Abstract
Glucokinase (GK) is an important enzyme for regulating blood glucose levels and a potentially attractive target for diabetes of the young type 2 and persistent hyperinsulinemic hypoglycemia of infancy. To characterize the conformational transition of GK from the closed state to the superopen state, a series of conventional molecular dynamics (MD) and target MD (TMD) simulations were performed on both the wild-type enzyme and its mutants. Two 10-ns conventional MD simulations showed that, although the allosteric site of GK is ≈20 Å away from the active site, the activator is able to enhance the activity of the enzyme through conformational restriction. Fourteen TMD simulations on GK and five of its mutants revealed a reliably conformational transition pathway. The overall conformational transition includes three stages, and three likely stable intermediate states were identified by free energy scanning for the snapshots throughout the pathway. The conformational transition feature revealed by our TMD simulations rationalized several important mutagenesis and kinetic data. Remarkably, the TMD simulations predicted that Y61S, I159A, A201R, V203E, and V452S mutations, which have not been investigated so far, may facilitate the opening process of GK. These predictions also have been verified by mutagenesis and kinetic analyses in this study. These observations are beneficial to understanding the mechanism of GK regulation and designing the compounds for treating metabolic diseases.
Keywords: molecular dynamics, mutagenesis
Glucokinase (GK) (hexokinase IV or D) is a glycolytic enzyme that plays an important role in blood sugar regulation related to the glucose utilization and metabolism in the liver and pancreatic β-cells. Serving as a glucose sensor, GK controls plasma glucose levels (1). GK plays a dual role in reducing plasma glucose levels: Glucose-mediated activation of the enzyme in hepatocyte facilitates hepatic glucose update and glycogen synthesis, while that in pancreatic β-cells ultimately induces insulin secretion. Both of these effects in turn reduce plasma glucose levels (2, 3). Clinical evidence has shown that GK variants with decreased and increased activities are associated with diabetes of the young type 2 (MODY2) and persistent hyperinsulinemic hypoglycemia of infancy (PHHI), respectively (4–7). Therefore, GK might be an important target for these two metabolic diseases.
GK belongs to the hexokinase family (8). Although GK shares a high degree of sequence homology with other family members such as hexokinase I (≈54.4%), it differs from other members in various aspects, such as binding glucose with lower affinity, positive cooperative behavior with regard to glucose binding, and lack of inhibition for glucose-6-phosphate (9). Remarkably, the activity of GK is represented by a sigmoidal glucose kinetic curve instead of the Michaelis–Menten kinetic curve of nonallosteric hexokinases (10). Structurally, GK is a monomeric enzyme containing a single active site. Therefore, it must behave as a distinct allosteric mechanism differing from that of other well studied allosteric enzymes, which consist of several subunits that are activated and inactivated in a concerted manner (11). To illustrate the allosteric mechanism of GK, numerous mutagenesis studies have been conducted (9, 12–14). Recently, several activators of GK with the same effect as the activating mutations were also discovered (15, 16). The x-ray crystallographic determination addressed the crystal structures of GK at the closed (active) and superopen (inactive) states, implying that GK exhibits a global conformational transition between these two states, and such a global alteration in enzyme conformation may be associated with the special allosteric characteristics of GK (17). Thus, a rigorous mechanistic study of the global conformational transition is critical to understanding the regulation mechanism of GK and for development of new therapeutic approaches for type 2 diabetes and persistent hyperinsulinemic hypoglycemia of infancy.
Although the crystal structures of GK have been determined, and facilitated a proposal for a mnemonical mechanism for the cooperativity of GK, several fundamental questions are still unanswered. The allosteric site is ≈20 Å away from the active site, so how does the activator enhance the GK activity? How does GK make progress in the large-scale conformational change from the closed to the superopen state? Why can some mutations activate the allosteric traits? Unfortunately, because it is difficult to catch the structures of the intermediate states in the pathway for the global conformational transition of GK, the investigation of the above problems in atomic detail through experimental methods is still intractable. Computational simulation, with its extremely high time resolution and atomic level representation, has been increasingly used in understanding the complex conformational features of proteins and predicting structural preferences (18–24). To our knowledge, the global alteration in GK conformation has not been computationally simulated.
