Abstract
Alveolar rhabdomyosarcoma (ARMS) is an aggressive myogenic-type tumor and a gain-of-function disease, caused by misexpression of the PAX3-FKHR or PAX7-FKHR fusion oncoprotein from structurally rearranged chromosomes. PAX3-FKHR misexpressed in terminally differentiating mouse myofibers can cause rhabdomyosarcoma at a low frequency, suggesting that skeletal muscle is an ARMS tissue of origin. Because patterned muscle is widely viewed as irreversibly syncytial, questions persist, however, regarding this potential pathogenetic mechanism for ARMS tumor initiation. To further explore this issue, we generated transgenic Drosophila lines that conditionally express human PAX-FKHR. Here we show that PAX7-FKHR causes nucleated cells to form and separate from syncytial myofibers, which then spread to nonmuscular tissue compartments, including the central nervous system, and that wild-type PAX3 demonstrates similar potential. We further show that Ras, which is known to interfere with the differentiation of myogenic cells, genetically interacts with PAX7-FKHR: constitutively activated Ras enhances PAX7-FKHR phenotypes, whereas loss-of-function ras alleles dominantly suppress PAX7-FKHR activity, including rescue of lethality. These results show that PAX-FKHR can drive the generation of discrete nucleated cells from differentiated myofibers in vivo, argue for syncytial muscle as an ARMS tissue of origin, and demonstrate that Drosophila provides a powerful system to screen for genetic modifiers of PAX-FKHR.
Keywords: PAX3, PAX3-FKHR, skeletal muscle, chromosomal translocation, sarcoma
Despite many advances, cancer continues to be a critical cause of childhood disease and mortality (1). Of the typical childhood soft tissue malignancies (or sarcomas), the rhabdomyosarcoma (RMS) family of tumors, so named because of its primitive, embryonal skeletal muscle-type histology, is the most common, accounting for ≈50% of all such cases (2). The RMS family is typically subclassified into two general subtypes, embryonal and alveolar RMS (ARMS), based on differing histopathologic features (3). This distinction is clinically important, as the alveolar variant is notoriously aggressive and portends a poor prognosis due to early metastasis (2). Outcomes for advanced ARMS remain dismal despite intensive therapy (2, 4), underscoring the need for understanding the pathogenetic mechanisms underlying tumorigenesis.
The genetic lesions that underlie ARMS are well known. ARMS uniquely associates with two diagnostic balanced chromosomal translocations, t(2;13)(q35;q14) and t(1;13)(p36;q14) (5). Both translocations cause fusion of a PAX gene (PAX3 from chromosome 2, PAX7 from chromosome 1) to the FKHR (Forkhead in RMS; or FOXO1A) locus on chromosome 13. The gene fusions give rise to structurally equivalent, in-frame PAX-FKHR chimeric transcription factors, in which the PAX DNA-binding domains are fused to the transcriptional activation domain of FKHR (Fig. 6, which is published as supporting information on the PNAS web site). Because both PAX3 and PAX7 are transcription factors that participate in skeletal muscle development, PAX-FKHR, misexpressed from the rearranged chromosomes, has been presumed to misregulate some aspect(s) of the muscle development program and thereby drive neoplastic transformation of skeletal muscle precursor cells or myogenic stem cells, such as satellite cells.
A PAX3-FKHR transgenic mouse described recently, however, suggests an altogether different model for PAX-FKHR tumorigenesis. Keller et al. (6) generated a conditional PAX3-FKHR “knock-in” transgenic allele, using a large genomic fragment from the FKHR locus (thereby including potential 3′ cis FKHR regulatory elements) to better mimic the t(2;13) rearranged chromosome. This model, upon introduction of Myf6-driven Cre, demonstrates misexpression of PAX3-FKHR, starting in terminally differentiating myofibers, and the development of RMS at a low frequency. In contrast, targeted PAX3-FKHR expression in satellite cells or muscle precursor cells does not cause tumorigenesis (7–10). These observations suggested the intriguing possibility that PAX3-FKHR can promote discrete, malignant cells to form from postmitotic, syncytial muscular tissue. The ARMS mouse study, however, did not capture cells originating from differentiated muscle, leaving open the possibility that some unidentified cell type had been targeted or influenced by this system. Consequently, speculation has continued regarding this potential pathogenetic mechanism for tumorigenesis, because the generation of nucleated cells from differentiated muscle has been documented only in cell culture (11–13) and not in the context of either PAX3-FKHR or PAX7-FKHR.
To further explore the pathogenic consequences of PAX-FKHR expression in differentiated muscle, we generated transgenic fruit flies expressing human PAX-FKHR in somatic muscle. We chose Drosophila to take advantage of the fact that the entire somatic musculature of the living organism, when highlighted by fluorescent protein reporters, can be easily visualized through the animal’s transparent outer cuticle, thereby allowing for real-time detection of muscle abnormalities evoked by PAX-FKHR, even if subtle or focal. Here we show that PAX7-FKHR, which unlike the PAX3-FKHR gene fusion (which is the more commonly occurring ARMS initiator) has not been profiled in vivo, disrupts differentiated muscular tissue and causes individual nucleated cells to form from syncytial myofibers. Once liberated from the syncytia, these cells spread most prominently to the larval CNS. We further find that wild-type PAX3, when overexpressed, demonstrates similar, if not quite equal, activity. Activated Ras, a known regulator of muscle precursor cell differentiation, enhances PAX7-FKHR activity, whereas heterozygous ras loss-of-function suppresses the PAX7-FKHR muscle phenotype and associated lethality. These studies demonstrate that individual nucleated cells can generate from syncytial muscle in vivo and that PAX-FKHR can drive this process, supporting the hypothesis that ARMS tumorigenesis can originate from differentiated muscle.
