Abstract
The Cyclin E1 gene (CCNE1) is an ideal model to explore the mechanisms that control the transcription of cell cycle-regulated genes whose expression rises transiently before entry into S phase. E2F-dependent regulation of the CCNE1 promoter was shown to correlate with changes in the level of H3-K9 acetylation/methylation of nucleosomal histones positioned at the transcriptional start site region. Here we show that, upon growth stimulation, the same region is subject to variations of H3-R17 and H3-R26 methylation that correlate with the recruitment of coactivator-associated arginine methyltransferase 1 (CARM1) onto the CCNE1 and DHFR promoters. Accordingly, CARM1-deficient cells lack these modifications and present lowered levels and altered kinetics of CCNE1 and DHFR mRNA expression. Consistently, reporter gene assays demonstrate that CARM1 functions as a transcriptional coactivator for their E2F1/DP1-stimulated expression. CARM1 recruitment at the CCNE1 gene requires activator E2Fs and ACTR, a member of the p160 coactivator family that is frequently overexpressed in human breast cancer. Finally, we show that grade-3 breast tumors present coelevated mRNA levels of ACTR and CARM1, along with their transcriptional target CCNE1. All together, our results indicate that CARM1 is an important regulator of the CCNE1 gene.
Keywords: ACTR, CCNE1, histone, arginine methylation, breast tumor
Cyclin E1 (CCNE1) protein and mRNA levels are tightly regulated as an endpoint of several regulatory pathways that are critical for growth control and frequently altered in cancer cells (1, 2). CCNE1 gene transcription is undetectable in G0 and G1 phases of the cell cycle, whereas it rises sharply during a narrow window of time that precedes each entry into S phase. Several pieces of evidence suggest that the periodic association of activators E2Fs– and E2F–pocket protein complexes regulate CCNE1 gene expression (3–18). E2F complexes bound to this gene were found to recruit chromatin modifiers, including members of the SNF2-like helicase family, type I histone deacetylases, the acetyltransferase CBP/p300, the lysine methyl transferase SUVAR39H1, and the protein arginine N-methyltransferase (PRMT) 5 (7, 9–14, 17, 18), suggesting that they foster periodic chromatin remodeling of the CCNE1 promoter region (11, 12, 14). Notably, repression of the CCNE1 gene in G0-G1 correlates with the methylation of H3-K9 and H4-R3 on a single nucleosome positioned at the transcriptional start site (11–14). Conversely, the late G1 activation of the CCNE1 gene correlates with decreased H3-K9 methylation and with enhanced H3/H4 acetylation of the same chromatin region (11–14). Here, we reveal that this CCNE1 proximal promoter region is targeted by another histone arginine methyl-transferase, the type I enzyme PRMT4 [coactivator-associated arginine methyltransferase (CARM1)] (19–25). PRMT4/CARM1 was initially described as a transcriptional coactivator of the p160 family of nuclear receptor-associated factors (Src-1/NCoA1, GRIP1/TIF2/Src-2/NCoA2, ACTR/AIB1/SRC-3/NCoA3) and with p300/CBP (19–30). Consistent with this function, CARM1 recruitment at nuclear receptor-responsive genes was found to coincide with their activation (23, 24, 26–29) and with histone H3-R17- and H3-R26-specific methylations of their promoter region (23, 24, 26–29). More recently, CARM1 was also found to associate and cooperate with p53, NF-κB and LEF1/TCF4 (29, 31, 32), suggesting that this enzyme plays pleiotropic roles in cell proliferation and survival. Here, we show that CARM1 acts as a potent coactivator for the CCNE1 gene together with ACTR and through E2F sites. Accordingly, we show that CCNE1 gene expression is altered in CARM1-deficient (30) and CARM1-overexpressing cells, and we provide data suggesting that CARM1, ACTR, and CCNE1 overexpression might be linked in high-grade breast tumors.
Results
CARM1-Dependent Chromatin Modifications Are Growth-Stimulated at the CCNE1 Promoter.
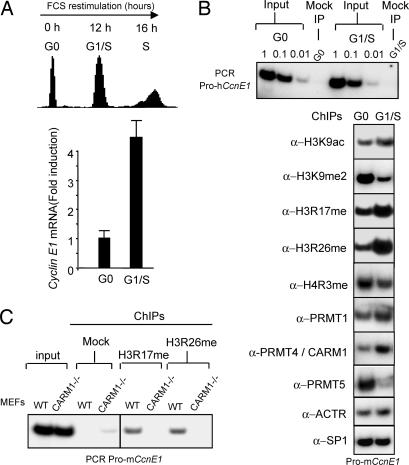
Consistent with previous reports (11–14), chromatin immunoprecipitation analysis of nucleosomal histones positioned within the transcriptional start site region of the CCNE1 gene reveals that repression of this gene in G0-arrested Swiss3T3 cells is associated with the dimethylation on K9 of H3, whereas its G1/S activation correlates with increased acetylation of this residue (Fig. 1A and B). In G0-arrested cells, we also previously observed that this region was associated with R3-methylated H4. Here, using antibodies that recognize both mono- and dimethylated forms of H4-R3, we extended this observation and found that these modifications, ascribed to PRMT1 and PRMT5, moderately decreased at the G1/S transition. Interestingly, H4-R3 methylation coincided with an accumulation of PRMT5 on this promoter region in G0 cells and an accumulation of PRMT1 in G1/S cells, suggesting that both type II and type I PRMTs, successively contribute to this modification. Aiming at identifying other PRMT-dependent marks present on the CCNE1 promoter, we detected greatly increased levels of H3-R17 and H3-R26 methylation in late G1/S samples (Fig. 1B), two modifications ascribed to PRMT4/CARM1. Consistently, ChIP assays showed a clear coincidental CARM1 accumulation on this promoter region in G1/S cells (Fig. 1B). To confirm CARM1’s involvement in these modifications, ChIP assays were performed on mouse embryo fibroblasts (MEFs) lacking CARM1 (CARM1−/−) (30). In these cells, H3-R17 and H3-R26 methylations were undetectable at the CCNE1 gene, whereas both modifications were observed in control MEFs (WT) isolated from CARM1−/+ littermates (Fig. 1C). All together, these results indicate a targeted recruitment of CARM1 at the transcription start site region of the CCNE1 gene, after growth stimulation.
