasis. Neither dppA nor mppA genes were identified by transcriptome analysis as being Fur regulated in E. coli (36). We constructed dppA-lacZ and mppA-lacZ transcriptional fusions and introduced them into the chromosome of E. coli. The initiation of transcription of neither was iron regulated (data not shown). However, several genes encoding proteins involved in energy metabolism (including cytochromes) are repressed under iron-restricted conditions, a mechanism for cells to minimize iron and heme requirements (36). Nevertheless, iron homeostasis presumably also depends on coordinated synthesis and/or export of heme and cytochromes. In most reported cases, export of apo-cytochromes and heme to the periplasm is independent (37). Heme and apo-cytochrome synthesis is coregulated in Bradyrhizobium japonicum, but there is less evidence for such control in other bacteria, including E. coli (28). Heme recycling by the MppA/DppA Dpp permease would prevent the accumulation of free, potentially toxic, heme in the periplasm during any drop in cytochrome content. However, coordinated expression of heme and cytochromes in E. coli may also be because of other as yet unidentified mechanisms.
During energetic growth, when apo-cytochromes are available in the periplasm, it is unlikely that MppA and DppA, which have relatively low affinity for heme, would sequester heme before its ligation to apo-cytochromes. Heme iron acquisition was inhibited in vivo by peptides at high concentrations. Bacteria do not encounter such conditions, physiologically. Thus, it is likely that peptides and heme might be transported simultaneously.
The presence of a system for endogenous heme recycling could explain how strains unable to use exogenous heme can import it if it reaches the periplasm. Strains able to use exogenous heme might have acquired either a whole uptake gene set encoding outer membrane receptors and an inner membrane permease by horizontal transfer. Alternatively, only the gene encoding the outer membrane receptor may have been acquired, and the housekeeping function of Dpp in endogenous heme transport might then have been extended to use exogenous heme. It would be interesting to determine whether other Dpp systems, present in most bacteria, are involved in heme uptake.
In conclusion, we identified a dipeptide permease as the specific E. coli heme permease. Our results invalidate the hypothesis of nonspecific partitioning of heme through the inner membrane. Although we only studied E. coli, it is likely that our conclusions are valid for other species. Second, our results suggest that many pathogenic strains able to acquire exogenous heme might have at least two heme permeases, one specific for heme and the other for peptides and heme. Such redundancy would explain the leaky phenotypes of specific heme-permease mutants. Finally, our results highlight the homologies between DppA and HbpA proteins, because we show that DppA and MppA are heme-binding proteins. These findings might have implications for the design of heme-permease inhibitors.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli strains FB8 (wild type, F−) and FB827 (entF::Tn10) are described in ref. 38. POP3 (araD139 ΔlacU169 rpsL relA thi), C600 (F− thr leu fhuA lacY thi supE), POP3 hemA, and SM10 λ pir (thi recA thr leu fhuA lacY supE RP4–2Tc::Mu KmR λ::pir) were from the laboratory collection. JWN 3513–1 (dppA::Km), JWN 3512–2 (dppB::Km), JWN 3511–2 (dppC::Km), JWN 3510–1 (dppD::Km), and JWN3509–1 (dppF::Km) (39) were obtained from the KO collection by means of the E. coli database “GenoBase” (http://ecoli.aist-nara.ac.jp/). TP985 (dapD2 relA1 spoT1 thi-1 Hfr mppA::miniTn10 Cm) is described in ref. 18. Mutations were introduced into strains by phage P1 transduction.
pSC189 carries the mariner transposon TnSC189 (40). pAM 238 and pTRC 99 were from the laboratory collection, and pAM 238-hasR is described in ref. 41. pTRC 99-dppA, pTRC 99-mppA, pTRC 99-dppCDF, pTRC 99-dppDF, pTRC 99-dppF, and pTRC 99-dppABCDF are described in this work.
Media and Growth Conditions.
