Abstract
Microtubules are indispensable dynamic structures that contribute to many essential biological functions. Assembly of the native α/β tubulin heterodimer, the subunit that polymerizes to form microtubules, requires the participation of several molecular chaperones, namely prefoldin, the cytosolic chaperonin CCT, and a series of five tubulin-specific chaperones termed cofactors A–E (TBCA–E). Among these, TBCC, TBCD, and TBCE are essential in higher eukaryotes; they function together as a multimolecular machine that assembles quasinative CCT-generated α- and β-tubulin polypeptides into new heterodimers. Deletion and truncation mutations in the gene encoding TBCE have been shown to cause the rare autosomal recessive syndrome known as HRD, a devastating disorder characterized by congenital hypoparathyroidism, mental retardation, facial dysmorphism, and extreme growth failure. Here we identify cryptic translational initiation at each of three out-of-frame AUG codons upstream of the genetic lesion as a unique mechanism that rescues a mutant HRD allele by producing a functional TBCE protein. Our data explain how afflicted individuals, who would otherwise lack the capacity to make functional TBCE, can survive and point to a limiting capacity to fold tubulin heterodimers de novo as a contributing factor to disease pathogenesis.
Microtubules are versatile and ubiquitous elements of the cytoskeleton, their dynamic filamentous arrays contributing to a remarkable diversity of biological functions that includes cell division, intracellular transport, and the maintenance of cell shape. The subunit that polymerizes to form microtubules is the tubulin heterodimer, which consists of one α- and one β-tubulin polypeptide. The tubulin heterodimer cannot associate spontaneously; both α- and β-tubulins require facilitated folding by one or more rounds of ATP-dependent interaction with the cytosolic chaperonin, CCT (1–3). Quasinative folding intermediates generated as a result of this interaction (4) are still not competent for assembly into heterodimers; they must first be captured and stabilized by a series of tubulin-specific chaperones known as cofactors A–E (TBCA–E; refs. 5–9). TBCA and TBCD interact with β-tubulin, whereas TBCB and TBCE interact with α-tubulin. There is free exchange of β-tubulin between TBCA and TBCD and of α-tubulin between TBCB and TBCE. TBCEα and TBCDβ interact with each other, forming a multimolecular complex; TBCC enters this complex, forming a supercomplex that releases native tubulin heterodimers upon GTP hydrolysis by β-tubulin (refs. 6 and 9; summarized in Fig. 1A). The participation of TBCC, TBCD, and TBCE in these reactions is essential in higher eukaryotes; genetic experiments in Saccharomyces pombe show the genes for the homologs of these proteins to be essential for life (10, 11), and in Arabidopsis thaliana, they are required for proper embryogenesis (12).
Fig. 1.
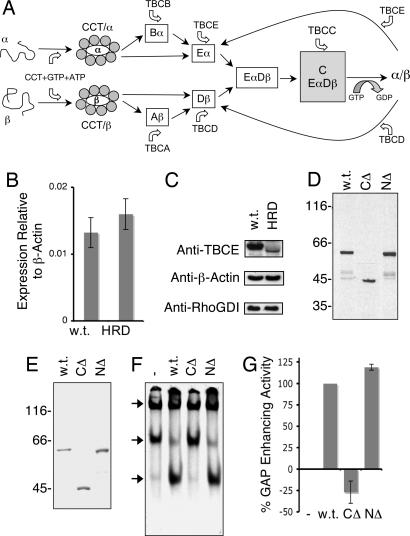
Expression and activity of wild-type and HRD mutant TBCE proteins. (A) Schematic representation of the tubulin heterodimer assembly pathway in higher eukaryotes (6, 9). (B) Expression levels of TBCE mRNA (relative to β-actin) in normal (w.t.) and compound heterozygote HRD patient-derived fibroblasts, determined by real-time PCR. (C) Abundance of TBCE protein determined by semiquantitative Western blotting by using an anti-TBCE antibody in postnuclear extracts of wild-type and HRD mutant (compound heterozygote) cells. Loading controls done with anti-β-actin and anti-RhoGDI antibodies are shown below. (D). Expression of wild-type (w.t.) and HRD mutant (CΔ, C371X; NΔ, deletion of nucleotides 66–67) forms of TBCE by transcription/translation in rabbit reticulocyte lysate. (E) Purification of recombinant wild-type and HRD mutant forms of TBCE expressed in insect sf9 cells. In D and E, location of molecular mass markers is shown on the left. (F) In vitro β-tubulin-folding assays (5, 6), in which bacterially expressed 35S-labeled urea-unfolded β-tubulin was suddenly diluted into a reaction containing the cytosolic chaperonin CCT, ATP, GTP, TBCC, and TBCD and either wild-type (w.t.) or compound heterozygous HRD mutant forms (NΔ, deletion of nucleotides 66–67) of TBCE. Reaction products were analyzed by nondenaturing gel electrophoresis. Arrows (top to bottom) denote the location of the CCT/β-tubulin binary complex, the β-tubulin/TBCD complex, and native tubulin heterodimers. (G) Relative tubulin GAP-enhancing activities (17) of compound heterozygous HRD mutant [C371X (CΔ) and deletion of nucleotides 66–67 (NΔ)] forms of TBCE protein relative to wild type (taken as 100%).
