Abstract
The current arsenal of tools and methods for the continuous monitoring and imaging of redox metabolic pathways in the context of intact cells is limited. Fluorogenic substrates allow for direct measurement of enzyme activity in situ; however, in contrast to proteases and exo-glycosidases, there are no simple guidelines for the design of selective probes for redox metabolic enzymes. Here, we introduce redox probe 1 and demonstrate its high selectivity in living cells for human hydroxysteroid dehydrogenases (HSDs) of the aldo-keto reductase (AKR) superfamily. AKR1C isoforms perform multiple functions among which the metabolism of potent steroid hormones is well documented. Moreover, expression of these enzymes is responsive to cellular stress and pathogenesis, including cancer. Our probe design is based on redox-sensitive optical switches, which couple a ketone–alcohol redox event to a profound change in fluorescence. The high selectivity of phenyl ketone 1 for AKR1C2 over the many endogenous reductases present in mammalian cells was established by a quantitative comparison of the metabolic rates between null control cells (COS-1) and AKR1C2-transfected cells. Phenyl ketone 1 is a cell-permeable fluorogenic probe that permits a direct, real-time, and operationally simple readout of AKR1C2 enzyme activity in intact mammalian cells. Furthermore, it was demonstrated that probe 1 enables the quantitative examination of physiological substrate 5α-dihydrotestosterone (“dark substrate”) in situ by means of a two-substrate competitive assay. Similarly, inhibitor potency of physiological (ursodeoxycholate) and synthetic inhibitors (flufenamic acid, ibuprofen, and naproxen) was also readily evaluated.
Keywords: aldo-keto reductases, fluorescent probes, hydroxysteroid dehydrogenase, metabolic reporters
The majority of studies aimed at molecular understanding of biological systems rely on the determination of gene and protein expression levels. Although these approaches are amenable to high-throughput modes and provide enormous amounts of information, they are discontinuous and destructive in nature and therefore poorly suited for the examination of dynamic properties of intact cells and tissues. Furthermore, protein function (e.g., activity of enzyme) may be controlled by posttranslational modifications and metabolic feedback mechanisms. Therefore, the next important step toward the noninvasive monitoring of complex metabolic networks requires a direct readout of the enzyme activity in situ.
One promising approach relies on small-molecule metabolic reporters, which provide a measurable signal for a particular enzymatic process (1–3). Fluorogenic or fluoromorphic probes are particularly suitable for this task because of the high sensitivity of fluorimetry and the potential for spatial resolution through fluorescent microscopy. Considering the rapid advancement of photon-capture hardware and pattern-processing software (4), progress in metabolic monitoring and imaging depends to a significant degree on the development of selective reporter probes.
Many fluorogenic probes consist of an organic dye attached at the periphery of a natural substrate wherein the emission change is usually achieved through FRET (5) or a phenol- or aniline-releasing reaction (6, 7). For instance, a short peptide equipped with an appropriate dye attached at the N terminus illustrates a common design for protease probes (8). In these cases, the enzyme recognizes the natural substrate while the organic dye resides outside the enzyme’s perimeter, thereby minimizing reporter–enzyme interactions. The activity of various proteases has been measured in intact cells, and good selectivity has been achieved primarily by the choice of peptide sequence (1).
In the case of redox metabolic enzymes, where these fluorogenic mechanisms are not applicable, the organic dye may become an integral part of the recognized substrate. In this instance, a synthetic molecule, bearing distant resemblance to a physiological substrate, would have to function as a competitive substrate. Thus, these approaches depend on significant substrate plasticity of the enzymes of interest (9–11). Furthermore, with activity comes the issue of selectivity, and as with all nonpeptidic ligand–protein interactions, there are no straightforward design guidelines.
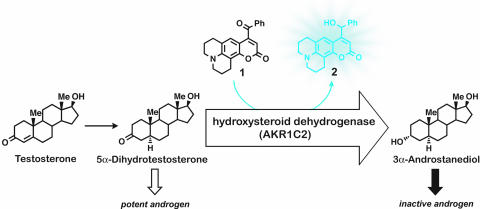
We recently described a novel fluorogenic substrate for human hydroxysteroid dehydrogenases (HSDs) of the aldo-keto reductase (AKR) superfamily (9). It has been proposed that these enzymes function as prereceptor switches by activating or deactivating steroid hormones through redox chemistry (12). For example, the occupancy of androgen receptors in the prostate may be regulated by reducing the highly potent androgen 5α-dihydrotestosterone (DHT) to the inactive metabolite 5α-androstane-3α,17β-diol (3α-diol) (Fig. 1). Similarly, reduction of 5α-dihydroprogesterone to 3α, 5α-tetrahydroprogesterone (allopregnanolone) produces an allosteric regulator of the GABA receptor in the brain (13, 14). Both reactions are catalyzed by human isoform AKR1C2, a multifunctional enzyme also known as 3α-HSD type III, dihydrodiol dehydrogenase (DDH), and bile acid binding protein (15–17). It has been reported that AKR1C2 expression levels are altered in prostate and breast tumors (18, 19). In an ovarian cancer cell line, it has been shown that resistance to cis-platin chemotherapeutics correlates with the induction of AKR1C2 expression (20). AKR1C2 is emerging as an important cellular-stress response marker in several different cell types (21, 22). Furthermore, it has been suggested that the antitumor effect of nonsteroidal anti-inflammatory drugs (NSAIDs) is mediated in part by the inhibition of certain AKR1C isoforms, in addition to inhibition of cyclooxygenases (COXs) and other targets (23).