Fig. 1.
G0-G1/S activation of CCNE1 coincides with the CARM1/PRMT4-stimulated arginine-methylation of nucleosomal histones located at the CCNE1 promoter. (A) Serum-starved (G0) Swiss 3T3 cells were stimulated by serum and analyzed for progression into S by propidium iodide (PI) staining/FACScan analysis and for expression of CCNE1 mRNA by quantitative RT-PCR (Q-RTPCR). (B) ChIP analyses of the mouse CCNE1 promoter with antibodies against PRMT1, CARM1, PRMT5, SP1, and modified histone residues. Formaldehyde-cross-linked chromatin samples prepared from G0-arrested and G1/S 3T3 cells were immunoprecipitated with the indicated antibodies and analyzed by PCR for the presence of mouse CCNE1 promoter fragment. PCR on gradually diluted (1, 0.1, 0.01) input chromatin confirmed that equal amounts of materials were used. (C) ChIP analyses of the mouse CCNE1 promoter by using antisera directed against arginine-methylated histone H3 (H3R17me and H3R26me) and chromatin from exponentially growing CARM1−/− or control (WT, CARM1+/−) MEFs.
CCNE1 Gene Expression Is Impaired in CARM1-Deficient Cells.
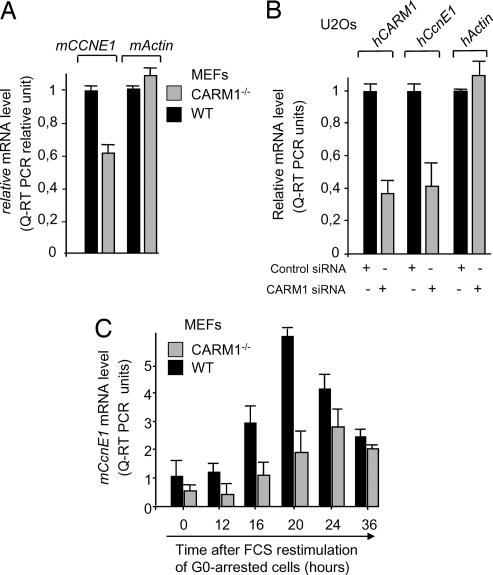
Consistent with a role of CARM1 in CCNE1 gene expression, CCNE1 mRNA levels were significantly lower (60%) in exponentially growing CARM1−/− primary MEFs than in CARM1-positive MEFs (Fig. 2A). A similar reduction of CCNE1 transcripts was observed in human U2Os cells transiently transfected with a synthetic siRNA directed against CARM1 (33) that lead to a 3-fold reduction in CARM1 mRNA levels (Fig. 2B). Importantly, mRNA levels of several other genes, exemplified here by Actin, (Fig. 1 A and B and data not shown) were found unaffected in the same cells, suggesting that CARM1 is involved in the transcriptional control of some, but not all, genes.
Fig. 2.
CARM1 inactivation impacts on the level and timing of expression of CCNE1. (A) mCCNE1 mRNA levels in exponentially growing CARM1−/− or control (WT, CARM1+/−) MEFs were determined by Q-RTPCR on total RNA. Histograms are the average of results obtained from two independent cell populations. (B) siRNA-mediated CARM1 knockdown leads to a decrease in CCNE1 mRNA level. Human U2Os cells were transfected either with a scrambled control siRNA or siRNA directed against human hCARM1 and analyzed 48 h later for hCCNE1, hCARM1, and hActin mRNA contents by Q-RTPCR. (C) Kinetics of expression of mCCNE1 mRNA after serum restimulation of G0-arrested CARM1−/− or control (WT, CARM1+/−) MEFs. Serum-starved MEFs were incubated with high serum for the indicated time, and mCCNE1 mRNA levels were determined by Q-RTPCR.
Considering that CCNE1 mRNA expression is temporally regulated during the first cell cycle after release from quiescence, we also tested whether CARM1 inactivation might have affected these kinetics. Quiescent CARM1−/− and control MEFs were stimulated to reenter the cell cycle by serum addition and were analyzed for CCNE1 mRNA levels. As shown in Fig. 2C, both the rate and kinetics of CCNE1 mRNA-expression were altered in CARM1-deficient cells, the latter showing a peak of CCNE1 expression with a delay of 4 h compared with control cells.
Collectively, these data support the notion that CARM1 is required for optimum activation and for proper timing of endogenous CCNE1 mRNA expression in mammalian cells.
CARM1 Acts as a Transcriptional Coactivator for the CCNE1 Promoter.