Results
Targeted Expression of PAX7-FKHR in Drosophila Causes Muscular Phenotypes.
To explore the hypothesis that misexpression of PAX-FKHR affects the biology of differentiated muscle, we used the bipartite Gal4-UAS expression system (14) to conditionally express PAX-FKHR in Drosophila. When crossed into genetic backgrounds where genomic enhancers temporally and spatially regulate Gal4 expression, UAS-transgene expression occurs in precise tissue-specific patterns. We generated sets of UAS-PAX3-FKHR and UAS-PAX7-FKHR transgenic lines incorporating human PAX-FKHR cDNA.
We predicted that flies expressing human PAX-FKHR would provide relevant phenotypes because: (i) The functional DNA-binding motifs present in PAX-FKHR originate from the PAX portion of the chimeric protein (Fig. 6). (ii) The PAX3/7 subfamily of PAX genes is conserved in Drosophila, represented by the gooseberry (gsb) and gooseberry-neuro (gsb-n) genes (Fig. 7, which is published as supporting information on the PNAS web site). Like mammalian PAX3 and PAX7, both gsb and gsb-n are expressed in embryonic ectodermal and mesodermal tissue (15), although the specific contribution of gsb/gsb-n to fly myogenesis has not been studied. (iii) Mammalian PAX3 can functionally substitute for Drosophila PAX (16, 17). Also, mammalian PAX6, which possesses the same structural organization of PAX3/7 with regard to the paired and homeodomain DNA-binding motifs, demonstrates functionally appropriate dominant phenotypes when misexpressed in fly tissues (18).
We used a Myosin heavy chain-Gal4 (MHC-Gal4) driver to express either PAX3-FKHR or PAX7-FKHR in syncytial muscle fibers. We identified three independent lines, all UAS-PAX7-FKHR, that displayed potent larval/pupal lethality when transheterozygous for one copy of both UAS-PAX-FKHR and MHC-Gal4 (additional UAS-PAX-FKHR lines, including PAX3-FKHR, exhibit lethality only when increased gene copies of UAS-PAX-FKHR and/or MHC-Gal4 are present; data not shown). We conducted a lethal-phase test for two of these lines, which showed that the lethal phases were late larval (third instar) and pupal (Table 1, which is published as supporting information on the PNAS web site).
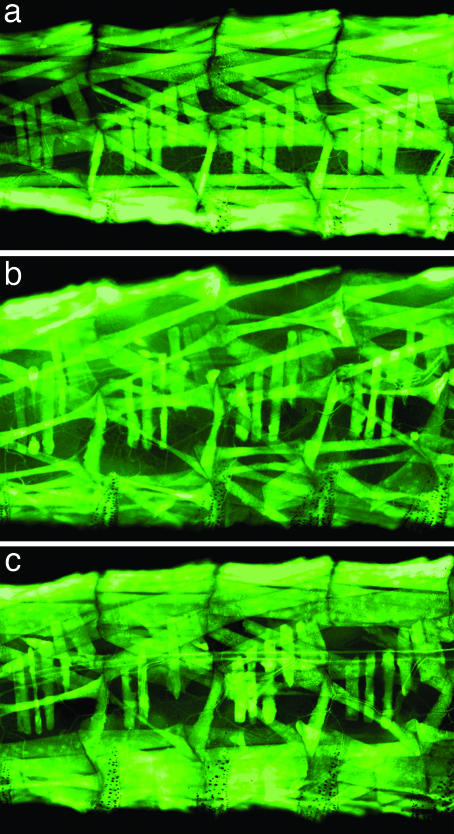
To specifically examine how PAX7-FKHR affects muscle in vivo, we included the UAS-2xeGFP transgene (19), which demonstrates bright fluorescence and allows the entire somatic body wall musculature to be visualized through the larval cuticle. PAX7-FKHR larvae exhibited abnormal muscle morphology (Fig. 1). Many individual fibers were absent, with all larval muscle groups appearing to be susceptible, although in a random distribution from animal to animal. Additional myofibers appeared wispy and hypotrophic (best seen in Fig. 1h). We observed these abnormalities in early third-instar larvae, documenting that the PAX7-FKHR muscle phenotype is unrelated to the physiologic histolysis of larval muscles that occurs during pupal metamorphosis.