Fig. 1.
Metabolic indicator 1 was designed to intercept a physiological pathway and report selectively on the activity of HSDs of the AKR superfamily. In the human prostate, AKR1C2 is implicated in the deactivation of potent androgen DHT. Probe 1 is a competitive substrate with a built-in mechanism for translating a redox chemical transformation to a measurable increase in fluorescent emission.
These findings provide a strong impetus for the development of selective metabolic probes for this enzyme. Although there are four known human isoforms (AKR1C1–4), we focus here on AKR1C2 because of its important metabolic role and the fact that the physiological substrate for this isoform has been well established (24). This latter point in turn enabled the comparison of the kinetic parameters obtained through the fluorimetric assay and the radiometric assay with [4-14C]DHT. Here we describe fluorogenic substrate 1 and demonstrate its high selectivity for AKR1C2 over the many oxidoreductases present in intact mammalian cells. This probe allows for noninvasive and continuous measurement of the AKR1C2 activity in live cells. As a competitive substrate, probe 1 also enables quantitative examination of physiological substrates (e.g., DHT) in situ through a two-substrate assay. Similarly, the kinetic characterization of inhibitors is readily available as demonstrated for both physiological and synthetic inhibitors.
Results
Development of a Metabolic Indicator for Selective Monitoring of AKR1C2 Activity in Intact Cells.
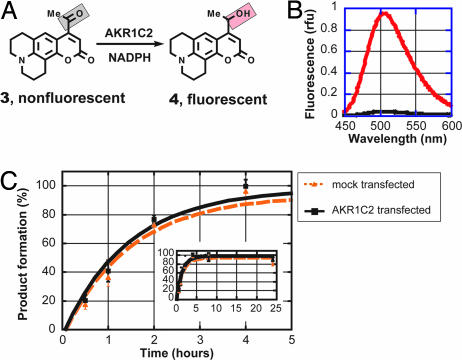
We recently described fluorogenic substrate methylketone 3 (9). This probe was developed in two major stages: first, by designing optical switches, which are organic compounds that translate a redox chemical transformation to a profound change in the emission profile, and second, by screening these compounds against a panel of purified dehydrogenases. Ketone 3 is virtually nonemissive (ketone quenching), while its reduction affords a highly fluorescent alcohol 4 with the emission maximum at 509 nm, constituting an excellent fluorogenic switch (Fig. 2A and B). We also showed that ketone 3 was an excellent substrate for purified HSDs of the AKR superfamily with the kinetic parameters comparable with the physiological steroid substrates.
Fig. 2.
Methyl ketone probe 3 shows no selectivity for AKR1C2 in intact cells. (A) Reduction of nonfluorescent methyl ketone 3 produces highly fluorescent alcohol 4, constituting an excellent fluorogenic switch. (B) Emission spectra of 3 (black) and 4 (red; emission maximum at 509 nm, excitation at 398 nm) expressed in relative fluorescence units (rfu). (C) Metabolic reduction of probe 3 in AKR1C2-transfected COS-1 cells compared with mock-transfected COS-1 cells. The high reduction rate in mock-transfected COS-1 cells (presumably by endogenous reductases) renders probe 3 unsuitable for selective readout of AKR1C2 enzyme activity. Data shown are the average ± SD of three independent experiments performed in duplicate.
Having identified a promising lead, we faced the next key question of whether our probe would be selective for AKR1C enzymes in intact mammalian cells. To this end, we incubated compound 3 at 5 μM concentration (KM,in vitro) with several common cell lines (COS-1, CHO, and HEK-293) that do not express AKR1C enzymes (null cells) and monitored the metabolic reduction by fluorimetric analysis of the growth medium. To our disappointment, the methyl ketone substrate was rapidly metabolized. For example, in null COS-1 cells (monkey kidney cells), complete conversion was observed in 4 h (Fig. 2C), presumably by endogenous reductases. The high endogenous metabolic reduction revealed the poor selectivity of this probe, rendering it unsuitable for selective readout of AKR1C2 activity in living cells.