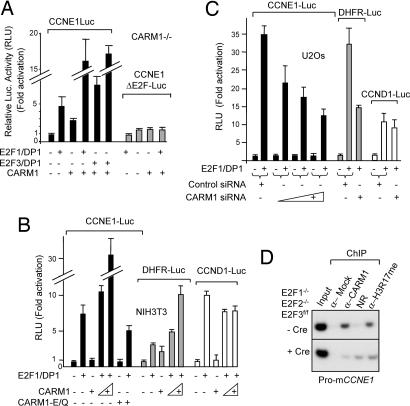
CARM1 has been described as a potent coactivator of transcription (19, 20). To evaluate its impact on CCNE1 promoter activity, reporter plasmids driven by the human CCNE1 promoter (4) were transfected in CARM1−/− 3T3 or NIH 3T3 cell lines, along with expression vectors for CARM1 and for the CCNE1 activators, i.e., the activating E2Fs, (E2F1, E2F3a) and their partner, DP1. CARM1 increased both E2F-stimulated (Fig. 3A and C) and basal (Fig. 3A) transcription of the CCNE1 reporter. Both effects depend on the presence of E2F sites in the CCNE1 promoter, because a promoter construct lacking these sites (4) was not stimulated by CARM1 (Fig. 3A). Notably, this stimulatory effect was not observed with CARM1-E/Q, a CARM1 mutant bearing a mutation (E267Q) in the S-adenosyl-methionine-binding domain that impairs its ability to methylate histones (Fig. 3B) (33). This indicates that CARM1 enzymatic activity is required for full enhancement of E2F-mediated CCNE1 gene transcription. It is worth noting that, during these assays, CCNE1 reporter activity [relative luciferase units (RLU) normalized to cotransfected β-gal activity] was always significantly lower in CARM1−/− 3T3 cells than in CARM1-positive NIH 3T3 cells (data not shown), suggesting that endogenous CARM1 also contributes to optimize CCNE1 reporter activation. Consistent with this hypothesis, siRNA-mediated depletion of CARM1 resulted in a reduction of the E2F-driven activation of the CCNE1 reporter gene in U2Os cells (Fig. 3C).
Fig. 3.
CARM1 acts as an E2F-dependent coactivator on CCNE1 promoter. (A) Ectopic expression of CARM1 stimulates the CCNE1 promoter and potentiates its E2F/DP-dependent transactivation in CARM1−/− cells. CARM1−/− 3T3 cells were transfected with a luciferase reporter gene driven by the WT human CCNE1 promoter (CycE1-luc) or the corresponding DNA bearing mutations within the six E2F sites (CycE1ΔE2F-Luc), together with CMV-β-gal and combinations of expression vectors encoding E2F1, E2F3a, DP1, or CARM1, as indicated. Results are expressed in relative luciferase units (RLU), normalized to β-gal. (B) CARM1 ectopic expression potentiates the transactivation of some, but not all, E2F1/DP1-responsive promoters. NIH 3T3 cells were transfected with luciferase reporter genes driven by human CCNE1 promoter, human DHFR promoter, or human CCND1 promoter, together with CMV-β-gal and vectors encoding E2F1, DP1, CARM1, or an enzymatically inactive CARM1-E/Q mutant, as indicated. (C) siRNA-mediated depletion of CARM1 inhibits the E2F1/DP1-dependent transactivation of the human CCNE1 and DHFR promoters but not of the CCND1 promoter. U2Os cells were transfected with the indicated luciferase reporters, CMV-β-gal and E2F1/DP1, together either with scrambled control siRNA or increasing amounts of siRNA directed against human hCARM1 and 48 h later were analyzed for luciferase activity normalized to β-gal (RLU). (D) pRb-associated E2Fs (E2F1, E2F2, and E2F3) are required for CARM1 recruitment and H3R17 methylation at the CCNE1 gene in vivo. ChIP analysis of the mouse CCNE1 promoter with antibodies to CARM1 and arginine-methylated histone H3 (H3R17me) was performed on chromatin samples prepared from exponentially growing E2F1−/−E2F2−/−E2F3flox/flox 48 h after infection either with Cre recombinase (+Cre) or empty control (−Cre) retroviruses. Immunoprecipitated chromatin samples were analyzed by PCR for mouse CCNE1 promoter fragment as in Fig. 1.
Significantly, CARM1 was also found to have an impact on other E2F-target genes. As shown in Fig. 3, ectopic expression or depletion of CARM1 had an effect on both E2F-stimulated and basal transcription of two other reporter constructs driven by the DHFR (Fig. 3B and C) and cdc6 (data not shown) promoters (34), i.e., two other E2F-responsive promoters for which expression rises at the G1/S transition. Accordingly, endogenous mRNA levels of DHFR and cdc6 were significantly lower in CARM1 siRNA-treated U2Os cells (Fig. 6, which is published as supporting information on the PNAS web site). Moreover, as described for the CCNE1 gene, ChIP assays showed increased levels of CARM1, H3-R17me, and H3-R26me at the DHFR promoter in late G1/S samples (Fig. 7, which is published as supporting information on the PNAS web site).
However, not all described E2F-responsive promoters appear to be sensitive to CARM1; we observed that neither CARM1 overexpression nor depletion had significant effects on the E2F-stimulted or basal transcription of the CCND1 promoter (35) (Fig. 3 B and C).
To further explore the involvement of E2Fs in the recruitment of CARM1 at the CCNE1 gene in vivo, ChIP assays were performed on cells deficient for the three pRB-associated E2Fs (15). CARM1 and H3-R17me associations with the CCNE1 promoter region were assessed in E2F1−/−E2F2−/−E2F3f/f MEFs [i.e., E2F1−/−E2F2−/− knockout MEFs with a conditional (floxed) E2F3 allele] (15) treated either with retroviruses coding for Cre recombinase (36) or with control empty viruses. As shown in Fig. 3D, Cre-mediated ablation of E2Fs resulted in significant, although uncomplete, reduction of the presence of CARM1 and H3-R17me with this DNA region, providing additional evidence that E2Fs participate to the recruitment of CARM1 at the CCNE1 promoter in vivo.