Here, we report the results of the conformational transition of GK addressed by a series of conventional molecular dynamics (CMD) and target molecular dynamics (TMD) simulations. By running two 10-ns CMD simulations in aqueous solution, we show that, although the allosteric site of GK is ≈20 Å away from the active site, the activator is able to enhance the activity of the enzyme through conformational restriction. Afterward, by running 14 TMD simulations on GK and five of its mutants, we find a reliably conformational transition pathway from the closed to the superopen state. The overall conformational transition includes three stages, and three likely stable intermediate states were identified by free energy scanning for the snapshots throughout the pathway. The important components relevant to the conformational change of GK were addressed by analyzing the detailed structures of the TMD trajectories. The simulation results are in accordance with the recent findings of mutagenesis experiments and related kinetic studies. In addition, the TMD simulations predicted that Y61S, I159A, A201R, V203E, and V452S mutations, which have not been investigated so far, may facilitate the opening process of GK. Remarkably, we verified these predictions by mutagenesis and kinetic analyses, which are also reported in this work. These observations are beneficial to understanding the mechanism of GK regulation and designing the compounds for treating metabolic diseases.
Results and Discussion
The main goal of this study was to investigate the allosteric process of human GK, which is more associated with the conformational change from the closed to the superopen state. To this end, four models for CMD and TMD simulations were designed. In model I, two 10-ns CMD simulations (A1 and A2) were conducted on the closed state and its complex with an activator of the free protein to probe how the activator affects the conformational change of GK. In model II, six TMD simulations (B1–B6) were performed by using a force constant of 0.1 kcal·mol−1·Å−2 with different initial velocities to explore the minimal energy pathway between the closed and superopen states. In model III, to verify the reliability of the pathway for the conformational alternation of GK, three additional TMD simulations (C1–C3) were carried out with force constants of 0.2, 0.5, and 1.0 kcal·mol−1·Å−2, respectively. In model IV, K56A, E256A, V62M, E158A, and I159A mutants were constructed for GK, and five TMD simulations were performed on these mutants. In total, 2 CMD and 14 TMD simulations were performed on GK and its mutants. More detailed information for these molecular dynamics (MD) simulations is provided in Table 2, which is published as supporting information on the PNAS web site.
Influence of Activator to the Conformational Dynamics of GK.
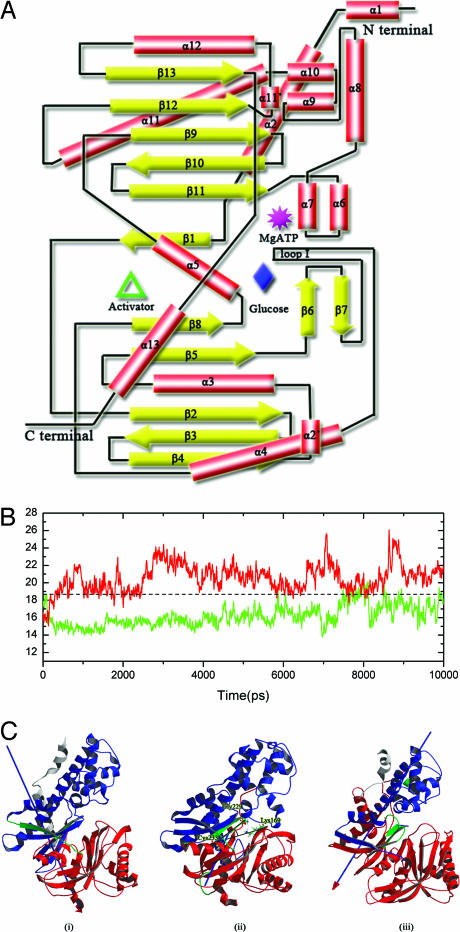
Structurally, GK is composed of a large and a small domain; the active site for phosphorylation is located in the deep cleft between these two domains, and on the back of the cleft is an allosteric site for activator binding (Fig. 1A). Two 10-ns CMD simulations for model I were performed to explore the influence of activator to the conformational dynamics of GK. The time evolutions of the angles between the two domains defined by two lines from the Cα atom of Cys-223 (a hinge residue) to the Cα atoms of Gly-229 and Lys-169 (denoted as cleft-angle in the following) were monitored during the simulations. The results are shown in Fig. 1B. Cleft-angle profile reveals a closed-and-open motion between the two domains for the free GK, indicating that GK is intrinsically dynamic. The principal component analysis (PCA) on the MD trajectory of free GK confirmed this assumption. PCA results revealed that the first two principal components correspond to twist and open-and-close motions between the two domains (Fig. 1C). Additionally, normal mode analysis on the free GK also detected the open-and-close motion (J.Z., unpublished data). At the utmost, the cleft-angle can open up ≈10° throughout the 10-ns CMD simulation (Fig. 1B). These results indicate that conformational dynamics of GK is its inherent property, which facilitates the allosteric function of this enzyme (16). However, the open-and-close motion was not detected in the CMD simulation on the activator–GK complex (A2 trajectory; Fig. 1B). The PCA results on the A2 trajectory revealed that only slight twist vibration exists between the large and small domains in the activator–GK complex (Fig. 1C), indicating that the activator may lock the conformation of GK into the closed state, which is beneficial to the catalytic activity of the enzyme. The mass weighted rmsds of the snapshots along the CMD trajectories (A1 and A2) relative to the x-ray crystal structure of the GK closed state also indicated the dynamics difference between free GK and GK in complex with the activator (see Fig. 5, which is published as supporting information on the PNAS web site). Accordingly, although the allosteric site is ≈20 Å away from the active site, the activator is still able to enhance the activity of the enzyme through conformational restriction. This result is in agreement with the kinetics analysis for GK that the activator changes the shape of the glucose saturation curve from sigmoidal to Michaelis–Menten hyperbolic curve and increases the Vmax value (15, 16, 25).