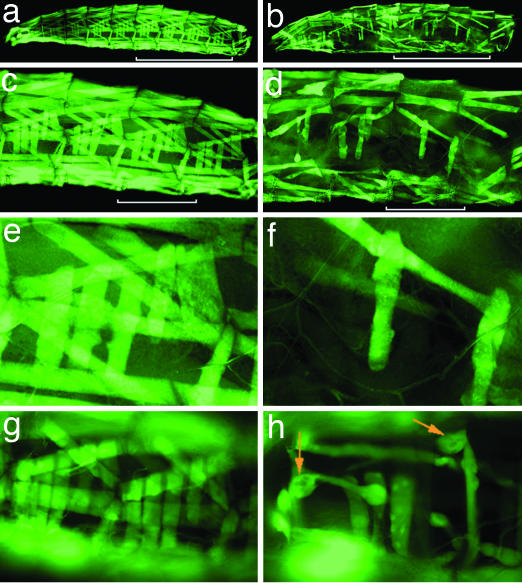
Fig. 1.
PAX7-FKHR causes muscle phenotypes in Drosophila. (a, c, and e) Wild type. (b, d, and f) PAX7-FKHR. (a) Body wall musculature from a control MHC-Gal4, UAS-GFP early third-instar larva. (b) An MHC-Gal4, UAS-GFP, UAS-PAX7-FKHR early third-instar larva. (c) Representative hemisegments of wild-type body wall musculature. The four hemisegments indicated by the white bar in a are shown. (d) Representative hemisegments of PAX7-FKHR musculature. The four hemisegments identified by the white bar in b are shown. (e) Abdominal hemisegment A6. (f) Abnormal musculature in abdominal hemisegment A6 of a PAX7-FKHR larva. (g) Representative hemisegments of an MHC-Gal4, UAS-GFP, UAS-PAX7-FKHR larvae at 2 days of age. (h) A representative image from the same PAX7-FKHR animal at 4 days of age. The orange arrows highlight dystrophic tissue. (Magnification: g and h, ×20.)
Because we observed no appreciable expression of the 2xUAS-eGFP (henceforth referred to as UAS-GFP) reporter in first-instar larvae (suggesting that Gal4-driven expression of PAX7-FKHR accumulates in postembryonic myofibers; Fig. 8, which is published as supporting information on the PNAS web site), we postulated that PAX7-FKHR specifically altered differentiated myofibers. To confirm this interpretation, we conducted a time-course study, during which we examined the musculature of living PAX7-FKHR larvae on sequential days of life. These studies showed that PAX7-FKHR larvae exhibit morphologically normal musculature up to day 2 of larval life (Fig. 1g). By day 4, however, the musculature had clearly deteriorated and was dysmorphic (Fig. 1h).
PAX7-FKHR Generates Nucleated Cells from Syncytial Myofibers.
Detailed analysis of PAX7-FKHR-expressing muscles in living animals revealed cellular-shaped tissue emanating from syncytial myofibers (Fig. 2b, compare with Fig. 2a), suggesting that new cells were forming from differentiated myofibers. Confocal microscopy of PAX7-FKHR larvae, partially dissected and stained with DAPI to highlight nuclei, showed individual nucleated cells separating from underlying myofibers (Fig. 2 c and c′) and separated mononuclear GFP-positive cells (Fig. 2 d and e).
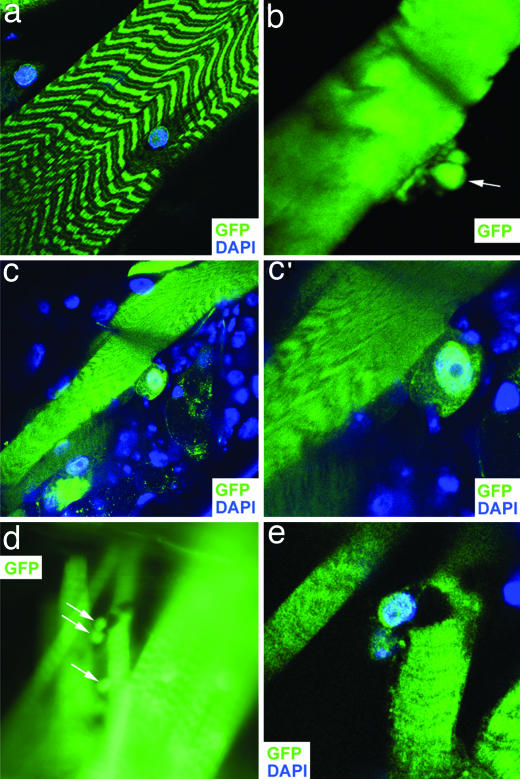
Fig. 2.
PAX7-FKHR generates nucleated cells from syncytial muscle. (a) Wild type. (b–e) PAX7-FKHR. (a) A confocal image of a myofiber from a control MHC-Gal4, UAS-GFP animal. (b) A putative cell generating de novo from syncytial muscle in a living PAX7-FKHR (MHC-Gal4, UAS-GFP, UAS-PAX7-FKHR) myofiber. The arrow highlights a spherical tissue element barely connected to the juxtaposed fiber. (c and c′) A PAX7-FKHR myofiber with a separating nucleated cell. Two different magnifications are shown. (d) An image of a myofiber having given rise to nucleated cells. Two cavities can be seen in this fiber, presumably corresponding to the deficits left behind by the newly separated or departing cells (white arrows). This profile shows surrounding myofibers (including fibers to the left and right, superficial and deep, respectively) that are intact, confirming that this area of tissue was not disrupted as a byproduct of animal dissection. (e) A confocal image of the same fiber shown in d. (Magnifications: a, ×60; b, ×252; c and d, ×63; c‘, ×152, e, ×160.)