Finally, coimmunoprecipitation experiments performed on nuclear extracts showed that endogenous E2F1 was detectable in an anti-CARM1 immunoprecipitate, indicating the existence of endogenous complexes containing E2Fs and CARM1 (Fig. 4A).
Fig. 4.
CARM1 and the p160 coactivator member ACTR/SRC3/AIB1 cooperate at the CCNE1 promoter. (A) Endogenous CARM1 coimmunoprecipitate with E2F1 and ACTR. A fraction of the input HeLa cellular extract (Input) and the proteins immunoprecipitated from this extract either by α-CARM1 or control Ab (IgG) were probed for the presence of E2F1 (Lower) or ACTR (Upper) by immunoblotting. (B) ACTR is required to detect an association between CARM1 and E2F1 in vitro. Equivalent amounts of GST-E2F1 proteins bound to beads (Bottom) were incubated with in vitro translated (IVT) 35S-labeled CARM1 in the presence (+) or absence (−) of a mixture of IVT unlabeled and 35S-labeled ACTR protein. GST-E2F1-pulled-down radiolabeled CARM1 (Top) and ACTR (Middle) proteins (P), and unbound proteins in the supernatant (S) were visualized by autoradiography. (C) ACTR is expressed in U20S and is required for CCNE1 mRNA full expression. Shown is Q-RTPCR analysis of the human hACTR, hCCNE1, and hActin mRNA levels in exponentially growing U2Os cells expressing either shRNA directed against hACTR (15) or scrambled control shRNA. (D) ACTR is required to CARM1 recruitment on the CCNE1 gene in vivo. ChIP analysis of the human CCNE1 promoter with antibodies to CARM1, ACTR, or modified histones (H3R17me and H4R3me) was performed on chromatin samples prepared from U2Os cells expressing either shRNA directed against hACTR or scrambled control shRNA. Immunoprecipitated chromatin samples were analyzed by PCR for human a CCNE1 promoter fragment (hCE1). (E) CARM1 and ACTR cooperatively stimulate the E2F1/DP1-mediated transactivation of the CCNE1 promoter. 3T3 CARM1−/− fibroblasts were transfected with hCCNE1-luc and CMV-β-gal reporters, together with combinations of E2F1, DP1, CARM1, or ACTR, as indicated. Results are expressed in RLU normalized to β-gal activity. (F) Cooverexpression of CARM1 and ACTR leads to an up-regulation of the endogenous CCNE1 mRNA level. NIH 3T3 cells were transfected with a plasmid encoding a puromycin-resistance gene (pBABEpuro), together with combinations of expression vectors encoding CARM1 or ACTR, as indicated. After 4 days of selection in the presence of puromycin, transfected cells were analyzed for mCCNE1 mRNA contents by Q-RTPCR.
Thus, although much of the previous evidence for the involvement of CARM1 in transcription has come from studies involving nuclear receptors and their p160 coactivators, our data suggest the existence of an E2F-dependent recruitment of enzymatically active CARM1 to the CCNE1 gene and to a subset of other E2F target genes (29, 31, 32).
ACTR Recruits and Cooperates with CARM1 at the CCNE1 Promoter.
Taking into account that CARM1 functional association with transactivators have been described to occur either through direct protein–protein interaction or through their common association with platform proteins, we next tested whether CARM1 interacted directly or indirectly with E2F/DP factors. Using various sources of recombinant CARM1 and DP1, E2F1, E2F3, and E2F4 proteins, we repeatedly failed to detect such direct interaction in vitro. This is illustrated by the experiment shown in Fig. 4B, in which recombinant GST-E2F1 protein failed to pull down in vitro translated CARM1.
Nevertheless, E2F1 was recently found to interact with ACTR/AIB1/SRC3/NcoA3 (18), a member of the p160 family of coactivators for nuclear hormone receptors, and because other p160 family members were shown to directly interact with CARM1 in vivo and in vitro (19–21), we hypothesized that ACTR might be needed to recruit CARM1 to E2F1 complexes. Consistent with this model, the addition of recombinant ACTR to CARM1 triggered its pull down by E2F1, providing evidence that an E2F1–ACTR–CARM1 trimeric complex can form, at least in vitro. (Fig. 4B). Importantly, coimmunoprecipitation experiments performed on cellular extracts confirmed that endogenous ACTR, like E2F1, was detectable in anti-CARM1 immunoprecipitates (Fig. 4A), indicating that these complexes might also exist in a cellular context. To test whether ACTR might be involved in the recruitment of CARM1 at the CCNE1 gene in vivo, ChIP assays were performed on cells treated with a selectable ACTR short hairpin RNA (shRNA). Efficient shRNA-mediated depletion of ACTR mRNA (Fig. 4C) and protein levels (Fig. 8, which is published as supporting information on the PNAS web site) were obtained in U2Os cells and resulted in significant inhibition of CCNE1 mRNA expression (Fig. 4C), indicating that ACTR, like CARM1, is required for full expression of the CCNE1 gene in these cells. ChIP assays performed with antibodies directed against human ACTR and CARM1 showed that both proteins are present at the human CCNE1 gene in this cell line (Fig. 4D, control shRNA-treated cells). Significantly, associations of these two proteins and of methylated H3R17 with this DNA region were strongly reduced in ACTR-depleted cells (Fig. 4D), providing evidence that ACTR is required to recruit CARM1 at the CCNE1 promoter in vivo.