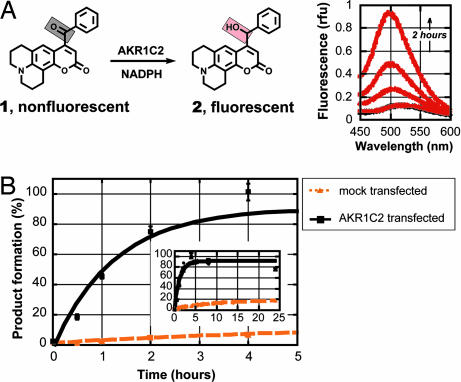
We hypothesized that better selectivity could be achieved by attaching a bulkier substituent to the ketone group. An optimization study identified phenyl ketone 1 as an excellent substrate with in vitro kinetic parameters superior to those for methyl ketone 3 (kcat/KM 7.8 vs. 2.1 min−1·μmol−1 for AKR1C2). Probe 1 was synthesized through a concise sequence (see Scheme 1, which is published as supporting information on the PNAS web site). Remarkably, this probe demonstrated high selectivity for AKR1C2 enzyme in living cells (Fig. 3). The mock-transfected cells gave a low background activity with <20% reduction after 24 h. On the other hand, AKR1C2-transfected cells rapidly metabolized probe 1 with a complete conversion occurring after 4 h. Thus, the first major goal of this enterprise was accomplished: namely, a metabolic indicator for direct and selective readout of AKR1C2 activity in intact cells was developed. This finding represents a notable achievement, considering the estimate of several hundred oxidoreductases existing in mammalian cells.
Fig. 3.
Phenyl ketone probe 1 is selective for AKR1C2 in intact cells. (A Left) Phenyl ketone 1 is an excellent substrate for AKR1C2 in vitro (KM = 3 μM; kcat/KM = 7.8 min−1·μM−1). (Right) Metabolic reduction was monitored by direct measurements of the fluorescence arising from the cell growth medium when excited at 385 nm [relative fluorescent units (rfu)]. (B) Probe 1 shows high selectivity for AKR1C2 in intact COS-1 cells over background (endogenous reductases). Probe 1 (5 μM) is completely reduced in 4 h by AKR1C2-transfected COS-1 cells. Mock-transfected cells show slow background metabolism. Data shown are the average ± SD of three independent experiments performed in duplicate.
The relevant redox metabolic abilities of the COS-1 cell line were examined with radiolabeled steroids, namely [4-14C]DHT. This cell line showed very low 3-keto- and 17-keto-reductase activity and significant 17β-oxidase activity. With regard to the latter activity, the stability of fluorescent alcohol 2 (reduced probe) was examined in cells, which showed that no oxidation of the probe alcohol occurred in the time frame of these experiments. This result in turn permitted a quantitative measurement of the metabolic reduction, which was significant only after AKR1C2 transfection into COS-1 cells, as described above. It should be noted, however, that mapping the complete redox metabolism of human and mammalian tissues is an ongoing process, and therefore it is reasonable to anticipate that there may be other redox enzymes, expressed in certain cell types, capable of reducing probe 1. In a different cellular context, the selectivity of this probe may have to be reexamined; for example, by correlating the metabolic activity (probe reduction) with the expression level of AKR1C gene or another candidate gene, as shown below.
Correlation of Probe 1 Metabolism with AKR1C2 Gene Transcription.
The relationship between the metabolic rates (or the enzyme activity) and the gene expression levels was quantitatively examined. This assessment is relevant in the context of recent studies that point to increased expression of AKR1C2 RNA transcripts in prostate cancer relative to normal prostate tissues (24) and in response to cellular stress (25). Also, this correlation may reveal any potential posttranscriptional mechanisms involved in controlling enzymatic activity.
COS-1 cells, transiently transfected with varying amounts of AKR1C2 cDNA (from 0 to 2 μg), were incubated with probe 1, and the growth medium was monitored fluorimetrically. Subsequent to the metabolic rate measurements, the amount of mRNA was determined through a standard procedure involving extraction of the cell lysates, reverse transcription, and PCR amplification. This study established the linear relationship between the metabolic rate and the gene transcription over a relatively broad range of mRNA levels (see Fig. 5, which is published as supporting information on the PNAS web site). In control experiments (mock-transfected or nontransfected cells), <5% of the probe was metabolized in 1 h, whereas transfection with 2 μg of AKR1C2 plasmid (the highest amount of DNA used) led to 50% conversion of the probe in the same period. The real-time RT-PCR approximations detected 25,000-fold less RNA in mock- and nontransfected cells than in those transfected with 2 μg of AKR1C2 plasmid. This quantitative analysis demonstrates that probe 1 can be used for direct and continuous readout of AKR1C2 gene expression levels in intact cells.
Determination of Kinetic Parameters for Probe 1 in Intact COS-1 Cells.
The quantitative correlation of the enzyme activity with RNA transcript amount described above rigorously established the high selectivity of the phenyl ketone substrate for AKR1C2 and set the stage for the determination of the kinetic parameters for this probe in situ. The latter point represents an important goal considering the dramatic differences between the intracellular environment and the dilute and controlled conditions of the in vitro experiment.
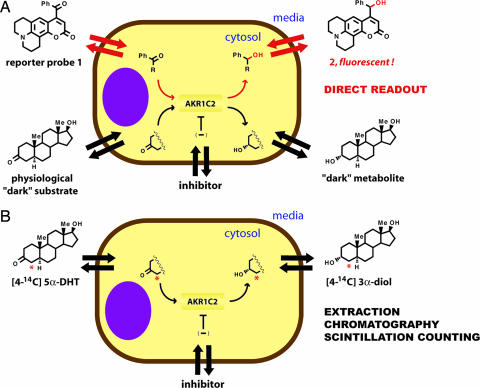
The apparent Michaelis constant (KM,app) was obtained by incubating AKR1C2-transfected cells with different concentrations of the probe (0.2–5KM,in vitro) and measuring the initial reaction rates. The exact nature of the assay must be considered for a clear understanding of this parameter (Fig. 4A). The KM,app is defined (and determined) as the extracellular concentration of the probe at which half the maximum metabolic rate is achieved. Assuming that the probe and the corresponding product readily permeate the cytoplasmic membrane, and thus the extracellular and intracellular concentration of both is similar, then KM,app equals (or closely approximates) the in situ parameters. Consequently, under such circumstances simple fluorimetric monitoring of growth medium provides valuable information about the enzyme activity inside the intact cell (the obtained value is an average over a measured cell population).