In addition to participating in the generation of tubulin heterodimers de novo, TBCD and TBCE can interact directly with the native heterodimer (Fig. 1A). When they do so, the dimer is disrupted; TBCD binds to the β-subunit, whereas TBCE binds to the α-subunit. In either case, the remaining subunit, bereft of its partner, decays to a nonnative state that is no longer capable of heterodimer formation and that can be captured by chaperonin (7). Thus, cells overexpressing either of these tubulin-specific chaperones lose most or all of their microtubules (7). TBCC and TBCD also act together as a GTPase activator [GTPase activating protein (GAP)] for native tubulin in a reaction that is further stimulated by TBCE (9, 13). This reaction is distinct from the GTP hydrolysis that accompanies microtubule polymerization, because it takes place with a Km for tubulin of ≈0.1 μM, which is ≈200-fold lower than the critical concentration for polymerization of heterodimers into microtubules. Because the cofactor-driven tubulin GAP reaction converts GTP tubulin to GDP tubulin, it may play a role in the cell as a regulator of microtubule dynamics, as well as a quality control mechanism that continuously monitors the ability of β-tubulin to hydrolyze its bound GTP (13).
Recently, a group of rare, devastating, and fatal inherited human diseases collectively termed HRD (for hyperparathyroidism, mental retardation, and facial dysmorphism) has been mapped and found to be caused by mutations in the gene encoding TBCE (14). Middle Eastern individuals with HRD carry a 12-bp in-frame deletion (at nucleotides 155–166) in the second coding exon of the gene encoding TBCE, resulting in the expression of a protein that lacks amino acids 52–55 (15) within the cytoskeleton-associated protein glycine-rich α-tubulin-binding domain (16). In addition, afflicted members of a Belgian family are compound heterozygotes: one allele contains a 2-bp deletion (at nucleotides 66–67) in the first coding exon that would be expected to yield only a very short (22-aa) N-terminal peptide fused to a “nonsense” sequence of 26 aa translated from the −1 reading frame; the second allele contains a termination codon in exon 12 (nucleotide 1113T→A) that would result in a protein lacking 157 (of a total of 527) amino acids at the C terminus. Thus, compound heterozygous HRD patients would not be expected to produce a functional TBCE protein. Here we show that these individuals do in fact express a functional TBCE polypeptide. We define the unique mechanism whereby this occurs and show that it involves cryptic translational initiation upstream of the 2-bp genetic lesion at three closely spaced out-of-frame methionine codons.
Results and Discussion
The HRD Allele with a Deletion at Nucleotides 66–67 Expresses a Functional TBCE Protein.
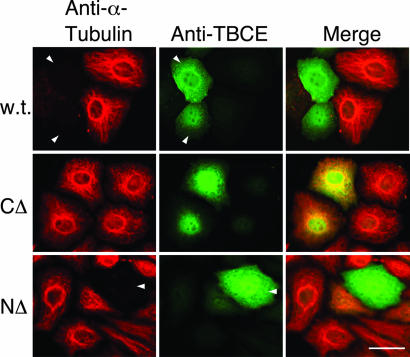
To investigate the mechanism of tubulin heterodimer formation in compound heterozygous HRD individuals, we first examined TBCE transcript and protein abundance. We found that TBCE mRNA levels were quite similar in mutant and wild-type cells (Fig. 1B); on the other hand, TBCE protein was apparently present in HRD disease cells at a greatly reduced level compared with wild-type controls and migrated with a slightly faster electrophoretic mobility (Fig. 1C). We next explored the consequences of expressing each of the two mutant TBCE forms by coupled in vitro transcription/translation (Fig. 1D). In these experiments, we found that the nucleotide 1113T→A mutant expressed a protein (CΔ) with an apparent molecular mass of ≈45 kDa, consistent with the predicted size of the encoded protein. However, in the case of the HRD allele containing the deletion at nucleotides 66–67 (NΔ), we were surprised to observe the expression of a polypeptide that, in common with the TBCE present in disease cells, migrated slightly faster than the wild-type TBCE polypeptide. To assess the functionality of these proteins, we expressed and purified each as an untagged recombinant protein (Fig. 1E). First, we tested their activities in in vitro CCT-mediated tubulin-folding assays (5, 6) containing purified TBCC, TBCD, and either wild-type or mutated forms of TBCE. In these experiments, a reaction containing the CΔ mutant polypeptide did not support productive folding to any greater extent than a parallel control done without TBCE, whereas the protein expressed from the NΔ HRD allele supported in vitro tubulin folding in a manner similar to the wild-type control (Fig. 1F). Second, we compared the activity of mutant and wild-type proteins