Fig. 1.
Analyses for the CMD simulations on GK (A1 trajectory) and its complex with the activator (A2 trajectory). (A) A 2D topological structure plot of GK. (B) Cleft-angle profiles along the A1 (red) and A2 (green) trajectories. The angle is defined by two lines from the Cα atom of Cys-223 (hinge residue) to the Cα atoms of Gly-229 and Lys-169 (see Cii). (C) Principle component modes of the motions in A1 (i and ii) and A2 (iii). Arrow colors denote the particular moving domain (right-hand rule). The fixed domains and the moving domain are in blue and red, respectively; residues for rotation in axis are in green.
Potential Pathway for Global Conformational Transition.
The CMD simulation on GK indicated that the closed state of the enzyme may open up in some degree without any action of outside force. However, current CMD methods cannot simulate such a large-scale conformational change like the global alternation of GK conformation. Accordingly, TMD, which can accelerate the process for large-scale conformational motion between two existing states, was used in this study to address the probable allosteric pathway of GK. To obtain a reliable pathway for the global conformational transition from the closed to superopen state, six TMD simulations (B1–B6) were performed on model II under a force constant of 0.1 kcal·mol−1·Å−2 with different initial velocities (Table 2). The six TMD simulations almost produced the same conformational transition pathway as demonstrated by the weighted rmsds among the six trajectories (see Table 3, which is published as supporting information on the PNAS web site), indicating that the initial velocities for TMD simulations did not affect the transition pathway. To probe the influence of the force constant on the conformational transition pathway, three additional TMD simulations (C1–C3) were conducted on model III under force constants of 0.2, 0.5, and 1.0 kcal·mol−1·Å−2, respectively. In comparison with trajectories B1–B6, trajectories C1–C3 did not give qualitatively different results (see Table 4 and Fig. 6, which are published as supporting information on the PNAS web site). All these data suggest that the conformational transition pathway is reliable. In the following, we use trajectory B1 for further discussion.
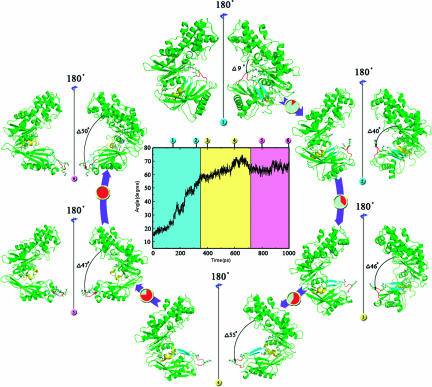
The TMD-simulated conformational transition pathway of GK from the closed to superopen state is shown in Fig. 2. Although the transition took place continuously, the overall conformational transition can be roughly divided into three stages (Fig. 2). The conformational transition begins with a stage in which the cleft-angle gradually opens up ≈9° during the time period from 0 to 160 ps. Above 10 ns, CMD simulation on free GK also addressed the conformational alternation of this stage (Fig. 1). Because it is difficult to overcome the energy barrier produced by the strong interaction of loop I between α4 and β7 in the small domain with β1 and β11 in the large domain (Fig. 1A), the CMD simulation is not able to go through the whole pathway of the conformational transition. As the TMD simulation goes on, the interaction between the small and large domains is ruptured, and the cleft-angle opens up ≈40°. At the second stage (360–700 ps), the β6 and β7 strands in the small domain continually relax their positions within the range of local space after GK undergoes a rapid opening process. At ≈550 ps, the β6 and β7 strands move away from the allosteric site, turn their orientation, and completely drift into the solvent. During this stage, these two β-strands begin to unfold their second structures into coils. In the third period (700–1,000 ps), the cleft-angle reaches the superopen state, and the β6 and β7 strands completely convert into coil structures and expose the solvent. Meanwhile, the α13 helix departs from the small domain. In general, the large domain moves almost as a rigid body during the conformational transition; however, the movement for the small domain is more complicated, which is associated with the biological function of GK as we will discuss below.
Fig. 2.