Fig. 4.
Comparison of fluorimetric and radiochemical methods for quantitative measurement of activity and inhibition of AKR1C2 in cells. Both methods depend on facile membrane permeability of the substrate and metabolic products. (A) Continuous fluorimetric method. Fluorogenic substrate 1 permeates the cell membrane and is selectively reduced by AKR1C2 enzyme. The (apparent) intracellular kinetic parameters are determined directly through fluorimetric analysis of the cell medium. Furthermore, a two-substrate competition assay enables the determination of KM,app for the physiological substrate (dark substrate), in this case DHT, through a real-time measurement. (B) The standard radiochemical method for determining cellular kinetics relies on the analysis of metabolites after incubation with a radiolabeled physiological substrate. This method is discontinuous; it requires extraction of cell medium, separation of products by chromatography, and analysis by scintillation counting.
Remarkably, the apparent Michaelis constant for AKR1C2 (KM,app = 3.0 ± 0.4 μM) was in excellent agreement with the value found for a purified enzyme (KM,in vitro = 3.0 ± 0.2 μM). This result suggests that in this particular system the enzyme’s activity is not modulated by intracellular factors (e.g., posttranslational modification, association with cellular components). Also, it supports the notion that probe 1 and product 2 readily diffuse across the cytoplasmic membrane. Indeed, preliminary fluorescent microscopy experiments support the claim that probe 1 readily permeates the cell membrane.
Fluorimetric Determination of KM,app for Physiological Substrate DHT in Intact Cells.
The previous results demonstrate that compound 1 is a cell-permeable probe with high selectivity for AKR1C2 enzyme. This probe enables quantitative evaluation of AKR1C2 activity in intact cells in a noninvasive and continuous manner.
The next key question was whether probe 1 could also provide quantitative information about a physiologically relevant enzymatic process, namely DHT reduction to 3α-diol, by means of a real-time measurement in intact cells.
To address this point, we explored the possibility of setting up a competition between reporter substrate 1 and DHT in living cells (Fig. 4A). This two-substrate assay is based on the premise that conversion of the reporter substrate, which can be monitored in real-time (e.g., change in fluorescence intensity), decreases by increasing the concentration of the “dark substrate.” In this case, the rate of reduction of probe 1 is attenuated by DHT. Applying the steady-state kinetics to this system, KM for the dark substrate can be derived if KM for the reporter substrate and the concentration of both substrates is known (26). The value of KM,app for substrate 1 was determined earlier, and the intracellular concentration of both substrates is controlled by adjusting their concentration in the growth medium. It is assumed that both the probe and the steroid substrate permeate the cytoplasmic membrane, and therefore the intracellular and extracellular concentrations are the same.
Indeed, DHT competed with the reporter substrate for AKR1C2, and comparison of the initial rates in the presence and absence of DHT allowed for the determination of KM,app for DHT (1.3 ± 0.2 μM; see Table 1). To the best of our knowledge, this result is a rare, if not previously undescribed, example of applying the two-substrate competitive assay to obtain the kinetic parameters for a physiological substrate in intact cells. Consequently, we compared the results of the fluorimetric real-time measurement with an independent control, the conventional radiochemical assay (Fig. 4B).
Table 1.
Probe 1 enabled the quantitative evaluation of a physiological process, DHT reduction to 3α-diol
| Substrate |
KM |
KM,app |
|
|---|---|---|---|
| Purified protein, fluorimetric | Intact cells, fluorimetric | Intact cells, radiometric | |
| Reporter probe 1 | 3.0 ± 0.2 | 3.0 ± 0.4 | — |
| DHT | 1.3 ± 0.2* | 1.3 ± 0.2 | 1.3 ± 0.4 |
All values are given as micromolar concentrations. Through a two-substrate competitive assay between physiological substrate DHT and reporter substrate 1, the KM of DHT was measured directly in living cells. The conventional assay for cellular kinetics that requires discontinuous analysis of radiolabeled metabolites is in excellent agreement with the fluorimetric measurement.
*In agreement with radiometric assay (1.4 ± 0.2 μM).
The standard method for determining kinetic parameters in intact cells relies on the use of radiolabeled physiological substrates. At various points during the incubation period, the radiolabeled metabolites are extracted from the cell growth medium, concentrated, separated by TLC, and finally analyzed by scintillation counting (27). In this case, [4-14C]DHT was used as the AKR1C2 substrate to afford [4-14C]-3α-diol as the main product. For experimental details, see Supporting Text, which is published as supporting information on the PNAS web site.