in tubulin GAP assays; in these reactions, TBCE acts as a stimulator of TBCC and TBCD-driven tubulin GTP hydrolysis at a heterodimer concentration that is several hundred-fold lower than that required for polymerization into microtubules (13). We found that the NΔ mutant protein enhanced TBCC and TBCD-driven tubulin GAP activity to a similar extent as the wild-type TBCE counterpart; in contrast, the CΔ mutant had no GAP-enhancing activity (Fig. 1G). Third, we transfected cultured cells with constructs engineered for the expression of the mutated proteins. In these experiments, we detected no perturbation of the microtubule cytoskeleton in the case of the CΔ truncation, which partitioned to the nucleus as well as appearing in the cytoplasm. On the other hand, parallel transfections done with either the wild-type or the NΔ form of TBCE both resulted in the obliteration of microtubules as a result of heterodimer disruption and sequestration of α-tubulin by excess TBCE (ref. 7; Fig. 2). We conclude that deletion of the C-terminal 157 aa of the protein renders it functionally incompetent. On the other hand, the NΔ HRD allele expresses a functional TBCE protein, even though the mutant sequence would not be expected to express a protein with any “correct” residues beyond amino acid 22.
Fig. 2.
Overexpression of wild-type and compound heterozygous HRD mutant forms of TBCE in HeLa cells. HeLa cells were transfected with constructs engineered for the expression of wild-type, CΔ, or deletion of nucleotides 66–67 (NΔ) mutant forms of TBCE. Cells were fixed 24 h posttransfection and analyzed by indirect immunofluorescence by using antisera for α-tubulin (shown in red) and TBCE (ref. 6; shown in green). Arrows highlight TBCE-expressing cells showing microtubule obliteration as a result of expression of the transgene. (Scale bar, 10 μm.)
TBCE Expression from the NΔ HRD Allele Occurs by Cryptic Out-of-Frame Upstream Translational Initiation.
We considered the possibility that out-of-frame initiation (17) upstream of the genetic lesion might be responsible for the expression of a functional polypeptide from the NΔ allele. Inspection of the nucleotide sequence in the vicinity of the deletion showed the presence of three closely spaced upstream AUG codons located in the −2 reading frame at nucleotides 23–25, 50–52, and 59–61, respectively (Fig. 3A). Translational initiation at these codons would result in the expression of a “nonsense” polypeptide consisting of 15, 6, or 3 aa fused to amino acids 24–527 of the wild-type protein. We found that introduction of a termination codon at a location immediately upstream of the 2-bp deletion-containing sequence and in-frame with respect to the three potential cryptic AUG initiation sites completely abolished translational activity (Fig. 3B, compare lanes 1 and 2). Moreover, changing the authentic in-frame initiation codon to a termination codon had no effect on the translation of the NΔ mutant (Fig. 3B, lane 3). These data define the site(s) of translational initiation of this HRD allele in the −2 reading frame at a point located between the authentic initiator methionine and the genetic lesion. Systematic alteration of each of the out-of-frame AUG codons failed to abrogate translation (Fig. 3B, lanes 4–6), but simultaneous alteration of all three AUG codons completely abolished translational activity (Fig. 3B, lane 10). These data, together with a series of mutations in which pairs of out-of-frame AUG codons were altered to GCG (Fig. 3B, lanes 7–9), demonstrate that translational initiation can occur at each of the out-of-frame methionine codons located at nucleotides 23–25, 50–52, and 59–61 (designated M3′, M12′, and M15′ in Fig. 3A). We also analyzed a tryptic digest of the purified protein expressed from the NΔ allele by MS and identified an N-terminally acetylated peptide with the sequence MENMQLR that was absent from corresponding digests of the wild-type protein (Fig. 6, which is published as supporting information on the PNAS web site). The location and close proximity of the three out-of-frame translational initiation sites explains the slightly faster mobility on SDS/PAGE of the polypeptide(s) expressed from this sequence compared with wild-type TBCE, as well as its appearance as a less sharply defined entity (Fig. 1 C and D). We conclude that initiation at any of three upstream out-of-frame AUG codons in patients harboring the NΔ deletion results in the expression of a fusion protein, and that one or more of these proteins has the functional properties of wild-type TBCE.
Fig. 3.