The conformational transition pathway of GK from the closed to the superopen state. The center image is the time dependence of the cleft-angle between the two domains. Around the center image, six snapshots are extracted from the B1 trajectory at the times of 150 (1), 320 (2), 400 (3), 600 (4), and 800 (5) ps, and superopen state (6), respectively. The loop I between α4 and β7 segments is in red; the β6 and β7 strands are in cyan; helix α13 is in yellow.
Possible Intermediate States in the Transition Pathway.
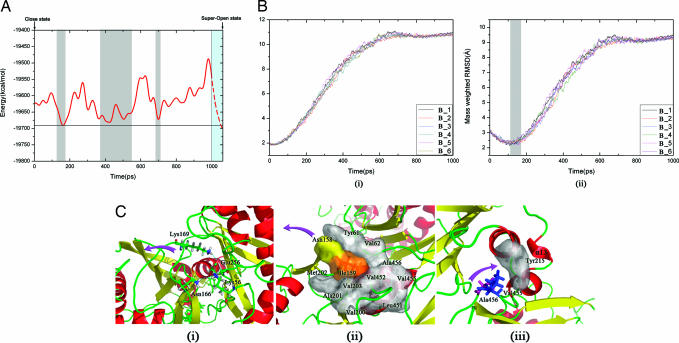
The x-ray crystal structures of GK indicate that the conformational transition from the closed to superopen state is a complicated process. According to the mnemonical mechanism of GK regulation proposed by Kamata et al. (17) and the behaviors of other hexokinases (25), stable intermediate states should exist in the conformational transition pathway to decrease cost for the large-scale conformational change and increase the opportunity of regulation. Nevertheless, such kinds of intermediate states of GK have not been detected by experimental methods (17). To explore the possible intermediate states for GK, the free energy landscape along the conformational transition pathway was calculated by using the MM-PBSA-Nmode method encoded in AMBER (Version 7.0) (26). The result is shown in Fig. 3A, which indicates that there are three energy wells (denoted P1, P2, and P3) in the pathway at time periods of ≈130–160, ≈370–550, and ≈695–705 ps. Conformations in these energy wells are possibly stable intermediate states. To verify the possibility of these intermediate states further, snapshots were isolated from the B1–B6 trajectories and superimposed with the crystal structures of other hexokinases. The result indicated that conformations in the P1 well are very similar to the x-ray crystal structure of the open state of hexokinase I (Fig. 3B) (25). This result shows that the P1 state may be a real stable intermediate state of GK, i.e., the open state of GK, which is a desired outcome (17). Conformations dropped into the second and third wells (P2 and P3 states) might also be the intermediate states in the conformational transition pathway, as we will discuss below.
Fig. 3.
Free energy landscape and intermediate states along the conformational transition pathway. (A) Free energy profiles of the conformations along the B1 trajectory. The shadows indicate three minimal free energy wells. (B) rmsd evolutions of the conformations along the six TMD trajectories (B1–B6) with the closed (i) and the open (ii) states of hexokinase I. (C) Important interactions for the three possible intermediate states [P1 (i), P2 (ii), and P3 (iii)] on the conformational transition pathway.
Important Components Along the Transition Pathway and Mutagenesis Validation.
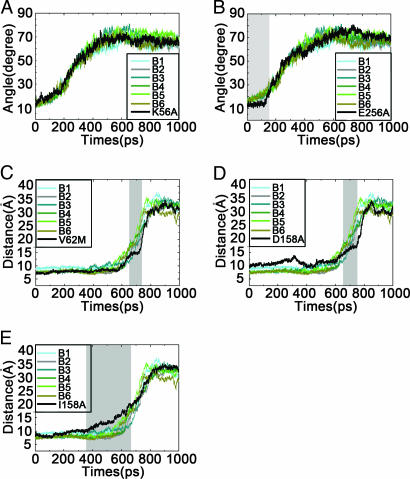
The x-ray crystal structure of the closed state indicates that there are two direct hydrogen bonds (H-bonds) between the small and large domains, i.e., Lys-56···Asn-166 and Glu-256···Lys-169. TMD simulation revealed that these two H-bonds are stable before 160 ps (the first stage in the pathway of conformational transition) (Fig. 3Ci and see also Fig. 7, which is published as supporting information on the PNAS web site). As the TMD went on, these two H-bonds ruptured, and the cleft of GK opened up more. Interestingly, the point of the H-bonds breaking (≈160 ps) exactly corresponds to the predicted intermediate (P1) state, as indicated by the free energy calculation (Fig. 3A). This result highlights that these two H-bonds play important roles in the conformational transition. Most likely, rupture force needed to break these two H-bonds caused the energy barrier from P1 to the superopen state. To map the functionality of these two H-bonds, two additional TMD simulations (D1 and D2) on K56A and E256A mutants were performed. The results indicated that K56A mutation did not change the conformational transition pathway, whereas E256A mutation delayed the opening process of the two domains (Fig. 4A and B). These results are consistent with the mutagenesis studies, which revealed that K56A has little impact on the kinetic property of GK, but E256A severely decreases the affinity and cooperativity of GK for glucose (27). Structural analysis indicated that, although K56A mutation abolished the H-bond of Lys-56···Asn-166, the H-bond of Glu-256···Lys-169 was still maintained before 160 ps during the TMD simulation. However, E256A mutation changed the local structure of the interaction interface between the two domains. In addition, Asn-166 formed a new H-bond with Ala-256, and the side chain of Lys-169 deeply inserted into the cleft between the two domains (see Fig. 8, which is published as supporting information on the PNAS web site). This structural adjustment enhanced the interaction between the two domains, slowing down the opening process of the cleft and discouraging glucose from binding with GK.