To further define the functional relevance of this ACTR–CARM1 association for CCNE1 gene expression, a CCNE1 promoter reporter construct was transfected in U2Os, NIH 3T3 (data not shown), or CARM1−/− 3T3 cells (Fig. 4E), together with E2F1, DP1, hACTR, and CARM1. In the absence of the ACTR construct, the results were essentially the same as those described in Fig. 3, i.e., CARM1 acted as a potent coactivator of the E2F-driven induction of the CCNE1 reporter. Significantly, this synergistic activation was enhanced upon cotransfection of ACTR and CARM1, suggesting that these two factors cooperate to coactivate the CCNE1 gene (Fig. 4E). Interestingly, when performed in CARM1−/− cells, this reporter gene assay also revealed that ACTR alone did not coactivate E2F1-stimulated transcription of the CCNE1 reporter gene (Fig. 4E), suggesting that ACTR requires CARM1 to function as a coactivator of the CCNE1 gene.
Accordingly, we also observed, that endogenous CCNE1 mRNA levels were significantly increased in 3T3 mouse fibroblasts transfected and drug-selected to cooverexpress hACTR and CARM1, whereas this increase was barely detectable in cells transfected with either hACTR or CARM1 alone (Fig. 4F).
All together, these data strongly suggest that CARM1 is recruited by ACTR at the CCNE1 gene, where they cooperate to enhance its E2F1-mediated transactivation.
Coincidental Expression of ACTR, CARM1, and CCNE1 in Breast Tumors.
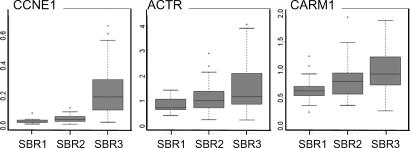
Having shown the direct impact of CARM1 and ACTR overexpression on CCNE1 gene expression in cultured cells, we next investigated whether the expression levels of CARM1 and ACTR might help to explain CCNE1 overexpression observed in various tumors (1, 2, 37). In a pilot screen, we addressed this question in breast tumors, in which CCNE1 overexpression is frequently observed in tumors of high grade and of high proliferation index (37–39). Likewise, ACTR/AIB1/SRC-3, is frequently amplified and overexpressed in human breast cancers of various grades (40, 41). mRNA expression levels of CCNE1, ACTR, and CARM1 were monitored by quantitative RT-PCR (Q-RTPCR) in a cohort of 81 human breast carcinomas of various types (42) that were ordered in three groups according to their Scarff-Bloom and Richardson (SBR) grades 1, 2, and 3. This analysis revealed a clear codistribution of the mRNA levels of the three genes, notably in grade-3 tumors that showed coelevated mRNA levels of ACTR and CARM1, along with their transcriptional target, CCNE1 (Fig. 5).
Fig. 5.
Codistribution of ACTR/SRC3/AIB1, PRMT4, and CCNE1 mRNA expression levels in breast tumors. RNA samples from 81 human breast tumors were analyzed for mRNA levels of these genes by normalized Q-RTPCR and ordered in three groups according to SBR grades 1, 2, and 3. Distribution of mRNA expression values for the three genes and for a given grade are shown as box plots. The line in the center of each box represents the median value of the distribution, and the upper and lower ends of the box are the upper (25th) and lower (75th) quartiles, respectively. The whiskers show the minimum and maximum data values.
Discussion
In earlier studies, we revealed that nucleosomes positioned in the transcription start site region of the CCNE1 gene were undergoing cell cycle variations in histone acetylation and methylation that correlate with CCNE1 expression (11, 12, 14). Here, we found that the same regulatory region interacts with nucleosomes methylated on H3-R17 and H3-R26. Methylation of histones by PRMTs is increasingly being found to play an important and dynamic role in gene regulation (19, 20). Thus, PRMT5, a type II PRMT, was shown to catalyze histone H4 mono- and symmetrical NG,NG-dimethylarginine methylation and was found to coincide with gene repression, in particular of the CCNE1 gene (14, 43). In contrast, PRMT1 and CARM1, two type I PRMTs, have been proposed to play a role in gene activation and were shown to catalyze mono- and asymmetrical NG,NG-dimethylarginine methylation on histone H4 (R3) and H3 (R2, R17, and R26), respectively (19, 20). Our ChIP assays clearly show that CARM1 is present at the CCNE1 proximal promoter and that its recruitment increases during the G0 to G1/S progression, coinciding with enhanced levels of H3-R26 and H3-R17 methylation. Accordingly, these modifications of nucleosomes positioned at the start site region of the CCNE1 gene are undetectable in CARM1−/− fibroblasts, indicating that CARM1 is the major or sole enzyme responsible for the serum-stimulated methylation of H3R26 and H3R17 in this promoter region. Consistent with a role for this enzyme in endogenous CCNE1 gene activation, we found decreased CCNE1 mRNA levels in CARM1−/− MEFs and in CARM1 siRNA-depleted U2Os cells. Accordingly, reporter gene assays showed that CARM1 acts as a potent transcriptional coactivator of the CCNE1 gene and of other G1/S-regulated genes (DHFR, cdc6), together with E2F/DP and by way of the E2F sites present in these promoters. This result is consistent with recent reports showing that the CARM1 transcriptional coactivating function is not restricted to nuclear receptors, but that it also associates and cooperates with p53, NF-κB, and LEF1/TCF4 (29–31). Thus, this ubiquitously expressed enzyme is likely to have a more pleiotropic function than was originally thought, notably in the control of the cell cycle and survival. Consistent with this hypothesis, mouse embryos with a targeted disruption of CARM1 are small in size and die perinatally (30). Moreover, in agreement with its role in CCNE1 gene expression during cell cycle reactivation (from G0 to S phase), we observed that CARM1−/− MEFs have altered capacity to reenter the cell cycle from quiescence, whereas they grow as fast as WT cells when they are maintained in high serum and exponentially growing conditions (Fig. 9, which is published as supporting information on the PNAS web site). Interestingly, this phenotype resembles that of MEFs genetically null for CCNE genes (44).