The NΔ TBCE HRD allele is expressed as a result of translational initiation at three upstream out-of-frame AUG codons. (A) Nucleotide and corresponding amino acid sequences of wild-type TBCE (Upper) and the allele with the HRD deletion of nucleotides 66–67 (Lower) in the vicinity of the 2-bp deletion. Deleted nucleotides in HRD are shown (Upper) in red. Potential translational reading frames in the mutant HRD allele are shaded in gray and yellow. Numbering of amino acids in the −2 reading frame (1′, 2′, etc.) starts at the first residue (Q) of this open frame preceding the 2-bp deletion. Three M residues in this reading frame are highlighted in turquoise. Note that translational initiation at these methionines in the NΔ HRD allele leads to a transition to translation from the correct ORF, whereas translation initiated at the “correct” methionine in the mutant sequence (amino acids highlighted in brown) leads to termination at a stop codon located at nucleotides 148–150 (not shown). A polypeptide (MENMQLR) identified by mass spectroscopy in a tryptic digest of the protein expressed from the mutant HRD allele (see Fig. 6) is boxed. (B) Translational analysis of the NΔ HRD allele by site-directed mutagenesis. The mutated forms shown (corresponding triplets altered to alanine or termination codons are highlighted in A in pink and blue, respectively) were expressed as 35S-labeled proteins by coupled transcription/translation in rabbit reticulocyte lysate and the reaction products detected by autoradiography following resolution by SDS/PAGE.
Cryptic Initiation Occurs at Multiple AUG Codons in Vivo.
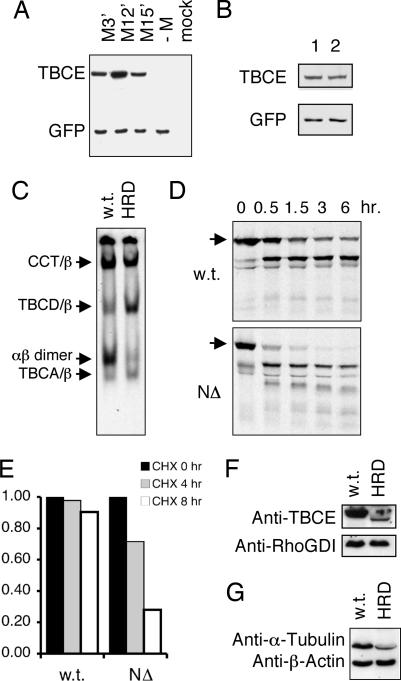
To determine the relative contribution of each of these codons to the translation of TBCE in vivo, we transfected cultured cells with constructs engineered so that only a single AUG triplet in the −2 reading frame remained as a potential initiation codon in each case. We then measured the resulting levels of TBCE expression by Western blotting (Fig. 4A). CHO cells were selected for this experiment, because the anti-human TBCE antibody does not crossreact with the corresponding Chinese hamster protein (see mock-transfected control). Although none of the nucleotide sequences flanking the three cryptic initiation codons conform to the suggested canonical motifs for translational initiation (18), we found that all three AUG triplets at nucleotides 23–25, 50–52, and 59–61 were capable of initiation, with the highest level of expression apparently occurring by the AUG at nucleotides 50–52. Furthermore, transfection of these constructs into HeLa cells showed that overexpression of any of the three cryptically initiated proteins was capable of causing microtubule destruction (data not shown). These observations are consistent with our in vitro translation data and with our sequence analysis of the tryptic N-terminal peptide derived from the recombinant protein expressed from the NΔ HRD allele.
Fig. 4.