Hemin, Hb, δ ALA, Dip, and isopropyl-β-d-thiogalactopyranoside were obtained from Sigma Chemical Company (Lyon, France). The hemoglobin concentration was calculated on the basis of the heme monomer. Hemoglobin and δ ALA solutions were sterilized by passage through 0.45-μm pore-size filters. Hemin was dissolved immediately before use in a minimal volume of 0.1 M NaOH, filtered, and diluted with the appropriate buffer to the desired concentration. Bacteria were grown aerobically at 37°C in LB rich medium, M63, or M63* (without added iron salt). All minimal media were supplemented with 0.4% glucose (not indicated in the text). When required, Dip was added to a final concentration of 100 μM to M63*. Antibiotics were added to the following final concentrations: μg ml−1 ampicillin, 25 μg ml−1 Km, 50 μg ml−1 spectinomycin, 10 μg ml−1 tetracycline, and 15 μg ml−1 Cm. For strain TP985, diaminopimelic acid was added to a final concentration of 50 μg ml−1. For each strain, the carbon source and the appropriate antibiotics were added to solid and liquid media but are not indicated in the text. All cultures were grown with aeration at 37°C, and optical density was measured at 600 nm (OD600).
Growth Promotion Assays.
The ability of the strain FB827 (pAM 238-hasR) carrying the various mutations to use heme as an iron source was tested on M63* Dip plates containing the appropriate antibiotics and supplemented with 40 μM Hb. Colony size was measured after 72 h of incubation at 37°C. We used FB827 mutants in all tests, but similar results were obtained with other strains carrying the entF mutation (such as MG1655 entF::Tn10) to inactivate enterobactin synthesis. The ability to use δ ALA was tested on M63 plates containing the appropriate antibiotics and supplemented with various concentrations of δ ALA. Colony sizes of POP3 hemA carrying the various mutations were measured after 48 h of incubation at 37°C.
Genetic Techniques.
P1 lysates and transductions were as described by Miller (42). Competent cells were prepared by a calcium chloride method.
Extraction and Manipulation of Plasmids.
Standard methods were used for isolation of plasmid DNA, cloning, restriction enzyme analysis, and transformation.
Plasmid Constructions.
Plasmids encoding DppABCDF, DppCDF, DppDF, DppF, DppA, and MppA were constructed by amplification of E. coli FB8 genomic DNA by using complementary oligonucleotides (sequences available on demand). Amplified fragments with appropriate restriction nuclease recognition sites were inserted into pTRC 99. Amplified gene sequences were checked by DNA sequencing.
Transposon TnSC189 Mutagenesis.
The E. coli donor strain SM10λpir carrying pSC189 and the recipient strain FB827 (pAM 238-hasR) were grown separately in LB to the exponential phase. The cells were washed and resuspended to an OD600 of 0.6 and mixed at a ratio of 3:1. The mixture was spotted onto 0.4-μm pore-size filters placed on an LB plate (no selection). After overnight incubation at 37°C, the bacterial cells were suspended in LB, and appropriate dilutions were plated on counter selecting tetracycline, Spc, Km LB media. Ten thousand clones were picked and distributed with a Q pix genetix instrument into 96-well microplates that contained 100 μl of M63 with appropriate antibiotics. Microplates were replica-plated on M63 and on M63* Glu (0.1 mM Dip, 40 μM Hb, and appropriate antibiotics).
Mapping of Transposon Insertion Sites.
Genomic DNA was isolated from transposants by using the Wizard Plus SV Minipreps kit (Promega, Madison, WI) and sequenced (MillegenR, Labege, France) by using the primer TCTAGGCGGCCGCGAAGTTCCTAT complementary to the 3′ end of the Km gene in the transposon.
Production and Purification of DppA and MppA Proteins.
The same procedure was used for MppA and DppA. Two liters of each POP3 (pTRC 99-dppA) and POP3 (pTRC 99-mppA) cell cultures were grown at 37°C in M63 medium and subjected to osmotic shock as described in ref. 43 with the following modifications. When the cultures reached an OD600 of 0.1, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 5 mM. The cultures were grown for an additional 3 h at 37°C to an OD600 of 1 and harvested by centrifugation for 15 min at 8,000 × g at 4°C. Cell pellets were washed once in TE buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA), and each pellet was resuspended at room temperature in 200 ml of 20% (wt/vol) sucrose and 30 mM EDTA and incubated for 10 min at room temperature. The cells were again pelleted by centrifugation and then rapidly resuspended in 200 ml of ice-cold 0.5 mM MgCl2. The suspension was incubated for 5 min at 0°C and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant, containing the periplasmic shock fluid, was concentrated by 80% ammonium sulfate precipitation and then extensively dialyzed against 50 mM Tris·HCl, pH 7.5, 80 mM NaCl (TN) at 4°C.