Our data suggest that CARM1 does not directly interact with activator E2F/DP proteins. Consistent with what was observed with nuclear receptors, its recruitment to E2F in vitro and to the CCNE1 proximal promoter in vivo seems to require the presence of a member of the p160 family of coactivators, ACTR. Accordingly, ChIP analyses indicate that ACTR, like CARM1, is associated in vivo with the proximal promoter region of the CCNE1 gene. This result is in agreement with a recent report showing that, in estrogen receptor-negative breast cancer cells, ACTR associates specifically with E2F and potentiates its activity on target genes, including CCNE1 (18). Interestingly, our reporter assays indicate that ACTR overexpression does not potentiate the E2F-mediated activation of the CCNE1 promoter in CARM1−/− cells, suggesting that CARM1 and ACTR are part of the same coactivator complex whose effector activity is carried out by the arginine methyl-transferase activity of CARM1.
The fact that CARM1-mediated H3R26 and H3R17 methylations increase at the CCNE1 and DHFR proximal promoters and coincide with their expression in G1/S cells suggests that these chromatin modifications are, at least in part, responsible for the ACTR/CARM1-positive effects on the transcription of these genes. However, the molecular mechanism by which arginine methylation of histones by CARM1 contributes to chromatin remodeling and transcription remains unknown. Thus, although recent reports clearly demonstrate that histone H4 modification by PRMT1 is essential both in vivo and in vitro, for many subsequent histone modifications (29, 45), this remains unclear for CARM1-mediated histone modification on H3. Recent reports provide evidence that CARM1-mediated methylation occur after acetylation and PRMT1-mediated methylation (27, 29, 45), suggesting a role for CARM1 in the reinforcement or stabilization of the transcriptional response rather than in its initiation. Consistent with this model, our ChIPs show that PRMT1 also is present at the CCNE1 promoter in G1/S cells. Methylation of non-histone proteins by CARM1 might also be involved in CCNE1 gene activation. Indeed, several nuclear proteins are methylated at various stages of gene regulation, namely, transcription initiation and elongation, splicing, and mRNA transport (19, 20, 46). Notably, CARM1 was shown to methylate and modulate the transcriptional activity of the acetyltransferases CBP/p300 (33, 47), factors that have been described as potent coactivators of the CCNE1 gene at the G1/S transition (13). Interestingly, CBP/p300 can also associate with the p160 platform protein family, including ACTR (20, 23, 26). Thus, the G1/S activation of the CCNE1 gene may depend on the recruitment of a CBP/p300–ACTR–CARM1 coactivator complex by E2Fs or E2F-associated factors at the proximal promoter region of the CCNE1 gene, where possibly it would trigger chromatin modifications compatible with transcription.
Our findings that CARM1 overexpression increases endogenous CCNE1 mRNA levels led us to speculate that increased level of this enzyme could be involved in the aberrant CCNE1 and other E2F-stimulated gene expression observed in various types of tumors. The increased level of cyclin E1 protein likely accelerates tumor progression through multiple mechanisms, including increased proliferation, alteration of the fidelity of DNA replication, increased genetic instability, and tumor suppressor inactivation (1, 2, 37–39). In particular, deregulated expression of the CCNE1 gene has been correlated with aggressive tumor characteristics in high-grade breast cancer (37–39); the hazard ratio for this type of cancer being lethal is >10 times higher for patients with high total cyclin E1 protein levels compared with those with low cyclin E1 levels (39). This overexpression is likely to result from various alterations, including an inactivation of the Rb pathway, which leads to the up-regulation of the CCNE1 promoter activity (1, 2, 38). However, it is possible that coactivators, such as ACTR and CARM1, also participate in this process. Thus, ACTR/AIB1 is amplified and/or overexpressed in various carcinomas, including 30% of breast tumors of various grades, where it affects both estrogen-receptor- and E2F-dependent transcriptions (18, 40, 41). Similarly, CARM1 was recently found to be overexpressed in androgen-independent prostate carcinomas (48). Here, we conducted a pilot experiment on breast tumor specimens of various grades and found cooverexpression of CCNE1, ACTR, and CARM1 genes in grade-3 tumors. Although one should be cautious in the interpretation of this type of multivariate models, this result, together with functional evidence showing that CARM1 and ACTR belongs to the same transcriptional complex, supports the notion that cooverexpression of these factors might play an important role in the aberrant CCNE1 expression observed in aggressive tumors.
In conclusion, we identified the type I arginine methyl-transferase CARM1 as an important positive regulator of the CCNE1 gene.
Collectively with our earlier report showing that a distinct PRMT, the type II PRMT5, is a negative regulator of the CCNE1 gene (14), these results open previously undiscovered directions that aim to evaluate the role of these enzymes and of protein methylations in cell proliferation and oncogenesis.
Materials and Methods
Cell Culture, siRNA and shRNA Transfections, and Reporter and Proliferation Assays.
All cells were cultured in DMEM with 10% FBS. Mouse fibroblasts were rendered quiescent by incubation in DMEM containing 0.1% FBS for 48 h. For reporter gene assays, exponentially growing cells plated in six-well plates were transfected with a total of 1.5 μg of indicated plasmids by using lipofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Luciferase and β-gal activities were measured on 1 × 105 cells as described in refs. 8 and 14. siRNA-mediated depletions of human CARM1 and ACTR in U2OS cells were obtained as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, by transfection (oligofectamine; Invitrogen) of 1 × 105 cells with 50 pmol of double-stranded synthetic oligonucleotides whose sequences are described in Supporting Materials and Methods (Eurogentec, Brussels, Belgium).