Expression and stability of wild-type and HRD mutant forms of TBCE. (A) A cloned full-length TBCE cDNA sequence containing the NΔ deletion was altered by site-directed mutagenesis to change pairs of methionine codons (shown in turquoise at positions 3′, 12′, and 15′ in Fig. 3A) upstream of the genetic lesion to alanine codons (GCG), thus leaving only a single AUG triplet as a potential initiation codon. A construct (−M), in which all three AUG codons were altered to GCG, was also generated as a control. Each plasmid was used to transfect CHO K1 cells together with a plasmid engineered for the expression of GFP as a marker for transfection efficiency. Twenty-four hours posttransfection, cell extracts were prepared, and aliquots containing equal amounts of total protein were analyzed by Western blotting. (B) Analysis by Western blotting of CHO cells transfected with constructs with the NΔ deletion containing either an intact (lane 1) or an altered (ATG→TAG; lane 2) authentic initiation codon. (Lower) Shown is detection of cotransfected GFP as a control for transfection efficiency. (C) In vitro chaperonin-mediated β-tubulin-folding assays done with purified CCT, TBCC, and TBCD and supplemented with soluble extracts from wild-type or compound heterozygous HRD patient cells. Reaction products were resolved by nondenaturing PAGE and detected by autoradiography. Arrows (top to bottom) denote the location of the CCT/β-tubulin binary complex, the β-tubulin/TBCD complex, native tubulin heterodimers, and the complex formed between CCT-discharged β-tubulin-folding intermediates and TBCA contained in the cell extracts (5, 6). (D) Sensitivity to proteolysis of wild-type (w.t.) and HRD mutant (NΔ) forms of TBCE. Proteins were 35S-labeled by coupled transcription/translation and incubated with a soluble extract prepared from HRD patient-derived fibroblasts for the times shown. The reaction products were analyzed by SDS/PAGE. Arrows denote location of intact proteins. (E) Half life of wild-type and cryptically initiated TBCE in vivo. HEK293 cells transfected with constructs engineered for the expression of wild-type and cryptically initiated (NΔ) TBCE were treated with cycloheximide and their TBCE content measured by Western blotting at the times shown. (F) Semiquantitative Western blot analysis of the tubulin content in normal and compound heterozygous HRD patient-derived fibroblasts. The blot is identical to that shown in Fig. 1C. Loading controls (done with an anti-β-actin antibody) are shown below. (G) Expression levels of TBCE in normal (w.t.) and compound heterozygous HRD patient cells determined by using an anti-TBCE antibody affinity-purified by using the cryptically initiated HRD NΔ protein expressed in E. coli as the selecting antigen. Use of this antibody avoids bias in favor of the wild-type protein as a result of potential recognition of epitopes in the wild-type protein that lie N-terminal to the genetic lesion. Loading controls (using an anti RhoGDI antibody) are shown below.
To see whether ribosomes initiating at the authentic (M1) initiation codon might interfere with initiation at the three AUG codons in the −2 reading frame, we compared the level of expression of TBCE in transfected CHO cells using NΔ constructs containing either an intact or an altered authentic initiation codon, We found that changing M1 to a termination codon had little, if any, effect on the expression of cryptically initiated TBCE (Fig. 4B). Thus, the low level of expression of TBCE in compound heterozygous HRD cannot be ascribed to ribosomes initiating at M1 and terminating downstream of M15′.
We also determined the activity of TBCE in compound heterozygous HRD patient cells. In these experiments, we did in vitro CCT-mediated β-tubulin-folding assays containing purified TBCC and TBCD and then added soluble extracts from primary patient-derived fibroblasts as a source of TBCE activity. We found that extracts from control wild-type cells were able to supply TBCE activity to produce de novo assembled tubulin heterodimers, whereas identically prepared extracts of compound heterozygous HRD patient cells had a greatly reduced capacity to do so (Fig. 4C). This result suggested that the relatively low level of TBCE protein expressed by cryptic initiation from the NΔ HRD allele might be insufficient to drive the in vitro folding reaction efficiently, or that the mutant protein(s) might be unstable.
Cryptically Initiated TBCE Is Vulnerable to Proteolysis, Is Expressed in Patient Cells at Greatly Reduced Abundance, and Leads to a Reduction in Tubulin Levels.
To address the issue of the stability of cryptically initiated TBCE, we labeled the mutant protein by coupled in vitro transcription/translation and incubated it with an unfractionated extract derived from fibroblast cells. We found that the 35S-radiolabeled protein expressed from the NΔ HRD deletion allele was much more sensitive to proteolysis than the corresponding wild-type control (Fig. 4D). Similar data were obtained by using proteinase K as an exogenous source of protease (Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, measurement of the half-life of wild-type and cryptically initiated TBCE in vivo showed a significant reduction (compared with wild type) in the stability of cryptically initiated polypeptides (Fig. 4E). The instability of the mutant protein is presumably a consequence of structural disruption caused by substitution of the authentic N-terminal sequence by “nonsense” polypeptides. We also measured the level of TBCE in patient cells using an anti-TBCE antibody affinity purified to avoid bias that might be conferred by recognition of epitopes in the wild-type protein that lie N-terminal to the genetic lesion. The result shows a level of TBCE in disease cells that is reduced by a factor of ≈10 compared with wild-type cells (Fig. 4F). This is explained by both the relative inefficiency of cryptic out-of-frame translational initiation at AUG codons lacking favorable flanking nucleotides (Fig. 3A), as well as the enhanced proteolytic sensitivity of the HRD mutant proteins.
Because TBCE is an essential component of the pathway leading to de novo tubulin heterodimer formation (6, 9), we reasoned that the dramatically reduced level of functional TBCE in compound heterozygous HRD cells might lead to a corresponding reduction in cellular tubulin levels. We therefore measured the overall tubulin level in patient cells by Western blotting and found a substantial reduction relative to normal control cells (Fig. 4G). Presumably, the modest amount of cryptically initiated TBCE protein in disease cells is sufficient to support a minimal requirement for de novo tubulin heterodimer assembly in vivo but inadequate to support this reaction efficiently in in vitro experiments (Fig. 4C) because of the inevitable dilution that accompanies the preparation of cell extracts.
