Abstract
Understanding the mechanisms of Salmonella virulence is an important challenge. The capacity of this intracellular bacterial pathogen to cause diseases depends on the expression of virulence factors including the second type III secretion system (TTSS-2), which is used to translocate into the eukaryotic cytosol a set of effector proteins that divert the biology of the host cell and shape the bacterial replicative niche. Yet little is known about the eukaryotic functions affected by individual Salmonella effectors. Here we report that the TTSS-2 effector PipB2 interacts with the kinesin light chain, a subunit of the kinesin-1 motor complex that drives anterograde transport along microtubules. Translocation of PipB2 is both necessary and sufficient for the recruitment of kinesin-1 to the membrane of the Salmonella-containing vacuole. In vivo, PipB2 contributes to the attenuation of Salmonella mutant strains in mice. Taken together, our data indicate that the TTSS-2-mediated fine-tuning of kinesin-1 activity associated with the bacterial vacuole is crucial for the virulence of Salmonella.
Keywords: SifA, molecular motor
The virulence of intracellular pathogens relies on their capacity to survive inside host cells. Phagocytic compartments in which pathogens are engulfed are poor in nutrients, are acidic, and contain degradative agents, thus confronting the pathogen with the problem of surviving in a hostile environment. Salmonella enterica circumvents these difficulties by secreting effector proteins that alter the host cell and turn the phagocytic compartment into a replicative niche. After entry into eukaryotic cells Salmonella resides in a membranous compartment named the Salmonella-containing vacuole (SCV) (1). Replication in the SCV is associated with expression of the second type III secretion system (TTSS-2) (2, 3), a sophisticated needle-like structure that allows the translocation of >15 effectors from the bacteria into the cytosol of infected host cells. These effectors facilitate the establishment of the intracellular replication niche. Their importance has been established by using Salmonella strains expressing a nonfunctional TTSS-2 and thereby unable to translocate any TTSS-2-dependent effectors. TTSS-2-null mutants are defective for replication in tissue culture cells and have a severe attenuation of virulence in vivo (4–6). TTSS-2 effector functions are required for many aspects of the maturation of the SCV including alteration of the cholesterol content (7) and the integrity of the SCV membrane (8), evasion of NADPH oxidase-dependent killing (9), and the formation of an actin nest (10). The establishment and maintenance of the SCV are also highly dependent on microtubules and microtubule motors (reviewed in ref. 11). We recently showed that the TTSS-2 effector SifA is involved in the regulation of kinesin-1 activity on the SCV. Indeed, through its binding to the host protein SKIP, SifA down-regulates the recruitment of kinesin-1 to the SCV. Thus, a sifA− mutant recruits high levels of this molecular motor whereas kinesin-1 is barely detected on the SCV of a wild-type Salmonella (12). Yet the absence of SifA per se is not sufficient to recruit this motor because kinesin-1 is not detected on the SCV of a TTSS-2-null mutant. Therefore, we have proposed that another TTSS-2 effector is necessary to link kinesin-1 onto the SCV membrane (12).
Results
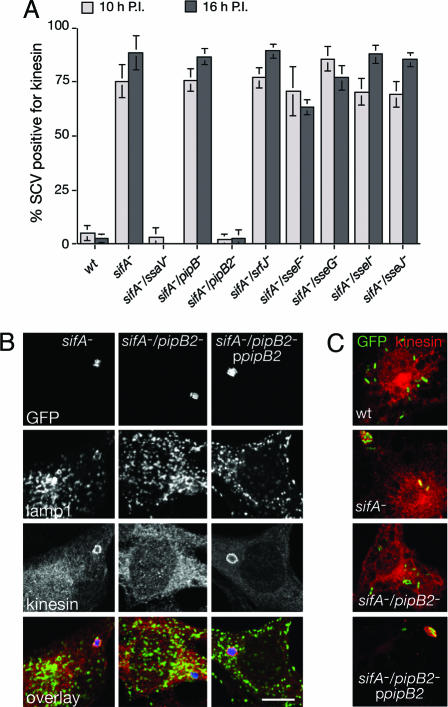
Here we confirmed this hypothesis using a double sifA−/ssaV− mutant, which is defective for the translocation of all TTSS-2 effectors (Fig. 1A). To investigate whether any single effector was required for kinesin-1 recruitment onto the SCV we individually tested the effect of their deletion in a sifA− strain. Most double mutants behaved similarly to the sifA− mutant with respect to kinesin-1 recruitment onto the SCV in HeLa cells (Fig. 1A). However, deletion of pipB2 in a sifA− background completely suppressed kinesin accumulation on the SCV (Fig. 1 A and B). This result, observed with Salmonella strain 12023 and confirmed in a SL1344 background (data not shown), is highly specific because deletion of pipB, which encodes a homologue of PipB2 (13), had no effect (Fig. 1A). Recruitment of kinesin-1 onto the sifA−/pipB2− mutant SCV could be rescued by complementing the mutant with a plasmid encoding PipB2 (Fig. 1 B and C). A similar PipB2-dependent phenotype was also observed in mouse macrophages (Fig. 1C), which are considered an important replicative niche for Salmonella in vivo (14, 15). As previously described (12), no specific enrichment of kinesin-1 on SCVs containing wild-type bacteria was observed (Fig. 1 A and C). Accordingly, no difference in the kinesin-1 labeling of the SCV was observed for a wild-type strain and a pipB2− mutant (data not shown). Together, these results clearly demonstrate that PipB2 is necessary for recruitment of kinesin-1 onto sifA− mutant vacuoles.