Samples of each concentrated osmotic shock fluid were first purified by cation and then anion exchange chromatography as described for DppA in ref. 20. This step was followed by gel filtration on a Superose 6HR 10/300 (GE Healthcare Europe, Orsay, France) column preequilibrated with 20 mM Tris·HCl, pH 7.8/ 80 mM NaCl buffer. Fractions were collected, and their DppA or MppA contents and purity were evaluated by SDS/PAGE. Fractions containing pure DppA or pure MppA were pooled.
Nondenaturing PAGE and Heme Detection by Chemiluminescence.
Twenty microliters of each purified DppA and MppA protein (4 × 10−5 M) were incubated at room temperature for 30 min with heme (10−4 M), with a mixture of heme (10−4 M) and dipeptide AA (10−4 M) or heme (10−4 M) and tripeptide PFK (10−4 M), or only buffer. Mixtures were separated by PAGE (4°C in the absence of SDS), and the proteins were transferred to nitrocellulose filters. Heme complexed with protein bands on the gel retains intrinsic peroxidase activity, which was detected by chemiluminescence (ECL+; Amersham Pharmacia) as described in ref. 21. The signal was measured either by autoradiography on films or with a Storm Imager.
N-terminal amino acid sequences were determined using the heme-loaded samples blotted onto nitrocellulose that were positive by ECL. N-terminal sequencing was performed by the Plateforme d’Analyze et de Microséquence des Protéines de l’Institut Pasteur.
Absorption Spectroscopy of DppA and MppA Proteins.
Heme-complexed protein bands were cut out of nondenaturing PAGE gels, placed into a cuvette, and directly examined by absorption spectroscopy from 250 nm to 700 nm; results were recorded on a Beckman DU 800 spectrophotometer.
Acknowledgments
We thank Dr. J. Park (Tufts University, Boston, MA), Dr. F. Biville (Institut Pasteur, Paris, France), and Dr. M. Simonet (Institut Pasteur, Lille, France) for providing strains. We especially thank Dr. F. Biville for many useful suggestions and advice. We also thank Dr. E. Dassa and Dr. J. M. Ghigo for carefully reading the manuscript.
Abbreviations
- AA
Ala–Ala
- ABC
ATP-binding cassette
- ALA
aminolevulinic acid
- Cm
chloramphenicol
- Dip
2,2′dipyridyl
- Hb
bovine hemoglobin
- Km
kanamycin
- PBP
periplasmic substrate-binding protein
- PFK
Pro–phe–Lys
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wandersman C., Delepelaire P. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 2.Stojiljkovic I., Hantke K. Mol. Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson A. D., Deisenhofer J. Cell. 2004;116:15–24. doi: 10.1016/s0092-8674(03)01030-4. [DOI] [PubMed] [Google Scholar]
- 4.Bracken C. S., Baer M. T., Abdur-Rashid A., Helms W., Stojiljkovic I. J. Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wandersman C., Stojiljkovic I. Curr. Opin. Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 6.Boos W., Lucht J. M. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F., editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1175–1209. [Google Scholar]
- 7.Stojiljkovic I., Perkins-Balding D. DNA Cell Biol. 2002;21:281–295. doi: 10.1089/104454902753759708. [DOI] [PubMed] [Google Scholar]
- 8.Hantke K. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 9.Thompson J. M., Jones H. A., Perry R. D. Infect. Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Occhino D. A., Wyckoff E. E., Henderson D. P., Wrona T. J., Payne S. M. Mol. Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 11.Rohde K., Dyer D. Infect. Immun. 2004;72:2494–2506. doi: 10.1128/IAI.72.5.2494-2506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morton D. J., Smith A., Ren Z., Madore L. L., VanWagoner T. M., Seale T. W., Whitby P. W., Stull T. L. Microbiology. 2004;150:3923–3933. doi: 10.1099/mic.0.27238-0. [DOI] [PubMed] [Google Scholar]