TBCE as an Essential Protein: A Paradox Resolved.
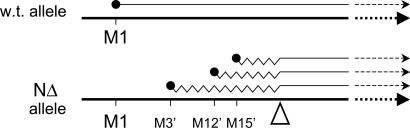
Genetic experiments in model organisms (i.e., S. pombe and A. thaliana) have demonstrated that expression of TBCE is essential either for life (10, 11) or for early embryonic development (12). Despite an extensive search using in vitro tubulin-folding assays done in the absence of TBCE, no alternative activity has been found that could contribute to de novo tubulin assembly by circumventing the need for TBCE. The existence of viable individuals who harbor mutated alleles, each of which apparently lacks the capacity to make a functional TBCE protein, therefore seemed paradoxical. This paradox is resolved by our discovery that the allele with the HRD deletion of nucleotides 66–67 can, in fact, express cryptically initiated TBCE polypeptides that are functional in terms of their ability to support de novo tubulin-folding and tubulin GAP activity (Fig. 5). Taken together, our data imply that a restricted availability of TBCE in these mutant cells leads to a limiting supply of tubulin heterodimers, resulting in a critical lowering of microtubule density that could contribute to disease pathogenesis.
Fig. 5.
Schematic illustration (not drawn to scale) of cryptic out-of-frame translational initiation of mutant (deletion of nucleotides 66–67) TBCE. M1, authentic initiator methionine codon leading to translation of the wild-type sequence. M3′, M12′, and M15′ denote three AUG codons in the −2 reading frame (see Fig. 3A), each of which serves to initiate translation of the mutant allele. Note the transition of all three out-of-frame nonsense translation products (shown as jagged lines) to the correct ORF at the point of mutation (marked by a triangle). Initiation at the authentic M1 AUG codon in the mutant allele leads to translation (not shown) of a peptide consisting of the first 22 aa of TBCE fused to 26 nonsense amino acids translated from the −1 reading frame downstream of the mutation.
Experimental Procedures
Protein Expression and Purification.
Variant forms of human TBCE (5) were generated by site-directed mutagenesis by using a QuikChange II kit (Stratagene, La Jolla, CA) and checked by DNA sequencing. These sequences and a wild-type TBCE sequence were expressed in insect sf9 cells by using the BacPAK baculovirus expression system (Clontech, Mountain View, CA). Infected cells were harvested at 50 h postinfection, washed in ice-cold buffer (10 mM Tris·HCl, pH 8.0/10 mM KCl/1 mM MgCl2/1 mM EGTA/1 mM 2-mercaptoethanol), and resuspended in 10 volumes of this buffer containing a mixture of protease inhibitors (Roche, Basel, Switzerland). The cells were lysed in a glass Dounce homogenizer and a particle-free supernatant prepared by two successive centrifugation steps at 5,000 × g and 200,000 × g (15 min each). Supernatants were fractionated by FPLC on a 10/10 QHP anion exchange column (Amersham Pharmacia, Piscataway, NJ) that was developed with a linear gradient (60 ml) of the same buffer containing 0.25 M NaCl. Fractions containing the recombinant protein were identified either by their immunoreactivity with an anti-TBCE antibody or by SDS/PAGE. These were pooled, concentrated by ultrafiltration using a Centricon 30 device (Millipore, Billerica, MA), exchanged into 10 mM NaPO4 buffer, pH 7.4, containing 1 mM 2-mercaptoethanol by passage over a column (PD10, Amersham Pharmacia) of Sephadex G25, and applied to a 5/5 column of ceramic hydroxylapatite (American Chemical, Natick, MA). This column was washed with 10 mM NaPO4 buffer, pH 7.4, and developed with a linear gradient (30 ml) containing 0.25 M NaPO4 buffer, pH 7.4. Wild-type TBCE or its mutant homologues emerged from this column as distinct absorbance peaks at a buffer conductivity in the range 11–12 mS per centimeter. This material was pooled, concentrated by ultrafiltration, and applied to a Superdex 75 column (Amersham Pharmacia) run in 10 mM Tris·HCl, pH 7.5/0.15 M NaCl/1 mM EDTA/1 mM 2-mercaptoethanol. The purified proteins emerged as symmetrical peaks. The cytosolic chaperonin CCT and cofactors C (TBCC) and D (TBCD) were prepared as described (5, 6, 17).
In Vitro Folding and Tubulin GAP Assays.