CMV-driven expression constructs for full-length CARM1, ACTR, E2F1, DP1, and DHFR-, CCND1-, and human CCNE1-luciferase reporter constructs have been described elsewhere (4, 8, 14, 18, 33, 34, 35).
ChIP Assays.
Swiss 3T3, WT MEFs, CARM1−/−, E2F1−/−E2F2−/−E2F3f/f, or U2Os cells were used for ChIP and processed essentially as described in refs. 12 and 14. Removal of the E2F3 alleles of E2F1−/−E2F2−/−E2F3f/f MEFs (15, 16) was achieved by infecting cells with retrovirus particles encoding a “self-excising” CRE recombinase (36) and was used for ChIP 2 days later. Chromatin was immunoprecipitated with histone tail-specific modifications antibodies (#06942, #07213, #07214, #07215, #07404, #07080, #07405, and #07212; Upstate Biotechnology, Lake Placid, NY), α-CARM1 (#07080; Upstate Biotechnology), α-PRMT1 (AB7027; Abcam, Cambridge, MA), α-PRMT5 [mix of BD#611539 (Becton Dickinson, Erembodegem, Belgium) and #07405 (Upstate Biotechnology)], α-SP1 (sc59; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-ACTR/AIB1 (BD#611105; Becton Dickinson). The immunoprecipitated DNA was analyzed by PCR for the presence of fragments corresponding to the transcription start site region of the mouse CCNE1, mouse DHFR, human CCNE1, or human CCND1 genes by using primers whose sequence is listed in Supporting Materials and Methods.
RNA Isolation and Q-RTPCR Analysis.
Total RNA was isolated from cells by using the High Pure RNA isolation kit (Roche, Meylon, France). Reverse transcription was performed by using the MMLV reverse transcriptase (Invitrogen) and random hexamers.
All these steps were performed as described in ref. 40. Quantitative PCR parameters are available from the authors upon request. Target gene quantities were normalized to S26 and RPLPO RNAs. Primers to amplify human DHFR and hcdc6 genes are described elsewhere (14, 18, 34).
Coimmunoprecipitation Assays.
GST-E2F1 pull down assay was performed as described in ref. 18 in the presence of S35-labeled in vitro translated (TNT; Promega, Madison, WI) full-length ACTR (18) and CARM1 (33). Coimmunoprecipitations of endogenous CARM1 with E2F and ACTR were performed on HeLa whole cell extracts by using a rabbit polyclonal α-CARM1 Ab (49) for immunoprecipitations and α-E2F1 Ab (C20#sc193; Santa Cruz Biotechnology) and α-ACTR Ab (BD#61104; Becton Dickinson) for immunoblotting.
Tumor Samples and Clinical Material.
Eighty-one untreated breast tumor samples were obtained from the pathology department at the Val d’Aurelle Cancer Center of Montpellier (Centre Régional de Lutte Contre le Cancer, Montpellier, France) (39). Tumor biopsies were snap-frozen in liquid nitrogen upon surgical removal and stored at −80°C until RNA extraction. The tumor cohort was composed of 73% invasive ductal carcinoma, 13% invasive lobular carcinomas and 14% of invasive adenocarcinoma and breast carcinomas of unspecified type. The mean age of the patient was 58. Tumors were mostly SBR grades 2 and 3 (46% and 35%, respectively), whereas 16% were grade 1.
Distribution of mRNA expression values for CARM1, CCNE1, and ACTR genes, according to SBR grades, were organized as box plots.
Supplementary Material
Acknowledgments
We thank M. Benkirane, R. Feil, R. Hipskind (all from Centre National de la Recherche Scientifique), V. Cavaillès (Institut National de la Santé et de la Recherche Médicale), K. Helin (Biotech Research and Innovation Centre, Copenhagen, Denmark), G. Leone (Ohio State University, Columbus, OH), H. W. Chen, and M. C. Louie (both from University of California, Davis, CA) for reagents and R. Hipskind, J. M. Blanchard, L. Le Cam, A. Le Cam, and E. Julien for critical readings. This work was supported by an Équipe Labellisée 2003 Grant (to C.S.) and fellowships (to S.E.M.) from La Ligue Contre le Cancer and from the French governmental program l’Action Concertée Incitative and with the institutional support of the Centre National de la Recherche Scientifique. M.T.B. was supported by National Institutes of Health Grant DK62248.