We considered that within Drosophila larvae, a sequestered population of “adult myoblasts” is present that, during metamorphosis, migrates, fuses, and forms the adult muscles. These myoblasts, which are generated during embryogenesis and proliferate during larval development, are only partially differentiated and located in association with the imaginal discs and in clusters along the peripheral nerves (20, 21). We performed immunocytochemistry with D-MEF2 antisera to highlight these cells and found that in both PAX7-FKHR animals and control animals (including late third-instar larvae containing two copies of the MHC-Gal4 driver and UAS-GFP reporter) the adult myoblasts remain partially differentiated and, unlike the cells observed in PAX7-FKHR animals, do not express GFP (Fig. 9, which is published as supporting information on the PNAS web site). Furthermore, adult myoblasts were present and properly positioned in PAX7-FKHR animals. Thus, we conclude that individual myogenic-type cells can generate de novo from differentiated muscle in response to PAX7-FKHR expression.
Liberated PAX7-FKHR Myogenic Cells Demonstrate Invasive Behavior.
We next considered the possibility that newly generated PAX-FKHR cells might enter the larval hemolymphatic circulatory system and spread to nonmuscular organs. Indeed, ectopic GFP-positive tissue was located within the internal larval soft tissue, which includes the imaginal discs, gut, CNS, fat body, and salivary glands. Adult myoblasts, however, are found in the imaginal discs and the gut contains visceral muscle. Therefore, we focused on the CNS, which normally contains no myogenic tissue. We identified larval CNS organs studded with GFP-positive tissue (Fig. 3a′) in 18 of 30 third-instar larvae. Confocal microscopy confirmed that the CNS GFP-positive tissue was nucleated (Fig. 3c). No similar tissue was seen in MHC-Gal4, UAS-GFP control animals (n = 30; Fig. 3b′). These findings show that PAX7-FKHR tissue disseminates and infiltrates non-native tissue compartments.
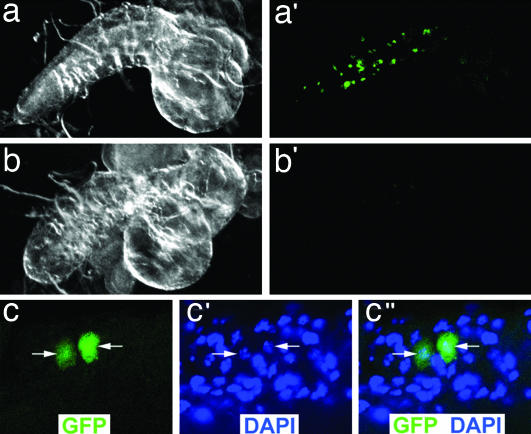
Fig. 3.
PAX7-FKHR cells enter the larval CNS. (a and a′) A dark field (a) and GFP fluorescent (a′) image of a PAX7-FKHR (MHC-Gal4, UAS-GFP, UAS-PAX7-FKHR) CNS organ with GFP-positive tissue. (b and b′) A dark field (b) and GFP fluorescent (b′) image of a wild-type (MHC-Gal4, UAS-GFP) CNS organ. No GFP-positive cells are identified, although dim, nonspecific, small foci of background autofluorescence can be seen. (c–c″) Confocal images from a GFP-positive PAX7-FKHR larval CNS organ. The white arrows highlight the individual nuclei that correspond to the GFP-positive cells. (Magnifications: c–c”, ×126.)
Wild-Type PAX3 Expressed in Drosophila Muscle.
To determine whether the formation of nucleated cells from differentiated tissue is a unique response to PAX7-FKHR, we generated transgenic flies expressing wild-type PAX3, because PAX3 has been previously shown to functionally substitute for Drosophila PAX orthologues (16, 17). In the presence of PAX3 expression, we again observed muscular phenotypes similar to PAX7-FKHR, with muscle dymorphology and myogenic tissue in the CNS (Fig. 10, which is published as supporting information on the PNAS web site), although this process was significantly less efficient than with PAX7-FKHR and required two copies of both UAS-PAX3 and the MHC-Gal4 driver. These data, nonetheless, suggest that PAX-FKHR disrupts myofibers through mechanisms that are functionally shared with wild-type PAX.
Ras Is a PAX7-FKHR Genetic Modifier.
In the transgenic ARMS mouse model, PAX3-FKHR is a poor oncogene and requires crippled tumor surveillance by Trp-53 or Ink4a/ARF deficiency for appreciable levels of tumorigenesis (6). We tested whether PAX7-FKHR activity would likewise be enhanced by Drosophila “tumorigenic” alleles. Although typically associated with embryonal RMS and not ARMS, we focused on a constitutively active allele of Ras (Ras85DG12V or “RasV12”) for the following reasons: (i) The canonical tumor surveillance function performed by tumor suppressor proteins such as p53 is not conserved in Drosophila. (ii) The RasV12 allele has been shown to augment cancer-related phenotypes in flies (22). (iii) The generation of individual cells suggests that PAX-FKHR reverses the terminally differentiated state of syncytial myofibers. Because activated Ras is known to interfere with the fate specification and terminal differentiation of both fly and mammalian muscle precursors (23–29), we hypothesized that, if interference with terminal differentiation was involved in PAX-FKHR muscular activity, Ras and PAX-FKHR might genetically interact.