Notably, an excellent agreement between the fluorimetric competitive assay and the standard radiochemical method was found (Table 1). This comparison not only validated the reliability of the fluorimetric method but also clearly highlighted its practical advantages: it is direct, continuous, and operationally simple.
Thus, selective reporter substrates such as probe 1 enable quantitative real-time examination of not only the enzyme (e.g., expression level, enzyme activity, and metabolic control mechanisms) but also the physiological substrate, in the relevant milieu of living cells.
Quantitative Measurement of AKR1C2 Inhibition in Intact COS-1 Cells.
The fluorimetric assay for direct measurement of AKR activity developed herein should also enable a quantitative evaluation of inhibitors in living cells. To demonstrate the utility of probe 1 in this regard, we examined a potent physiological inhibitor of AKR1C2, ursodeoxycholic acid (14). Metabolic rates were measured fluorimetrically as described above in the presence of ursodeoxycholate. The apparent IC50 value was obtained by holding the concentration of probe 1 constant while varying the concentration of the inhibitor (10 nM to 6 μM). The availability of KM,app for fluorogenic substrate 1 allowed the calculation of Ki,app using the Cheng–Prusoff equation (competitive inhibition mechanism for this inhibitor was established) (28). Indeed, the enzyme was inhibited by ursodeoxycholate; however, apparent IC50 in intact cells was 5-fold higher in comparison with the value obtained with purified AKR1C2 enzyme (IC50,app = 0.24 μM vs. IC50,in vitro = 0.049 μM; see Table 2). These results were also in excellent agreement with the radiochemical control assay. Both methods showed that the intracellular inhibition of AKR1C2 by ursodeoxycholic acid was in the high nanomolar range. It should be pointed out that in each assay the inhibitor competes with a different substrate; nonetheless, the kinetic parameters of inhibition match very well.
Table 2.
Apparent in situ and in vitro kinetic parameters for physiological and synthetic inhibitors of human AKR1C2
| Inhibitor | Monkey kidney cells (COS-1) |
In vitro |
|
|---|---|---|---|
| Fluorimetric, IC50, μM (Ki,app, μM) | Radiometric, IC50, μM (Ki,app, μM) | Fluorimetric, IC50, μM (Ki, μM) | |
| Natural inhibitor | |||
| Ursodeoxycholate | 0.24 ± 0.03 (0.11) | 0.14 ± 0.03 (0.070) | 0.049 ± 0.005 (0.012) |
| NSAIDs | |||
| Naproxen | 9.4 ± 0.9 | 16 ± 2 | 2.7 ± 0.2 |
| Flufenamic acid | 4.0 ± 0.6 (2.2) | — | 0.31 ± 0.03 (0.11) |
| Ibuprofen | 17 ± 2 | — | 9 ± 1 |
| Celecoxib | >50* | — | 50 ± 10 |
The fluorimetric method based on the novel metabolic probe 1 was validated by the standard radiochemical assay (see Fig. 4). Data shown are the average ± SD of three independent experiments run in triplicate.
*Concentrations of celecoxib >50 μM resulted in noticeable cell detachment.
There may be many reasons for a 5-fold difference between the in situ and in vitro inhibition potency for ursodeoxycholic acid (Table 2). Considering the fact that the KM,app and KM,in vitro for both the fluorogenic and physiological substrate were comparable, a most likely explanation is the lower effective concentration of the inhibitor inside cells. A low concentration of serum was present in the cellular assay medium, and therefore, binding of the inhibitor to serum albumin (and thus decreasing the concentration of the free compound) also may contribute to the observed difference in potency.
Recent studies have suggested that inhibition of AKR1Cs may contribute to the anticancer activity of NSAIDs (19, 23). Specifically, it is hypothesized that the inhibition of these enzymes modulates the formation of proliferation-inducing steroids and prostaglandins (29).
Consequently, the fluorimetric assay was applied to examine common anti-inflammatory drugs and their inhibition potency in intact COS-1 cells. Naproxen was investigated in detail as an example of a synthetic inhibitor with low micromolar potency (Table 2). Once again, good agreement between the two measurements reconfirmed the reliability of the fluorimetric method, in this case in the context of a weaker inhibitor. Flufenamic acid showed the best potency out of examined NSAIDs, whereas celecoxib was inactive against AKR1C2 within a nontoxic concentration range.
In analogy to ursodeoxycholate, synthetic NSAIDs were less potent in living cells (2- to 10-fold). As mentioned above, we ascribed these differences to the membrane permeability and bioavailability properties of the individual compounds. However, the comparative potency in cells followed the same trend found with the purified enzyme.
In summary, probe 1 and the fluorimetric assay described here provides a practical platform for the examination of AKR1C2 inhibitors in cells. We confirmed that in addition to cyclooxygenase (COX)-1 and -2 enzymes, the common anti-inflammatory drugs naproxen, flufenamic acid, and ibuprofen inhibit AKR1C2 in intact cells at micromolar concentrations. These results further substantiate the claim that AKRs are off-targets for NSAIDs in vivo.