Fig. 4.
Impacts of mutations K56A (A), E256A (B), V62M (C), D158A (D), and I159A (E) to the conformation transition pathway of GK. Angles in A and B are defined in the same way as in Fig. 1B. Distances in C–E are measured between the Cα atoms of I159 and V452.
Several experiments indicated that the catalytic cycle of GK is a slow process, implying that the conformational transition corresponding to this cycle goes slowly (28, 29). Remarkably, the free energy calculation captured another probable intermediate state from the second stage in the conformational transition pathway (P2 state in Fig. 3A). Accordingly, the structural details of the P2 state were carefully analyzed. The result revealed that, at the second stage, Tyr-61, Val-62, Ala-201, Val-203, and Val-452 form a “hydrophobic pocket,” and the side chains of Asp-158 and Ile-159 in the β6 strand insert into the hydrophobic pocket through hydrophobic interactions, which lock the movement between the two domains of GK (Fig. 3Cii). Therefore, the energy barrier, caused by the P2 state, might be the reason underlying the slow process of the catalytic cycle. To confirm this finding, three additional TMD simulations (D3–D5) on V62M, D158A, and I159A mutants of GK were conducted. The first two mutations enhanced the hydrophobic interactions, and the last mutation reduced the hydrophobic interactions between the two domains at the second stage. Indeed, V62M and D158A mutations delayed the opening process from the closed to the superopen state, as indicated by the profiles of the distance between Ile-159 and Val-452 vs. simulation time (Fig. 4 C and D). This result is in agreement with the mutagenesis results, which revealed that V62M and D158A are activating mutations with decreased S0.5 values (concentration of glucose at which GK shows the half activity of Vmax) (9, 30). TMD simulation predicted that I159A mutation might facilitate the opening process of GK (Fig. 4E). However, there are no experimental data for this mutation. We constructed the clone of this mutant and obtained the pure protein by expression (detailed procedures for the protein expression, purification, and enzymatic assay are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Kinetic assay indicated I159A is an inactivating mutation: Its S0.5 increased ≈8 fold, and Vmax decreased in comparison with the wild-type GK (Table 1). The TMD simulations also indicate the importance of residues Tyr-61, Ala-201, Val-203, and Val-452 in stabilizing the structure of P2 (Fig. 3Cii). Also, no mutation data have been reported for these residues. To verify the simulation result, these residues were mutated into polar amino acids, resulting in Y61S, A201R, V203E, and V452S mutants, for weakening the hydrophobic interactions of Asp-158 and Ile-159 with the hydrophobic pocket (Fig. 3Cii). Indeed, kinetic determinations revealed that all these mutations reduced the enzymatic activity of GK with increased S0.5 and decreased Vmax values (Table 1), suggesting that, similar to I159A, these mutations facilitate the opening process of GK. These mutagenesis analyses verified the TMD prediction, demonstrating again the reliability of the conformational transition pathway and corresponding intermediate states derived from the TMD simulations.
Table 1.
Kinetic analyses for GK and its mutants
| Mutants | Protein conc., mg/ml | Vmax, mOD/min | Km for ATP, mM | S0.5 for glucose, mM | nH† |
|---|---|---|---|---|---|
| LGK2-WT | 2.24 | 181.51 ± 6.78 | 0.14 ± 0.01 | 11.64 ± 0.83 | 1.40 ± 0.12 |
| Y61S mutant | 2.24 | 60.33 ± 3.83 | 0.08 ± 0.01 | 116.86 ± 7.46 | 1.23 ± 0.10 |
| I159A mutant | 2.24 | 23.32 ± 1.20 | 0.05 ± 0.01 | 95.16 ± 7.32 | 0.96 ± 0.04 |
| A201R mutant | 2.24 | 149.62 ± 8.50 | 0.20 ± 0.01 | 18.14 ± 0.26 | 0.76 ± 0.06 |
| V203E mutant | 2.24 | 39.26 ± 2.85 | 0.55 ± 0.07 | 201.23 ± 18.76 | 0.58 ± 0.04 |
| V452S mutant | 2.24 | 86.61 ± 1.78 | 0.23 ± 0.01 | 28.11 ± 0.73 | 0.96 ± 0.02 |
Data are means ± SEM for wild-type hLGK2 and hLGK2 mutants. The results are means of kinetic analyses of three independent experiments of wild-type hLGK2 and hLGK2 mutants.