In the presence of PAX7-FKHR and RasV12, we observed larval muscular patterns different from those of either wild-type or PAX7-FKHR animals. PAX7-FKHR, RasV12 animals exhibited a pattern of GFP fluorescence that was disorganized and, by comparison, difficult to discern at low magnification (Fig. 4b). Even at high magnification, the contours of many myofibers were vague and suggested an ill-defined appearance (Fig. 4b′). In contrast, the muscle pattern in control RasV12 animals was microscopically normal (Fig. 4 a and a′; of note, MHC≫RasV12 expression causes pupal lethality) (data not shown), demonstrating that the PAX7-FKHR, RasV12 muscular phenotype depends on PAX7-FKHR. Histologic examination of PAX7-FKHR, RasV12 paraffin-embedded tissue suggested that collections of mononuclear cells were present at the expense of syncytial myofiber tissue (Fig. 11, which is published as supporting information on the PNAS web site). Confocal microscopy demonstrated mononuclear cells forming from syncytial myofibers at a frequency significantly higher than with PAX7-FKHR alone (Fig. 4 d and e), establishing activated Ras as a modifier of the PAX7-FKHR phenotype.
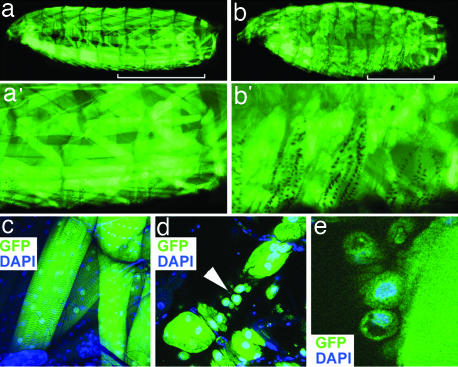
Fig. 4.
Activated Ras enhances PAX7-FKHR muscular activity when coexpressed in syncytial muscle. (a and a′) An MHC-Gal4, UAS-RasV12 (MHC≫RasV12) larva. The three hemisegments indicated by the white bar are shown in a′. (b and b′) An MHC-Gal4, UAS-RasV12, UAS-PAX7-FKHR (MHC≫RasV12, PAX7-FKHR) larva. The three hemisegments indicated by the white bar are shown in b′. (c) Representative, stereotactically normal myofibers from an MHC-Gal4, UAS-RasV12 larva. (d and e) Abnormal myofibers from MHC-Gal4, UAS-RasV12, UAS-PAX7-FKHR larvae showing individual nucleated cells (arrowhead in d). Muscle tissue is highlighted by GFP expressed from the UAS-GFP transgene. (Magnifications: c, ×20; d, ×40; e, ×126.)
To further explore the Ras, PAX7-FKHR genetic interaction, we tested three lethal or semilethal loss-of-function ras alleles (rasE2f, rasE1FB, and ras06677) (30, 31) to determine whether diminished Ras activity would suppress PAX7-FKHR activity. These alleles dominantly suppressed the PAX7-FKHR muscular phenotype to varying degrees and also allowed for viable adult escapers (data not shown). Focusing on the stronger ras suppressor, rasE2f, we confirmed suppression of the muscular phenotype in a blinded study (Fig. 5). Additionally, at the lower temperature of 23°C, we observed that, with this allele, 83% of larval animals (n = 300) were rescued to adult viability, whereas only 15% of PAX7-FKHR larvae (n = 300) survived to adulthood. Therefore, diminished Ras activity dominantly suppresses PAX7-FKHR activity, including rescue of PAX7-FKHR-related lethality. These data confirm that Ras is a genetic modifier of PAX7-FKHR.
Fig. 5.
A loss-of-function ras allele dominantly suppresses the PAX7-FKHR muscular phenotype. (a) Representative hemisegments of wild-type (MHC-Gal4, UAS-GFP) musculature (same animal as shown in Fig.1a). (b) Representative hemisegments of abnormal MHC-Gal4, UAS-PAX7-FKHR musculature. (c) Representative hemisegments of MHC-Gal4, UAS-PAX7-FKHR musculature with one loss-of-function rasE2f allele. The number of intact myofibers is significantly increased in this genetic background. The musculature is highlighted by the UAS-GFP reporter.
Discussion
We have used the fruit fly Drosophila melanogaster as a model organism to explore the pathogenicity of the ARMS initiator PAX7-FKHR, which has not previously been profiled in an animal model system to our knowledge. We have shown that (i) PAX7-FKHR, when expressed in differentiated muscle, causes discrete nucleated cells to form from synyctial myofibers, (ii) these cells, freed from myofiber attachment, spread to the CNS, (iii) these properties are not unique to the PAX7-FKHR chimera, as human wild-type PAX3 demonstrates similar activity in fly muscle, and (iv) activated Ras enhances and diminished Ras activity suppresses the PAX7-FKHR muscular phenotype and associated lethality.