Discussion
To gain new insights into metabolism and its dynamic properties, we need new tools for the noninvasive monitoring of enzyme activity in the context of metabolic networks, that is, in intact cells, tissues, and organisms. Protein expression may in some cases be misleading because the activity of proteins is often controlled by metabolic regulatory mechanisms such as covalent modification or metabolite allosterism. Indeed, there are numerous examples that show poor correlation between protein expression and protein activity (30, 31).
For decades, the field of enzyme histochemistry has centered on the development of quantitative methods for measuring enzyme activity in tissue sections (32). These methods rely on reagents that capture the product of an enzymatic reaction to form a colored precipitate. A common histochemical method for oxidoreductases utilizes tetrazolium salts as the terminal oxidant, which upon reduction yields a colored insoluble product formazan (ref. 32, pp. 21–22). The selectivity is achieved by the addition of a large excess of the physiological substrate and a single electron carrier to transfer electrons from NAD(P)H produced by the enzyme of interest to the tetrazolium reagent. Although quantitative measurements of enzyme activity can be obtained, these protocols typically require tissue fixation or freezing, or cell membrane perforation, resulting in significant cellular damage.
These limitations are eliminated by the use of membrane-permeable fluorogenic substrates, which allow for highly sensitive monitoring of enzyme activity in intact cells by either fluorimetry or fluorescent microscopy. Design of such reporter substrates must combine an optical switching mechanism with an enzyme recognition unit to assure selective and sensitive read out of desired enzyme(s). For example, constructing fluorogenic substrates for sequence-selective proteases is relatively straightforward: a fluorophore is attached at the terminus of a selectivity-determining peptide sequence (33).
This modular design is not readily applicable to the development of dehydrogenase probes because a clear separation of the recognition and reporting elements may not be possible. Consequently, we approached this challenge by developing robust emission switches based on ketone–alcohol redox transformation wherein the ketone group is directly attached to the aromatic system of the fluorophore (9). This step was followed by searching for promiscuous enzymes in the context of important metabolic pathways. In addition to systematic screening, the search for promiscuous enzymes may be focused by a close examination of physiological substrates. For example, enzymes involved in lipid metabolism are likely to have large hydrophobic active sites, which may accommodate aromatic “nonphysiological” substrates. In many instances, the metabolic role of an enzyme requires significant substrate plasticity (10, 11, 34).
This work illustrates the potential of this emerging design algorithm. Methyl ketone 3 was identified as an excellent reporter substrate for human AKR1Cs in vitro; however, it showed very low selectivity for these enzymes in intact mammalian cells. This result exposed yet another key challenge in the development of metabolic indicators, namely the issue of achieving selectivity in living cells containing a plethora of reductase enzymes. Remarkably, a simple structural alteration, exchanging the methyl group for the phenyl ring at the ketone moiety, gave highly selective probe 1. Apparently, a varying degree of substrate fidelity among functionally related enzymes can be exploited to achieve high selectivity, in this case favoring AKR1C2 over other endogenous reductases. It is notable that a synthetic compound with no apparent structural similarities to physiological steroid substrates can function as a selective substrate for this enzyme.
In summary, we describe cell-permeable fluorogenic probe 1 that permits direct, continuous, and operationally simple readout of the AKR1C2 activity in intact mammalian cells. It was also demonstrated that probe 1 enables the quantitative examination of a physiologically relevant process, namely reduction of steroid hormone DHT by means of a two-substrate competitive assay. This previously undescribed probe will enable a spectrum of research efforts including the development of selective inhibitors, drugs, and imaging agents.
Materials and Methods
Chemicals.
8-Hydroxyjulolidine and PPh3 were purchased from Sigma-Aldrich. The nonradioactive steroids used in the study, 3α-diol, 5α-androstane-3β,17β-diol (3β-diol), 5α-androstan-3α-ol-17-one (androsterone), 5α-androstan-3,17-dione (androstanedione), and DHT, were all purchased from Steraloids. Radioactive steroid 5α-[4-14C]DHT (57.3 mCi/mol; 1 Ci = 37 GBq) was purchased from PerkinElmer. Celecoxib was purchased from ChemPacific USA, ibuprofen and naproxen were from Cayman Chemicals, and flufenamic acid was from Lancaster Synthesis. Ursodeoxycholic acid was purchased from ICN.
Cell Culture.
CHO, COS-1, and HEK-293 cells were purchased and maintained according to the protocols provided by American Type Culture Collection. COS-1 cells were grown in DMEM with 100 units/ml penicillin, 100 μg/ml streptomycin, 4 mM l-glutamine (Invitrogen), and 10% heat-inactivated FBS (HyClone). For metabolism experiments, COS-1 cells were plated in six-well dishes at a density of 2.5 × 105 cells and were grown at 37°C in 5% CO2. Approximately 3 h before transfection, the medium was washed twice with PBS and replaced with fresh growth medium. Cells were transfected 2 days after they were plated using FuGENE6 (Roche) at a ratio of 6 μl of FuGENE6 to 1 μg of DNA (pcDNA3, pcDNA3-AKR1C2). Approximately 3 h before metabolism studies, the medium was changed to DMEM minus phenol red (Invitrogen) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 4 mM l-glutamine, and 1% charcoal/dextran-treated FBS (HyClone). Metabolism studies were initiated 24 h after cellular transfection.