CCT-mediated in vitro folding assays containing purified TBCC, TBCD, and TBCE were done by using a bacterially expressed, urea-unfolded [35S]methionine-labeled β-tubulin probe and the reaction products analyzed by electrophoresis on native polyacrylamide gels as described (5, 6). In some experiments (shown in Fig. 4C), unfractionated soluble extracts of wild-type or HRD patient-derived fibroblasts containing 20 μg of total protein were substituted for purified TBCE. These extracts were prepared by homogenization of harvested cultured primary fibroblasts (grown in DMEM supplemented with 10% FBS) in ice-cold 10 mM Tris·HCl, pH 8.0/1 mM EDTA containing a mixture of protease inhibitors (Roche) and centrifugation at either 2,000 × g for 5 min (for the preparation of postnuclear extracts) or 100,000 × g for 10 min (for the preparation of a cleared lysate for in vitro folding reactions). The total protein concentration in these extracts was adjusted to 2.5 mg/ml. Tubulin GAP activity was measured by using purified TBCC, TBCD, and recombinant wild-type or HRD mutant forms of TBCE by the method described (13).
Transcription/Translation and Protein Stability Assays.
Coupled transcription/translation reactions were done in rabbit reticulocyte lysate (TNT; Promega, Madison, WI) containing [35S]methionine [specific activity, 1,000 Ci/mmol; 10 μCi per μl (1 Ci = 37 GBq)] by using either a full-length cDNA encoding human TBCE (5) or corresponding sequences harboring various HRD mutations (see text) cloned in the pET23b vector at the NdeI and XhoI sites. Reaction products were detected by autoradiography after resolution on 8.5% SDS/PAGE. In experiments to determine protein stability, products of translation reactions (20 μl) were exchanged into buffer (10 mM NaP04, pH 7.4/1.0 mM MgCl2) by passage through a microcolumn (3.0 × 0.4 cm) of Sephadex G25. Labeled proteins emerging in the void volume were incubated at 30°C with either proteinase K (60 nM) or extracts from HRD patient-derived fibroblasts. These extracts were prepared as described above but without the 100,000 × g centrifugation step; EDTA and the protease inhibitor mixture were also omitted from the buffer. At various intervals, aliquots were withdrawn and (in the case of experiments done with proteinase K), the proteolytic reaction quenched by the addition of PMSF to 5 mM. Reaction products were analyzed by resolution on 10% SDS/PAGE. For determination of protein stability in vivo, HEK293 cells were treated 48 h posttransfection with 1 μg/ml of cycloheximide.
MS.
Wild-type and mutant TBCE protein (the latter expressed from the NΔ HRD allele) were excised as Coomassie blue-stained bands from SDS/PAGE and reduced, alkylated, and digested with trypsin in a ProGest workstation (Genomic Solutions, Cambridgeshire, U.K.). Reaction products were analyzed by nano liquid chromatography tandem MS (MS/MS) on a Micromass (Manchester, U.K.) Q-TOF 2. De novo sequencing of selected peptides was done by using PEAKS software (Bioinformatic Solutions, Waterloo, ON, Canada) followed by manual validation.
Transfection Experiments and Immunofluorescence Microscopy.
A full-length cDNA encoding human TBCE (5) and corresponding sequences harboring various HRD mutations contained in pET vectors were subcloned (using the flanking XbaI and XhoI sites) into the pcDNA3.1(+) vector. HeLa cells or CHO K1 cells grown in DMEM supplemented with 10% FBS on glass cover slips were transfected with these constructs using the FuGENE6 transfection reagent (Roche). In some experiments, a construct engineered for the expression of GFP was cotransfected at one-tenth the total DNA amount as a marker for transfection efficiency. Cells were fixed with 4% paraformaldehyde 24 h posttransfection and analyzed by indirect immunofluorescence using a mouse monoclonal anti-α-tubulin antibody and a rabbit polyclonal antibody raised against human TBCE (5).
Real-Time PCR.
Total RNA was prepared from wild-type or HRD patient-derived fibroblasts by using an Aurum Total RNA Mini Kit (BioRad, Hercules, CA) according to the manufacturer’s instructions. Reverse transcription was done by using 0.1 μg of RNA and the resulting cDNA subjected to real-time PCR analysis with gene-specific primer pairs using the SuperScript III Platinum SYBR Green Two-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Relative quantification was assessed by using β-actin as the internal control. The following primers were used in the amplification reactions: TBCE: 5′-GCTTTTGGAAATGAGTGGA-3′ (sense), 5′-GGAACTGGTATCTGGGATGG-3′ (antisense); β-actin: 5′-AAGGATTCCTATGTGGGCGA-3′ (sense), and 5′-TCCATGTCGTCCCAGTTGGT-3′ (antisense).
Antibodies and Western Blotting.