Muscle as an ARMS Tissue of Origin.
Despite intensive study, the tissue of origin for ARMS has been puzzling. Because skeletal muscle is irreversibly postmitotic and syncytial, the origin had long been hypothesized to be a muscle precursor cell or stem-like cell. Yet, transgenic expression of PAX3-FKHR in muscle precursor cells or muscle-specific satellite stem cells demonstrates no evidence of tumorigenesis (7–10), whereas expression of PAX3-FKHR in terminal differentiating myofibers caused rhabdomyosarcomagenesis (6). Because no evidence existed, either in cell culture or in vivo, that PAX-FKHR can induce individual cells to form de novo from synytial tissue, questions persisted regarding whether the mouse model had undetectably generated tumors from an unknown cell.
Because most PAX3-FKHR mice in the ARMS model do not grow tumors (6), we predicted that exhaustively surveying mouse muscle for focal or subtle cellular changes would be difficult. In contrast, we postulated that Drosophila would provide a practical approach for this type of study, given the amenability of the organism to rapid, thorough, serial examination of living muscle. Additionally, because fly muscle contains no known mechanism for repair (including satellite cells), we predicted that muscle dysmorphology would not be obfuscated by physiologic regeneration. With this approach, we were able to document evidence of cells generating de novo from syncytial tissue. These results show that PAX-FKHR can specify this process and that cells can form from differentiated muscle in vivo. Thus, in a complementary fashion, the ARMS mouse and PAX7-FKHR fly strongly argue that muscle can serve as a RMS tissue of origin.
PAX and Terminal Differentiation.
How might PAX-FKHR cause muscle “dedifferentiation?” Because both wild-type PAX and PAX-FKHR demonstrate activity in fly muscle and, therefore, presumably share gene targets and cofactors, we can look toward our knowledge of wild-type PAX for mechanistic insight. Specifically, wild-type PAX performs at least two distinct developmental functions: (i) PAX proteins regulate early organogenesis and lineage determination (32); and (ii) PAX functions late in lineage development to repress the terminal differentiation of tissue-specific precursors. For example, in melanocyte development, PAX3 inhibits the terminal differentiation of maturing melanoblasts, thereby maintaining a population of partially differentiated precursors available for damage response (33). PAX7 likewise inhibits late steps in the terminal differentiation of muscle satellite cells, where PAX7 and myogenin expression are mutually exclusive, and overexpression of PAX7 interferes with MyoD-dependent transcriptional activation and down-regulates MyoD expression (34). PAX5 has also been characterized as a negative modulator of plasma cell terminal differentiation (35). We propose that PAX-FKHR, upon chromosomal rearrangement and misexpression, reacquires its inhibitory activity on terminal differentiation and interferes with the integrity of differentiated myofibers.
Ras and ARMS.
For ARMS tumorigenesis to originate from postmitotic muscle, newly generated PAX-FKHR cells would need to re-enter the cell cycle to proliferate. The fact that PAX3-FKHR in the ARMS mouse model requires loss of cell-cycle regulation for appreciable levels of tumorigenesis argues that PAX3-FKHR alone is not sufficient to cause cell-cycle reentry. Consistent with this observation, we found no evidence of cell-cycle reentry in myofiber nuclei or individual GFP-positive myogenic cells in either PAX7-FKHR or PAX7-FKHR, RasV12 animals after staining with phosphohistone H3 antibody. These results are similar to previous findings that activated Ras does not promote proliferation of either mammalian or fly myogenic precursor cells (23, 25, 27, 28, 36).
Our genetic gain-of-function and loss-of-function ras studies suggest, nonetheless, that Ras signaling is involved in PAX7-FKHR-mediated dedifferentiation of muscle. In this regard, it is noteworthy that constitutively activated Ras strongly interferes with the terminal differentiation of mammalian myoblasts (25, 27, 28), whereas ras gain-of-function or loss-of-function alters the differentiation of Drosophila founder cell myoblasts (24, 36, 37), the patterning pioneers of fly muscle. Therefore, we suspect the interaction observed between PAX7-FKHR and Ras results not from proliferation but from the fact that both molecules functionally perturb muscle differentiation.
A Genetic Approach to PAX-FKHR and ARMS.
Outcomes for advanced ARMS remain dismal despite intensive therapy (2, 4), underscoring the need to characterize the pathogenetic mechanisms underlying tumorigenesis. As presented above, the PAX7-FKHR Drosophila phenotypes are sensitive to both genetic suppression and enhancement. Therefore, this PAX-FKHR model is well situated for unbiased genetic screens. This approach, unexplored with regards to PAX-FKHR, should allow for the isolation of previously unknown PAX-FKHR gene targets and cofactors. Uncovering these molecular entities should represent a starting point for the conceptual development of therapeutics to target these interactions and poison ARMS.
Materials and Methods
Constructs.