†nH is the Hill number.
The α13 helix is surrounded by α5, β3, β4, and β5, and Tyr-215 on β5 serves as a gatekeeper to prevent the α13 helix from escaping the small domain. This process produced the third intermediate state in the conformational transition pathway (P3 in Fig. 3C). As the cleft opened more after the P2 state, the β6 and β7 strands gradually departed from the allosteric site, changed orientation, and drifted into the solvent. At this stage, the small domain twisted mildly relative to the large domain, vacating space for the α13 helix to depart from the small domain. The detailed process of the α13 helix being released from the small domain is shown in Fig. 9, which is published as supporting information on the PNAS web site. During this process, Tyr-215 on α5 prevents the α13 helix from leaving the small domain, and the residues in the α13 helix that directly contact Tyr-215 are Val-455 and Ala-456. It can be predicted from the TMD results that mutations by residues with bulkier side chains may inhibit the release of α13 helix, thus prolonging the conformational transition process. Two such mutants of GK (V455M and A456V) were found in the metabolic disease patients. In line with the TMD results, kinetic analysis of these two mutants showed substantially decreased S0.5 values (6, 7). Accordingly, the release of the α13 helix from the small domain, especially the contacts of Val-455 and Ala-456 of α13 with Tyr-215 of α5, is also an important component for the conformational transition.
Implications in the Allosteric Mechanism.
Based on the x-ray structures of GK, Kamata et al. (17) devised a kinetic model for the allosteric mechanism of GK regulation and predicted that intermediate states likely exist in the conformational transition pathway between closed and superopen states. However, these intermediate states of GK have not yet been detected by experimental methods. A very important mechanistic conclusion from the present TMD simulations of GK is that there are three intermediate states (P1–P3 in Fig. 3A) in the conformational transition pathway from the closed to superopen state. The interaction components that caused the energy barriers corresponding to the intermediate states have been addressed by the TMD simulations, and all of them are in agreement with the available data of mutagenesis and kinetic analyses. Remarkably, the TMD simulations revealed that Ile-159, which has not been investigated before, might play an essential role during the conformational transition of GK, and its mutation is an inactivating one. By using mutagenesis and kinetic analyses, we verified the TMD prediction.
Based on the present MD simulation analyses, we propose an atomic mechanism for the kinetic model of the GK allosteric process. As shown in Fig. 3A, GK may exist in three intermediate states (P1–P3) between the closed and superopen states. The TMD simulations revealed that GK undergoes conformational transition from closed to superopen state through three stages (Fig. 2). During the first stage, GK opens up from the closed state to P1 state, which is the open state as predicted by Kamata et al. (17). The 10-ns CMD simulation indicated that closed–open conformational change between the closed and P1 states is an intrinsic property of GK. This process corresponds to the “fast cycle” for GK catalytic mechanism proposed by Kamata et al. (17). Two H-bonds formed between Lys-56 and Asn-166 and between Glu-256 and Lys-169 are the major components to stabilize the structure of the P1 state. During the second stage, conformation changes from P1 to P2. At this stage, the important component to produce the energy barrier for the conformational change is the hydrophobic interactions between the side chains of Asp-158 and Ile-159 and the hydrophobic pocket rounded by Tyr-61, Val-62, Ala-201, Val-203, and Val-452. The third stage is the release of the α13 helix from the small domain, and the energy barrier of this step is mainly from the resistant interaction between the α13 helix (Val-455 and Ala-456) and the α5 helix (Tyr-215). The existence of the P2 and P3 states is in agreement with the conclusion deduced from the glucose phosphorylation that the conformational changes between the open and superopen states are slower than the closed–open conformational change (17).
Methods
Simulation Systems.
Initial coordinates for the closed and superopen states of GK were taken from the x-ray crystal structures (17) (Protein Data Bank ID codes 1V4S and 1V4T). The missing residues were repaired by using the loop search method in the Homology module of Insight II (Accelrys, San Diego, CA). For the simulations of GK (or mutated GK) in aqueous solution, the protein was first put into a suitably sized box, in which the minimal distance from the protein to the box wall was 1.5 nm. Then, the box was solvated with the SPC water model (31). The protein/water system was submitted to energy minimization. Afterward, counterions were added to the system to provide a neutral simulation system. The whole system was subsequently minimized again. The charges of the atoms of activator N-thiazol-2-yl-2-amino-4-fluoro-5-(1-methylimidazol-2-yl)thiobenzamide were calculated by using the RESP method (32) encoded in the AMBER program suite (26) at the level of RHF/6-31G*. Covalent and nonbonded parameters for the activator atoms were assigned by analogy or through interpolation from those already present in the AMBER force field (33).