Kinetics of Fluorogenic Substrate Metabolism.
Fluorogenic substrates 1 and 3 were prepared and stored as 5 mM stock solutions in acetonitrile or DMSO at 0°C. In vitro kinetics were determined as described (9). In short, 1-ml reaction mixtures containing 100 mM potassium phosphate buffer (pH 6), 0.250 mM NADPH (Roche), and 0.2KM to 5KM fluorogenic ketone 1 or 3 in acetonitrile (up to 4% vol/vol) were initiated by 2 μl of diluted AKR1C2. To determine the kinetics of metabolism of 1 in living cells, aliquots of the stock DMSO solution were diluted appropriately and added to the cells to give final concentrations of 0.2 to 5KM,app 1. The DMSO added (0.25% vol/vol) had no effect on cell viability. For fluorimetric analysis, aliquots (100 μl) of the culture medium were removed over time, collected on 96-well plates, and measured by a MicroMax 384 connected to a Jobin Yvon Fluorolog by detecting fluorescence upon excitation at 385 nm. Initial reaction velocities were obtained from plots of fluorescence vs. time by using only data points corresponding to <30% substrate reduction. The slope was divided by the change in fluorescence corresponding to complete reduction (24 h) and multiplied by the substrate concentration to obtain initial velocity in units of picomoles per hour. All fluorescence measurements were corrected for endogeneous cellular metabolism. Plots of velocity against substrate concentration were hyperbolic and could be iteratively fit to the Michaelis–Menten equation v = (Vmax × S)/(KM + S) by using KaleidaGraph (Synergy Software) to yield Vmax, KM, Vmax,intact, and KM,app and their associated standard errors. All reported enzymatic kinetic parameters are the average of three independent determinations (performed in duplicates).
Fluorimetric Determination of the KM,app of Dark Physiological Substrate, DHT.
The KM and KM,app of DHT for AKR1C2 in vitro or living cells were measured by varying the dark substrate concentrations 0.2–5KM (or KM,app), while holding fluorogenic substrate 1 concentration equal to its KM (or KM,app) for AKR1C2. DHT–probe cocktails were dissolved in acetonitrile (in vitro determinations) or DMSO (for determinations in living cells). The final concentration of acetonitrile in the in vitro assays did not exceed 4% (vol/vol), and the final concentration of DMSO in the cell medium did not exceed 0.5% (vol/vol). The presence of the acetonitrile and DMSO had no effect on initial reaction velocities. The competition reaction was followed by fluorimetric analysis as described above. The corresponding competitive substrate data, which described the fluorimetric rate in the presence and absence of varying dark substrate concentrations, were compared to give rates relative to the no-DHT control and fit to the equation vi/v0 = [(1 + KM/S)]/[1 + KM/S(1 + S′/K′M)] by using KaleidaGraph to yield K′M (the Michaelis constant for the dark substrate) as described (26). The reported Michaelis constants are the average of three independent determinations (performed in duplicates).
Supplementary Material
Acknowledgments
We thank Prof. Brent R. Stockwell for use of the tissue-culture facility. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation (D.S.) and National Institutes of Health Grant R01-CA097442 (to T.M.P.). D.J.Y. was supported by a predoctoral fellowship from the American Chemical Society Division of Medicinal Chemistry and Wyeth Research.
Abbreviations
- AKR
aldo-keto reductase
- DHT
5α-dihydrotestosterone (17β-hydroxy-5α-androstane-3-one)
- HSD
hydroxysteroid dehydrogenase
- NSAID
nonsteroidal anti-inflammatory drug
- 3α-diol
5α-androstane-3α,17β-diol.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Boonacker E., Van Noorden C. J. J. Histochem. Cytochem. 2001;49:1473–1486. doi: 10.1177/002215540104901201. [DOI] [PubMed] [Google Scholar]
- 2.Chen C. A., Yeh R. H., Lawrence D. S. J. Am. Chem. Soc. 2002;124:3840–3841. doi: 10.1021/ja017530v. [DOI] [PubMed] [Google Scholar]
- 3.Shults M. D., Imperiali B. J. Am. Chem. Soc. 2003;125:14248–14249. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]
- 4.Lichtman J. W, Conchello J. A. Nat. Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 5.Matayoshi E. D., Wang G. T., Krafft G. A., Erickson J. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman M., Ashe B., Yurewicz E. C., Patel G. Anal. Biochem. 1977;78:47–51. doi: 10.1016/0003-2697(77)90006-9. [DOI] [PubMed] [Google Scholar]