The pUAST (14) constructs were generated as follows: (i) UAS-PAX7-FKHR cDNA (100 bp of 5′ UTR, 2,490 bp of coding sequence, and 1,180 bp of 3′UTR) was cloned by flanking NotI (5′) and XbaI (3′) cleavage sites; (ii) UAS-PAX3-FKHR cDNA (24 bp of 5′ UTR, 2,508 bp of coding sequence, and 1,178 bp of 3′UTR) was cloned by flanking BglII (5′) and XbaI (3′) cleavage sites; and (iii) UAS-PAX3-cDNA (24 bp of 5′ UTR, 1,437 bp of coding sequence, and 173 bp of 3′UTR) was cloned by flanking EcoRI (5′) and XbaI (3′) cleavage sites.
D. melanogaster Stocks.
We generated UAS-PAX7-FKHR, UAS-PAX3-FKHR, and UAS-PAX3 transgenic lines by P-element-mediated gene transfer (38). The remaining stocks used were UAS-2xeGFP-AH2 (19), UAS-Ras85D.V12-TL1 (39), Ras85De2F (30), Ras85De1B (30), Ras85D0667 (31), and Myosin heavy chain-Gal4 (MHC-Gal4) (40).
Lethal-Phase Test and Time-Course Studies.
For the lethal-phase test study, conducted at 29°C, control (wild type) or PAX7-FKHR virgin females were mated to MHC-Gal4 males. Genotypes were as follows: wild type = w1118;+; +; PAX7-FKHR−2 = w1118,UAS-PAX7-FKHR−2;+; +; PAX7-FKHR−3D = w1118;UAS-PAX7-FKHR−3D; +; and MHC-Gal4 = w1118;UAS-2xeGFP;MHC-Gal4. Fertilized embryos were allowed to hatch on wet yeasted apple juice plates, with the animals transferred as first-instar larvae to prewarmed dry yeasted cornmeal agar vials. Dead larvae were staged as second or third instars by their anterior spiracles (41). The number of animals not scored represents animals injured or lost during the course of the study.
For the time-course study, newly hatched (day 0) UAS-PAX7-FKHR−2;UAS-2xeGFP-AH2;MHC-Gal4 larvae were obtained and individually placed into separate, wet yeasted grape juice plates and raised at 25°C or 29°C. The animals were examined daily for the next 4 days. When necessary, larvae were placed at −20°C for ≈5 min to temporarily immobilize the animals for photographic purposes.
Immunohistochemistry.
Antibodies used were rabbit anti-D-MEF2 (1:1,000) (42) and Alexa Fluor 568 goat anti-rabbit (1:500; Invitrogen, Carlsbad, CA). For these preparations, larvae were pinned dorsal-side down on silicone [Sylgard(R) 184; Dow Corning, Midland, MI] plates, and the cuticle was opened along the ventral surface. The larvae were formalin-fixed directly on the plates for 1 h at room temperature. The larvae were then briefly rinsed with PBS and exposed to antibody as described (43). Stained tissue was mounted in Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed on an LSM510-meta confocal microscope (Zeiss, Thornwood, NY).
Histopathology.
To prepare Drosophila larvae for histologic processing, the anterior-most and/or posterior-most aspect of the larval cuticle was breached with a razor blade to facilitate formalin penetration and fixation of internal tissue. The fixed tissue was then processed for routine histology and embedded in paraffin. Sections were cut at 3 μm and stained with hematoxylin and eosin.
Supplementary Material
Acknowledgments
We thank the Children’s Medical Center at Dallas (M. McNeese) for histopathology; M. Ramaswami (University of Arizona, Tucson, AZ) for the MHC-Gal4 stock; the Bloomington Drosophila Stock Center at Indiana University; F. Barr (University of Pennsylvania, Philadelphia, PA) for PAX-FKHR and PAX cDNAs; H. Kramer, Z. P. Liu, J. Maines, D. McKearin, D. Sosic, S. Wasserman, R. Cripps, R. Bassel-Duby, and S. Cameron for manuscript review; and the University of Texas Southwestern Medical Center invertebrate genetics group and J. Abrams and members of his laboratory for ongoing discussions. This work was funded in part by a Society for Pediatric Pathology Young Investigator Research Grant, University of Texas Southwestern Medical Center President’s Research Council Award, and University of Texas Southwestern Medical Center Physician Scientist Training Program Fellowship (to R.L.G.). Work in the laboratory of E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Cardiovascular Clinical Research Center, the Robert A. Welch Foundation, and the Nearberg Foundation.