CMD and TMD Simulations.
CMD simulations were carried out by using the AMBER package (Version 7.0) with constant temperature, constant pressure (NPT), and periodic boundary conditions. The Amber Parm99 force field (33) was applied for the proteins. The particle mesh Ewald method (34) was used to calculate the long-range electrostatics interactions. The nonbonded cutoff was set to 12.0 Å, and the nonbonded pairs were updated every 25 steps. The SHAKE method (35) was applied to constrain all covalent bonds involving H atoms. Each simulation was coupled to a 300 K thermal bath at 1.0 atm of pressure (1 atm = 101.3 kPa) by applying the algorithm of Berendsen et al. (36). The temperature and pressure coupling parameters were set as 0.2 and 0.05 ps, respectively. An integration step of 2 fs was set up for the MD simulations. The TMD simulations were performed by using the Sander module encoded in AMBER (Version 7.0) (26). Restraint force was applied onto each initial structure to bias the trajectories toward each respective target structure. The integration step for TMD simulations was set to 1 fs. The detailed procedure of TMD simulation can be found in ref. 37 and in several recent applications (38, 39).
Kinetic Analyses for GK and its Mutants.
The DNA sequence of human liver GK isoform2 (hLGK2) was prepared by total gene synthesis (Sangon, Shanghai, China), and then cloned into pQE-30 (Qiagen, Valencia, CA) to make the recombinant plasmid pQE-30-hLGK2. The Y61S, I159A, A201R, V203E, and V452S mutant clones were prepared with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) by using pQE-30-hLGK2 as the template. Expression and purification of the recombinant proteins were preformed according to His·Bind Kits (Novagen, San Diego, CA). The type hLGK2 and mutant proteins were then subjected to kinetic analysis as described by Grimsby et al. (16).
For further details, see Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Shanghai Supercomputing Center and Computer Network Information Center of Chinese Academy of Sciences for allocation of computing time. This work was supported by Special Fund for the Major State Basic Research Project of China Grant 2002CB512802, National Natural Science Foundation of China Grant 30525024, and Shanghai Science and Technology Commission Grant 03DZ19228.
Abbreviations
- GK
glucokinase
- MD
molecular dynamics
- CMD
conventional MD
- TMD
target MD.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Al-Hasani H., Tschop M. H., Cushman S. W. Mol. Interventions. 2003;3:367–370. doi: 10.1124/mi.3.7.367. [DOI] [PubMed] [Google Scholar]
- 2.Van Schaftingen E. Diabetologia. 1994;37(Suppl. 2):S43–S47. doi: 10.1007/BF00400825. [DOI] [PubMed] [Google Scholar]
- 3.Matschinsky F. M., Glaser B., Magnuson M. A. Diabetes. 1998;47:307–315. doi: 10.2337/diabetes.47.3.307. [DOI] [PubMed] [Google Scholar]
- 4.Vionnet N., Stoffel M., Takeda J., Yasuda K., Bell G. I., Zouali H., Lesage S., Velho G., Iris F., Passa P., et al. Nature. 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- 5.Froguel P., Zouali H., Vionnet N., Velho G., Vaxillaire M., Sun F., Lesage S., Stoffel M., Takeda J., Passa P., et al. N. Engl. J. Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 6.Glaser B., Kesavan P., Heyman M., Davis E., Cuesta A., Buchs A., Stanley C. A., Thornton P. S., Permutt M. A., Matschinsky F. M., et al. N. Engl. J. Med. 1998;338:226–230. doi: 10.1056/NEJM199801223380404. [DOI] [PubMed] [Google Scholar]
- 7.Christesen H. B., Jacobsen B. B., Odili S., Buettger C., Cuesta-Munoz A., Hansen T., Brusgaard K., Massa O., Magnuson M. A., Shiota C., et al. Diabetes. 2002;51:1240–1246. doi: 10.2337/diabetes.51.4.1240. [DOI] [PubMed] [Google Scholar]
- 8.Grossbard L., Schimke R. T. J. Biol. Chem. 1966;241:3546–3560. [PubMed] [Google Scholar]
- 9.Gloyn A. L., Odili S., Zelent D., Buettger C., Castleden H. A., Steele A. M., Stride A., Shiota C., Magnuson M. A., Lorini R., et al. J. Biol. Chem. 2005;280:14105–14113. doi: 10.1074/jbc.M413146200. [DOI] [PubMed] [Google Scholar]