- 13.Perkins-Balding D., Ratliff-Griffin M., Stojiljkovic I. Microbial. Mol. Biol. Rev. 2004;68:154–171. doi: 10.1128/MMBR.68.1.154-171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghigo J. M., Létoffé S., Wandersman C. J. Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills M., Payne S. M. J. Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abouhamad W., Manson M. Mol. Microbiol. 1994;14:1077–1092. doi: 10.1111/j.1365-2958.1994.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 17.Higgins C., Ames G. Proc. Natl. Acad. Sci. USA. 1981;78:6038–6042. doi: 10.1073/pnas.78.10.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J., Raychaudhuri D., Li H., Normark S., Mengin-Lecreulx D. J. Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verkamp E., Backman V. M., Bjorsson J., Soll D., Eggertsson G. J. Bacteriol. 1993;175:1452–1456. doi: 10.1128/jb.175.5.1452-1456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith M., Tyreman D., Payne G., Marshall N., Payne J. Microbiology. 1999;145:2891–2901. doi: 10.1099/00221287-145-10-2891. [DOI] [PubMed] [Google Scholar]
- 21.Vargas C., McEwan A. G., Downie J. A. Anal. Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- 22.Paquelin A., Ghigo J. M., Bertin S., Wandersman C. Mol. Microbiol. 2001;42:995–1005. doi: 10.1046/j.1365-2958.2001.02628.x. [DOI] [PubMed] [Google Scholar]
- 23.Wayne J., Marshall N. In: Microbial Transport Systems. Winkelmann G., editor. Weinheim, Germany: Wiley–Verlag Chemie; 2001. pp. 139–164. [Google Scholar]
- 24.Saier M. J., Beatty J., Goffeau A., Harley K., Heijne W., Huang S., Jack D., Jahn P., Lew K., Liu J., et al. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 25.Doeven M., Kok J., Poolman B. Mol. Microbiol. 2005;57:640–649. doi: 10.1111/j.1365-2958.2005.04698.x. [DOI] [PubMed] [Google Scholar]
- 26.Treptow N., Shuman H. J. Bacteriol. 1985;163:654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doring F., Walter J., Will J., Focking M., Boll M., Amasheh S., Clauss W., Daniel H. J. Clin. Invest. 1998;101:2761–2767. doi: 10.1172/JCI1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brian M., Thony-Meyer L. Adv. Microb. Physiol. 2002;46:257–318. doi: 10.1016/s0065-2911(02)46006-7. [DOI] [PubMed] [Google Scholar]
- 29.Hanson M., Slaughter C., Hansen E. Infect. Immun. 1992;60:2257–2266. doi: 10.1128/iai.60.6.2257-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton D. J., Madore L. L., Smith A., VanWagoner T. M., Seale T. W., Whitby P. W., Stull T. L. FEMS Microbiol. Lett. 2005;253:193–199. doi: 10.1016/j.femsle.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Dunten P., Mowbray S. L. Protein Sci. 1995;4:2335–2340. doi: 10.1002/pro.5560041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunten P., Mowbray S. L. Protein Sci. 1995;4:2327–2334. doi: 10.1002/pro.5560041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eakanunkul S., Lukat-Rodgers G., Sumithran S., Ghosh A., Rodgers K., Dawson J., Wilks A. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 34.Borths E., Locher K., Lee A., Rees D. Proc. Natl. Acad. Sci. USA. 2002;99:16642–16647. doi: 10.1073/pnas.262659699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson E., Dunyak D., Jurss L., Poorma R. J. Bacteriol. 1991;173:234–244. doi: 10.1128/jb.173.1.234-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHugh J., Rodriguez-Quinones F., Abdul-Tehrani H., Svistunenko D., Poole R., Cooper C., Andrews S. J. Biol. Chem. 2003;278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 37.Thony-Meyer L. Microbiol. Mol. Biol. Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrotte-Piquemal M., Danchin A., Biville F. Biochimie. 1999;81:245–253. doi: 10.1016/s0300-9084(99)80058-3. [DOI] [PubMed] [Google Scholar]
- 39.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. Mol. Syst. Biol. 2006;2:E1–E11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang S., Rubin E. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 41.Létoffé S., Debarbieux L., Izadi N., Delepelaire P., Wandersman C. Mol. Microbiol. 2003;50:77–88. doi: 10.1046/j.1365-2958.2003.03686.x. [DOI] [PubMed] [Google Scholar]
- 42.Miller J. H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Woodbury, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 43.Neu H., Heppel L. J. Biol. Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]