- 7.Rotman B., Zderic J. A., Edelstein M. Proc. Natl. Acad. Sci. USA. 1963;50:1–6. doi: 10.1073/pnas.50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris J. L., Backes B. J., Leonetti F., Mahrus S., Ellman J. A., Craik C. S. Proc. Natl. Acad. Sci. USA. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yee D. J., Balsanek V., Sames D. J. Am. Chem. Soc. 2004;126:2282–2283. doi: 10.1021/ja039799f. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Yee D. J., Gubernator N. G., Sames D. J. Am. Chem. Soc. 2005;127:4544–4545. doi: 10.1021/ja0428457. [DOI] [PubMed] [Google Scholar]
- 11.Froemming M. K., Sames D. Angew. Chem. Int. Ed. 2006;45:637–642. doi: 10.1002/anie.200502675. [DOI] [PubMed] [Google Scholar]
- 12.Penning T. M., Burczynski M. E., Jez J. M., Hung C. F., Lin H. K., Ma H., Moore M., Palackal N., Ratnam K. Biochem. J. 2000;351:67–77. doi: 10.1042/0264-6021:3510067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usami N., Yamamoto T., Shintani S., Higaki Y., Ishikura S., Katagiri Y., Hara A. Biol. Pharm. Bull. 2002;25:441–445. doi: 10.1248/bpb.25.441. [DOI] [PubMed] [Google Scholar]
- 14.Trauger J. W., Jiang A., Stearns B. A., LoGrasso P. V. Biochemistry. 2002;41:13451–13459. doi: 10.1021/bi026109w. [DOI] [PubMed] [Google Scholar]
- 15.Shiraishi H., Ishikura S., Matsuura K., Deyahiki Y., Ninomiya M., Kakai S., Hara A. Biochem. J. 1998;334:399–405. doi: 10.1042/bj3340399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takikawa H., Stolz A., Sugiyama Y., Yoshida H., Yamanaka M., Kaplowitz N. J. Biol. Chem. 1990;265:2132–2136. [PubMed] [Google Scholar]
- 17.Dufort I., Soucy P., Labrie F., Luu-The V. Biochem. Biophys. Res. Commun. 1996;228:474–479. doi: 10.1006/bbrc.1996.1684. [DOI] [PubMed] [Google Scholar]
- 18.Ji Q., Aoyama C., Nien Y. D., Liu P. I., Chen P. K., Chang L., Stanczyk F. Z., Stolz A. Cancer Res. 2004;64:7610–7617. doi: 10.1158/0008-5472.CAN-04-1608. [DOI] [PubMed] [Google Scholar]
- 19.Bauman D. R., Steckelbroeck S., Penning T. M. Drug News Perspect. 2004;17:563–578. doi: 10.1358/dnp.2004.17.9.872570. [DOI] [PubMed] [Google Scholar]
- 20.Deng H. B., Adikari M., Parekh H. K., Simpkins H. Cancer Chemother. Pharmacol. 2004;54:301–307. doi: 10.1007/s00280-004-0815-0. [DOI] [PubMed] [Google Scholar]
- 21.Lou H., Du S., Ji Q., Stolz A. Mol. Pharmacol. 2006;69:1662–1672. doi: 10.1124/mol.105.019794. [DOI] [PubMed] [Google Scholar]
- 22.Agapova O. A., Yang P., Wang W. H., Lane D. A., Clark A. F., Weinstein B. I., Hernandez M. R. Neurobiol. Dis. 2003;14:63–73. doi: 10.1016/s0969-9961(03)00101-3. [DOI] [PubMed] [Google Scholar]
- 23.Bauman D. R., Rudnick S. I., Szewczuk L. M., Jin Y., Gopishetty S., Penning T. M. Mol. Pharmacol. 2005;67:60–68. doi: 10.1124/mol.104.006569. [DOI] [PubMed] [Google Scholar]
- 24.Rizner T. L., Lin H. K., Peehl D. M., Steckelbroeck S., Bauman D. R., Penning T. M. Endocrinology. 2003;144:2922–2932. doi: 10.1210/en.2002-0032. [DOI] [PubMed] [Google Scholar]
- 25.Ciaccio P. J., Stuart J. E., Tew K. D. Mol. Pharmacol. 1993;43:845–853. [PubMed] [Google Scholar]
- 26.Xie D., Suvorov L., Erickson J. W., Gulnik A. S. Protein Sci. 1999;8:2460–2464. doi: 10.1110/ps.8.11.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizner T. L., Lin H. K., Penning T. M. Chem. Biol. Interact. 2003;143–144:401–409. doi: 10.1016/s0009-2797(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y.-C., Prusoff W. H. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 29.Desmond J. C., Mountford J. C., Drayson M. T., Walker E. A., Hewison M., Ride J. P., Luong Q. T., Hayden R. E., Vanin E. F., Bunce C. M. Cancer Res. 2003;63:505–512. [PubMed] [Google Scholar]
- 30.Swezey R. R., Epel D. Proc. Natl. Acad. Sci. USA. 1988;85:812–816. doi: 10.1073/pnas.85.3.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies S. E. C., Brindle K. M. Biochemistry. 1992;31:4729–4735. doi: 10.1021/bi00134a028. [DOI] [PubMed] [Google Scholar]
- 32.Van Noorden C. J. F., Frederiks W. M. Enzyme Histochemistry: A Laboratory Manual of Current Methods. New York: Oxford; 1992. [Google Scholar]
- 33.Boonacker E., Elferink S., Bardai A., Fleischer B., Van Noorden C. J. J. Histochem. Cytochem. 2003;51:959–968. doi: 10.1177/002215540305100711. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien P. J., Herschlag D. Chem. Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.