- 10.Storer A. C., Cornish-Bowden A. Biochem. J. 1976;159:7–14. doi: 10.1042/bj1590007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacRae I. J., Segel I. H., Fisher A. J. Nat. Struct. Biol. 2002;9:945–949. doi: 10.1038/nsb868. [DOI] [PubMed] [Google Scholar]
- 12.Xu L. Z., Harrison R. W., Weber I. T., Pilkis S. J. J. Biol. Chem. 1995;270:9939–9946. doi: 10.1074/jbc.270.17.9939. [DOI] [PubMed] [Google Scholar]
- 13.Xu L. Z., Zhang W., Weber I. T., Harrison R. W., Pilkis S. J. J. Biol. Chem. 1994;269:27458–27465. [PubMed] [Google Scholar]
- 14.Moukil M. A., Veiga-da-Cunha M., Van Schaftingen E. Diabetes. 2000;49:195–201. doi: 10.2337/diabetes.49.2.195. [DOI] [PubMed] [Google Scholar]
- 15.Brocklehurst K. J., Payne V. A., Davies R. A., Carroll D., Vertigan H. L., Wightman H. J., Aiston S., Waddell I. D., Leighton B., Coghlan M. P., et al. Diabetes. 2004;53:535–541. doi: 10.2337/diabetes.53.3.535. [DOI] [PubMed] [Google Scholar]
- 16.Grimsby J., Sarabu R., Corbett W. L., Haynes N. E., Bizzarro F. T., Coffey J. W., Guertin K. R., Hilliard D. W., Kester R. F., Mahaney P. E., et al. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 17.Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. Structure (London) 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Daura X., Jaun B., Seebach D., van Gunsteren W. F., Mark A. E. J. Mol. Biol. 1998;280:925–932. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]
- 19.Duan Y., Wang L., Kollman P. A. Proc. Natl. Acad. Sci. USA. 1998;95:9897–9902. doi: 10.1073/pnas.95.17.9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmerling C., Strockbine B., Roitberg A. E. J. Am. Chem. Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 21.Snow C. D., Nguyen H., Pande V. S., Gruebele M. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 22.Mayor U., Guydosh N. R., Johnson C. M., Grossmann J. G., Sato S., Jas G. S., Freund S. M. V., Alonso D. O. V., Daggett V., Fersht A. R. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 23.Zangi R., de Vocht M. L., Robillard G. T., Mark A. E. Biophys. J. 2002;83:112–124. doi: 10.1016/S0006-3495(02)75153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yechun X., Jianhua S., Xiaomin L., Weiliang Z., Kaixian C., Jianpeng M., Hualiang J. Proc. Natl. Acad. Sci. USA. 2005;102:5403–5407. [Google Scholar]
- 25.Aleshin A. E., Zeng C., Bartunik H. D., Fromm H. J., Honzatko R. B. J. Mol. Biol. 1998;282:345–357. doi: 10.1006/jmbi.1998.2017. [DOI] [PubMed] [Google Scholar]
- 26.Case D. A., Cheatham T. E., III, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilkis S. J., Weber I. T., Harrison R. W., Bell G. I. J. Biol. Chem. 1994;269:21925–21928. [PubMed] [Google Scholar]
- 28.Lin S. X., Neet K. E. J. Biol. Chem. 1990;265:9670–9675. [PubMed] [Google Scholar]
- 29.Neet K. E., Keenan R. P., Tippett P. S. Biochemistry. 1990;29:770–777. doi: 10.1021/bi00455a026. [DOI] [PubMed] [Google Scholar]
- 30.Miller S. P., Anand G. R., Karschnia E. J., Bell G. I., LaPorte D. C., Lange A. J. Diabetes. 1999;48:1645–1651. doi: 10.2337/diabetes.48.8.1645. [DOI] [PubMed] [Google Scholar]
- 31.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Hermans J. In: Intermolecular Forces. Pullman B., editor. Dordrecht, The Netherlands: Reidel; 1981. pp. 331–342. [Google Scholar]
- 32.Bayly C. I., Cieplak P., Cornell W. D., Kollman P. A. J. Phys. Chem. 1993;97:10251–10512. [Google Scholar]
- 33.Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 34.Darden T., York D., Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 35.Ryckaert J. P., Ciccotti G., Berendsen J. C. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 36.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 37.Schlitter J., Engels M., Kruger P., Jacoby E., Wollmer A. Mol. Simul. 1993;10:291–308. [Google Scholar]
- 38.Kong Y., Shen Y., Warth T. E., Ma J. Proc. Natl. Acad. Sci. USA. 2002;99:5999–6004. doi: 10.1073/pnas.092051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong Y., Ma J., Karplus M., Libscomb W. N. J. Mol. Biol. 2006;356:237–247. doi: 10.1016/j.jmb.2005.10.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.