Fig. 1.
PipB2 is required for kinesin-1 accumulation on sifA− mutant SCVs. (A) SCVs containing the sifA−/pipB2− mutant do not accumulate kinesin during infection. HeLa cells were infected with various GFP-expressing Salmonella strains. The percentage of kinesin-positive SCVs was scored at 10 and 16 h postinvasion (P.I.) after immunostaining for Lamp-1 as an SCV membrane marker and for kinesin heavy chain (HC). (B and C) In trans complementation of a sifA−/pipB2− mutant with ppipB2 restores kinesin accumulation onto the SCV in both HeLa cells (B) and bone marrow-derived macrophages (C). HeLa cells were infected for 10 h with GFP-expressing sifA−, sifA−/pipB2−, and sifA−/pipB2−ppipB2 mutant strains (blue), fixed, and immunostained for Lamp-1 (green) and kinesin HC (red). Bone marrow-derived macrophages were infected with the same strains and wild-type Salmonella and immunostained for kinesin and Lamp-1. Salmonella (green) and kinesin (red) are shown. (Scale bar: 10 μm.)
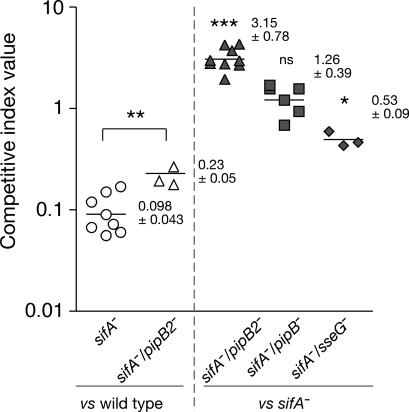
Deletion of sifA leads to several diverse phenotypes, including redistribution of SCVs from a juxtanuclear position to the cell periphery (12), loss of vacuole integrity (8), a replication defect in macrophages (8, 16), and a marked attenuation of virulence in mice (17). The first two phenotypes can be partially rescued by inhibiting host kinesin activity (12, 18), suggesting that the accumulation of this microtubule motor onto the sifA− SCV was responsible for its loss of positioning and membrane integrity. Because deletion of pipB2 suppressed the kinesin-1 accumulation on sifA− SCV, we tested whether it could also complement the other sifA− phenotypes. However, although the suppressive action of PipB2 in regard to kinesin-1 recruitment was very clear, no rescue in the maintenance of the sifA− vacuolar membrane was observed in HeLa cells, which is in agreement with previous data (13), and no difference in SCV positioning in HeLa cells or replication in RAW-264.7 macrophage-like cells was detected (Fig. 5, which is published as supporting information on the PNAS web site). We next tested whether deletion of pipB2 could rescue the virulence of the sifA− mutant in vivo. To address this issue we determined the competitive index (CI) of various strains during mixed infections in mice, a very sensitive technique to compare the virulence of different strains (19). A pipB2− mutant has been shown previously to have a slight attenuation compared with the wild-type strain (13), whereas a deletion of sifA strongly affects Salmonella virulence (17). Interestingly, a sifA−/pipB2− mutant was significantly less attenuated than the sifA− mutant when compared with the wild-type strain (P = 0.006) (Fig. 2). This result was confirmed by a direct comparison of the single sifA− and the double sifA−/pipB2− mutant strains. We found that deletion of pipB2 on a sifA− background significantly increased Salmonella virulence (P < 0.001). This shift is highly specific to pipB2 because deletion of genes encoding for the PipB2 homolog PipB or another TTSS-2 effector, SseG, in the sifA− background either did not change the virulence or rendered the strain even more attenuated (Fig. 2). Thus, although we were unable to detect a strong rescue of the sifA− mutant phenotypes in tissue culture cells by deleting pipB2, these in vivo results show that loss of PipB2 compensates in part for the virulence defect of a sifA− mutant. This finding demonstrates that PipB2-associated functions are contributing to attenuation of the sifA− mutant in mice.
Fig. 2.