Abbreviations
- RMS
rhabdomyosarcoma
- ARMS
alveolar RMS.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Smith M. A., Gloecker Ries L. A. In: Principles and Practice of Pediatric Oncology. Pizzo P. A., Poplack D. G., editors. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 1–12. [Google Scholar]
- 2.Wexler L. H., Crist W. M., Helman L. J. In: Principles and Practice of Pediatric Oncology. Pizzo P. A., Poplack D. G., editors. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 939–971. [Google Scholar]
- 3.Weiss S. W., Goldblum J. R. In: Enzinger and Weiss’s Soft Tissue Tumors. Weiss S. W., Goldblum J. R., editors. St. Louis: Mosby; 2001. pp. 785–835. [Google Scholar]
- 4.Arndt C. A., Crist W. M. N. Engl. J. Med. 1999;341:342–352. doi: 10.1056/NEJM199907293410507. [DOI] [PubMed] [Google Scholar]
- 5.Barr F. G. Oncogene. 2001;20:5736–5746. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 6.Keller C., Arenkiel B. R., Coffin C. M., El Bardeesy N., DePinho R. A., Capecchi M. R. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson M. J., Shelton G. D., Cavenee W. K., Arden K. C. Proc. Natl. Acad. Sci. USA. 2001;98:1589–1594. doi: 10.1073/pnas.98.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller C., Hansen M. S., Coffin C. M., Capecchi M. R. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagutina I., Conway S. J., Sublett J., Grosveld G. C. Mol. Cell. Biol. 2002;22:7204–7216. doi: 10.1128/MCB.22.20.7204-7216.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relaix F., Polimeni M., Rocancourt D., Ponzetto C., Schafer B. W., Buckingham M. Genes Dev. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo T., Nadal-Ginard B. J. Cell. Sci. 1998;111:1081–1093. doi: 10.1242/jcs.111.8.1081. [DOI] [PubMed] [Google Scholar]
- 12.Odelberg S. J., Kollhoff A., Keating M. T. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 13.Rosania G. R., Chang Y. T., Perez O., Sutherlin D., Dong H., Lockhart D. J., Schultz P. G. Nat. Biotechnol. 2000;18:304–308. doi: 10.1038/73753. [DOI] [PubMed] [Google Scholar]
- 14.Brand A. H., Perrimon N. Development (Cambridge, U.K.) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 15.Gutjahr T., Patel N. H., Li X., Goodman C. S., Noll M. Development (Cambridge, U.K.) 1993;118:21–31. doi: 10.1242/dev.118.1.21. [DOI] [PubMed] [Google Scholar]
- 16.Xue L., Noll M. EMBO J. 1996;15:3722–3731. [PMC free article] [PubMed] [Google Scholar]
- 17.Xue L., Li X., Noll M. Development (Cambridge, U.K.) 2001;128:395–405. doi: 10.1242/dev.128.3.395. [DOI] [PubMed] [Google Scholar]
- 18.Halder G., Callaerts P., Gehring W. J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 19.Halfon M. S., Gisselbrecht S., Lu J., Estrada B., Keshishian H., Michelson A. M. Genesis. 2002;34:135–138. doi: 10.1002/gene.10136. [DOI] [PubMed] [Google Scholar]
- 20.Bate M., Rushton E., Currie D. A. Development (Cambridge, U.K.) 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 21.Currie D. A., Bate M. Development (Cambridge, U.K.) 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- 22.Pagliarini R. A., Xu T. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 23.Artero R., Furlong E. E., Beckett K., Scott M. P., Baylies M. Development (Cambridge, U.K.) 2003;130:6257–6272. doi: 10.1242/dev.00843. [DOI] [PubMed] [Google Scholar]
- 24.Carmena A., Buff E., Halfon M. S., Gisselbrecht S., Jimenez F., Baylies M. K., Michelson A. M. Dev. Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- 25.Gossett L. A., Zhang W., Olson E. N. J. Cell Biol. 1988;106:2127–2137. doi: 10.1083/jcb.106.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halfon M. S., Carmena A., Gisselbrecht S., Sackerson C. M., Jimenez F., Baylies M. K., Michelson A. M. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 27.Konieczny S. F., Drobes B. L., Menke S. L., Taparowsky E. J. Oncogene. 1989;4:473–481. [PubMed] [Google Scholar]
- 28.Olson E. N., Spizz G., Tainsky M. A. Mol. Cell. Biol. 1987;7:2104–2111. doi: 10.1128/mcb.7.6.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyman C. M., Ramocki M. B., Taparowsky E. J., Wolfman A. Oncogene. 1997;14:697–704. doi: 10.1038/sj.onc.1200874. [DOI] [PubMed] [Google Scholar]
- 30.Simon M. A., Bowtell D. D., Dodson G. S., Laverty T. R., Rubin G. M. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 31.Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S., Rubin G. M. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi N., Epstein J. A. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 33.Lang D., Lu M. M., Huang L., Engleka K. A., Zhang M., Chu E. Y., Lipner S., Skoultchi A., Millar S. E., Epstein J. A. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 34.Olguin H. C., Olwin B. B. Dev. Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin K. I., Tunyaplin C., Calame K. Immunol. Rev. 2003;194:19–28. doi: 10.1034/j.1600-065x.2003.00040.x. [DOI] [PubMed] [Google Scholar]
- 36.Carmena A., Gisselbrecht S., Harrison J., Jimenez F., Michelson A. M. Genes Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee H. H., Norris A., Weiss J. B., Frasch M. Nature. 2003;425:507–512. doi: 10.1038/nature01916. [DOI] [PubMed] [Google Scholar]
- 38.Rubin G. M., Spradling A. C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 39.Lee T., Feig L., Montell D. J. Development (Cambridge, U.K.) 1996;122:409–418. doi: 10.1242/dev.122.2.409. [DOI] [PubMed] [Google Scholar]
- 40.Schuster C. M., Davis G. W., Fetter R. D., Goodman C. S. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 42.Lilly B., Zhao B., Ranganayakulu G., Paterson B. M., Schulz R. A., Olson E. N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 43.Patel N. H. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.