Abbreviations
- CARM1
coactivator-associated arginine methyltransferase 1
- MEF
mouse embryo fibroblast
- PRMT
protein arginine N-methyltransferase
- Q-RTPCR
quantitative RT-PCR
- RLU
relative luciferase units
- SBR
Scarff-Bloom and Richardson
- shRNA
short hairpin RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Moroy T., Geisen C. Int. J. Biochem. Cell Biol. 2004;36:424–439. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Hwang H. C., Clurman B. E. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 3.Ohtani K., Degregori J., Nevins J. R. Proc. Natl. Acad. Sci. USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geng Y., Eaton E., Picon M., Roberts J. M., Lundberg C., Sardet C., Weinberg R. A. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 5.DeGregori J., Kowalik T., Nevins J. R. Mol. Cell. Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen N. H., Emdin S. O., Cajander J., Landberg G. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- 7.Brehm A., Miska E. A., McCance D. J., Reid J. L., Bannister A. J., Kouzarides T. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 8.Le Cam L., Polanowska J., Fabbrizio E., Olivier M., Philips A., Ng Eaton E., Classon M., Geng Y., Sardet C. EMBO J. 1999;18:1878–1890. doi: 10.1093/emboj/18.7.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H. S., Gavin M., Dahiya A., Postigo A. A., Ma D., Luo R. X., Harbour J. W., Dean D. C. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 10.Polanowska J., Fabbrizio E., Le Cam L., Trouche D., Emiliani S., Herrera R., Sardet C. Oncogene. 2001;20:4115–4127. doi: 10.1038/sj.onc.1204514. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen S. J., Schneider R., Bauer U. M., Bannister A. J., Morrison A., O’Carrol D., Firestein R., Cleary M., Jenuwein T., Herrera R. E., Kouzarides T. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 12.Morrison A. J., Sardet C., Herrera R. Mol. Cell. Biol. 2002;22:856–865. doi: 10.1128/MCB.22.3.856-865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandyopadhyay D., Okan N. A., Bales E., Nascimento L., Cole P. A., Medrano E. E. Cancer Res. 2002;62:6231–6239. [PubMed] [Google Scholar]
- 14.Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J. H., Nègre V., Rousset M., Pestka S., Le Cam A., Sardet C. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Timmers C., Maiti B., Saavedra H. I., Sang L., Chong G. T., Nuckolls F., Giangrande P., Wright F. A., Field S. J., et al. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 16.Saavedra H. I., Maiti B., Timmers C., Altura R., Tokuyama Y., Fukasawa K., Leone G. Cancer Cell. 2003;3:333–346. doi: 10.1016/s1535-6108(03)00083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandel L., Nicolas E., Vaute O., Ferreira R., Ait-Si-Ali S., Trouche D. Mol. Cell. Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie M. C., Zou J. X., Rabinovich A., Chen H. W. Mol. Cell. Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wysocka J., Allis C. D., Coonrod S. A. Front. Biosci. 2006;11:344–55. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 20.Lee D. Y., Teyssier C., Strahl B. D., Stallcup M. R. Endocr. Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 21.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 22.Chen D., Huang S. M., Stallcup M. R. J. Biol. Chem. 2000;275:40810–4081. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 23.Ma H., Baumann C. T., Li H., Strahl B. D., Rice R., Jelinek M. A., Aswad D. W., Allis C. D., Hager G. L., Stallcup M. R. Curr. Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 24.Bauer U. M., Daujat S., Nielsen S. J., Nightingale K., Kouzarides T. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurter B. T., Koh S. S., Chen D., Bunick G. J., Harp J. M., Hanson B. L., Henschen-Edman A., Makay D. R., Stallcup M. R., Aswad D. W. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 26.Koh S. S., Chen D., Lee Y. H., Stallcup M. R. J. Biol. Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 27.Daujat S., Bauer U. M., Shah V., Turner B., Berger S., Kouzarides T. Curr. Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y. H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. Proc. Natl. Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An W., Kim J., Roeder R. G. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Yadav N., Lee J., Kim J., Shen J., Hu M. C., Aldaz C. M., Bedford M. T. Proc. Natl. Acad. Sci. USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh S. S., Li H., Lee Y. H., Widelitz R. B., Chuong C. M., Stallcup M. R. J. Biol. Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Covic M., Hassa P. O., Saccani S., Buerki C., Meier N. I., Lombardi C., Imhof R., Bedford M. T., Natoli G. O. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chevillard-Briet M., Trouche D., Vandel L. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hateboer G., Wobst A., Petersen B. O., Le Cam L., Vigo E., Sardet C., Helin K. Mol. Cell. Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herber B., Truss M., Beato M., Muller R. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- 36.Silver D. P., Livingston D. M. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 37.Keyomarsi K., Tucker S. L., Buchholz T. A., Callister M., Ding Y., Hortobagyi G. N., et al. N. Engl. J. Med. 2002;347:1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 38.Sutherland R., Musgrove E. A. J. Mammary Gland Biol. Neoplasia. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 39.Hunt K., Keyomarsi K. Semin. Cancer Biol. 2005;15:319–326. doi: 10.1016/j.semcancer.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 41.Bouras T., Southey M., Venter D. J. Cancer Res. 2001;61:903–907. [PubMed] [Google Scholar]
- 42.Rodriguez C., Hughes-Davies L., Valles H., Orsetti B., Cuny M., Ursule L., Kouzarides T., Theillet C. Clin. Cancer Res. 2004;10:5785–5791. doi: 10.1158/1078-0432.CCR-03-0410. [DOI] [PubMed] [Google Scholar]
- 43.Pal S., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng Y., Yu Q., Sicinska E., Das M., Schneider J. E., Bhattacharya S., Rideout W. M., Bronson R. T., Gardner H., Sicinski P. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 45.Huang S., Litt M., Felsenfeld G. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisvert F. M., Chénard C. A., Richard S. Sci. STKE. 2005;re2:1–10. doi: 10.1126/stke.2712005re2. [DOI] [PubMed] [Google Scholar]
- 47.Xu W., Chen H., Du K., Asahara H., Tini M., Emerson B. M., Montminy M., Evans R. M. Science. 2001;8:8–11. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 48.Hong H., Kao C., Jeng M. H., Eble J. N., Koch M. O., Gardner T. A., Zhang S., Li L., Pan C. X., Hu Z., MacLennan G. T., Cheng L. Cancer. 2004;101:83–89. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 49.Kim J., Lee J., Yadav N., Wu Q., Carter C., Richard S., Richie E., Bedford M. T. J. Biol. Chem. 2004;279:25339–25344. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.