A rabbit polyclonal anti-human TBCE antibody (5) was affinity-purified by using selecting antigen derived from inclusion bodies (purified from Escherichia coli BL21 DE3 cells expressing either wild-type or the HRD del66–67 mutant form of TBCE, as described (19). Protein contained in the inclusion bodies was resolved on 8% SDS/PAGE and the gel content transferred to nitrocellulose and blocked by incubation in 3% BSA in TBST. The band containing TBCE was excised, cut into small segments, and incubated for 1 h at 32°C with 1.5 ml of crude antiserum. The nitrocellulose was washed successively with 5 ml of the following buffers: PBS (twice), 0.1 M NaHCO3, 0.1 M NaHCO3 containing 0.5 M NaCl. Selected antibody was eluted by incubation with 1.5 ml of 0.1 M Tris-glycine, pH 2.6, for 5 min and the solution brought to pH 7.5 by addition of Tris base. Extracts of transfected cells or of wild-type or HRD patient-derived primary fibroblasts were prepared as described above for in vitro folding and tubulin GAP assays. Aliquots containing 20 μg of total protein were analyzed by Western blotting by using one or more of the following antisera: affinity-purified anti-TBCE; a mouse monoclonal anti-α-tubulin antibody (T5168; Sigma-Aldrich, St. Louis, MO); a rabbit polyclonal anti-GFP antibody (A6455; Molecular Probes, Carlsbad, CA); a rabbit polyclonal anti-RhoGDI antibody (A-20; Santa Cruz Biotechnology, Santa Cruz, CA); and a mouse monoclonal anti-β-actin antibody (A5541; Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank R. J. Schneider for stimulating discussions and D. L. Allen and R. Amunugama of Proteomic Research Services, Inc., for help with MS. This work was supported by National Institutes of Health Grant R01 DK47234 (to N.J.C.).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chen X., Sullivan D. S., Huffaker T. C. Proc. Natl. Acad. Sci. USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinh D. B., Drubin D. G. Proc. Natl. Acad. Sci. USA. 1994;91:9116–9120. doi: 10.1073/pnas.91.19.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis S. A., Tian G., Vainberg I. E., Cowan N. J. J. Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian G., Vainberg I. E., Tap W. D., Lewis S. A., Cowan N. J. J. Biol. Chem. 1995;270:23910–23913. doi: 10.1074/jbc.270.41.23910. [DOI] [PubMed] [Google Scholar]
- 5.Tian G., Huang Y., Rommelaere H., Vandekerckhove J., Ampe C., Cowan N. J. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 6.Tian G., Lewis S. A., Feierbach B., Stearns T., Rommelaere H., Ampe C., Cowan N. J. J. Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhamidipati A., Lewis S. A., Cowan N. J. J. Cell Biol. 2000;149:1087–1096. doi: 10.1083/jcb.149.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski D. Curr. Biol. 2002;12:R767–R769. doi: 10.1016/s0960-9822(02)01288-5. [DOI] [PubMed] [Google Scholar]
- 9.Cowan N. J., Lewis S. A. Adv. Protein Chem. 2002;59:73–104. doi: 10.1016/s0065-3233(01)59003-8. [DOI] [PubMed] [Google Scholar]
- 10.Radcliffe P. A., Hirata D., Vardy L., Toda T. Mol. Biol. Cell. 1999;10:2987–3001. doi: 10.1091/mbc.10.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grishchuk E. L., McIntosh J. R. J. Cell Sci. 1999;112:1979–1988. doi: 10.1242/jcs.112.12.1979. [DOI] [PubMed] [Google Scholar]
- 12.Steinborn K., Maulbetsch C., Priester B., Trautmann S., Pacher T., Geiges B., Kuttner F., Lepiniec L., Stierhof Y. D., Schwarz H., et al. Genes Dev. 2002;16:959–971. doi: 10.1101/gad.221702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian G., Bhamidipati A., Cowan N. J., Lewis S. A. J. Biol. Chem. 1999;274:24054–24058. doi: 10.1074/jbc.274.34.24054. [DOI] [PubMed] [Google Scholar]
- 14.Hershkovitz E., Parvari R., Diaz G. A., Gorodischer R. J. Pediatr. Endocrinol. Metab. 2004;17:1583–1590. doi: 10.1515/jpem.2004.17.12.1583. [DOI] [PubMed] [Google Scholar]
- 15.Parvari R., Hershkovitz E., Grossman N., Gorodischer R., Loeys B., Zecic A., Mortier G., Gregory S., Sharony R., Kambouris M., et al. Nat. Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 16.Pierre P., Pepperkok R., Kreis T. E. J. Cell Sci. 1994;107:1909–1920. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- 17.Maser R. S., Zinkel R., Petrini J. H. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y., Thomas J. O., Chow R. L., Lee G. H., Cowan N. J. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.