PipB2 is partially responsible for the attenuation of the sifA− mutant in vivo. Mice were inoculated i.p. with a mixture of two strains. At day 2 after injection, spleens were harvested for bacterial counts. CIs of sifA− (circles), sifA−/pipB2− (triangles), sifA−/pipB− (squares), and sifA−/sseG− (diamonds) against the wild-type 12023 strain (open symbols) or the sifA− mutant (gray symbols) were determined. Each symbol represents the CI from one mouse, and horizontal bars correspond to the geometric mean of all of the mice. The CIs of sifA− vs. wild type (n = 9) and sifA−/pipB2− vs. wild type (n = 3) are very significantly different (∗∗, P = 0.0063). The CI for sifA−/pipB2− vs. sifA− (n = 9) is extremely significantly greater than 1 (∗∗∗, P < 0.0001), whereas the CI for sifA−/pipB− vs. sifA− (n = 6) is not significantly (ns) different from 1 (P = 0.15). In contrast, the CI for sifA−/sseG− vs. sifA− (n = 3) is significantly lower than 1 (∗, P = 0.0095).
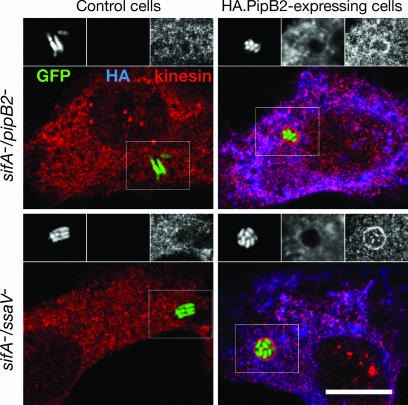
To further dissect PipB2 function we took advantage of the fact that, because Salmonella effectors are translocated into the host cell cytosol, ectopic expression of tagged effectors from plasmids transfected into the host cell can complement the respective bacterial mutants (8, 20). In HeLa cells expression of HA.PipB2 supported recruitment of kinesin-1 onto the sifA−/pipB2− mutant SCV even though the tagged effector itself could barely be detected on the SCV (Fig. 3). Although this experiment clearly confirmed that PipB2 was necessary for kinesin-1 recruitment to the SCV, it did not reveal whether PipB2 is sufficient because recruitment could result from the cooperative action of several bacterial effectors. To discriminate between these possibilities we infected HeLa cells with a sifA−/ssaV− mutant, which is defective for translocation of TTSS-2 effectors. In untransfected cells the SCVs containing this mutant did not show any specific kinesin-1 labeling. In contrast, ectopic expression of HA.PipB2 led to a strong accumulation of kinesin-1 onto the sifA−/ssaV− SCV (Fig. 3), unequivocally demonstrating that PipB2 acts independent of other TTSS-2 effectors.
Fig. 3.
PipB2 triggers kinesin accumulation onto the SCV independent of other TTSS-2 effectors. HeLa cells were infected with GFP-expressing sifA−/pipB2− or sifA−/ssaV− strains (green) and either untreated (control cells) or transiently transfected at 2 h after invasion with a plasmid encoding HA.PipB2. At 16 h after invasion (14 h after transfection) cells were fixed and immunostained for HA tag (blue) and kinesin HC (red). Grayscale images represent GFP, HA, and kinesin HC labeling of Insets. (Scale bar: 10 μm.)
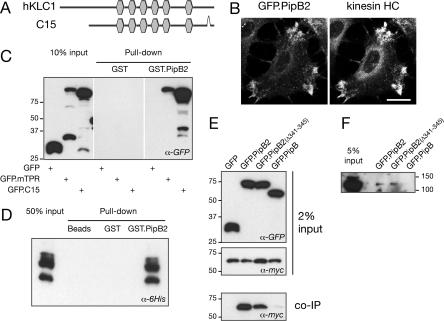
Although the above results show that cytosolic PipB2 is necessary and sufficient to trigger accumulation of kinesin-1 onto the SCV, the mechanisms and the host proteins involved were unknown. To gain insights into the molecular interactions underlying kinesin-1 recruitment we performed a yeast two-hybrid screen of a human fetal brain cDNA library using PipB2 as a bait. Two independent cDNA clones were isolated. C7 encoded a Golgi integral membrane protein for which the specificity of the interaction with PipB2 could not be confirmed biochemically, and C15 encoded an isoform of the kinesin light chain 1 (human KLC). C15 corresponds to the KLC1B splice form of the KNS2 gene (National Center for Biotechnology Information accession no. DAA01292) (21), missing the 94 N-terminal amino acid residues but containing the six tetratricopeptide repeat (TPR) motifs known to be important for binding cargoes (Fig. 4A). This finding suggested that PipB2 recruits kinesin-1 by directly binding the KLC. The specificity of the PipB2–KLC1B interaction was verified by immunofluorescence microscopy and biochemically. It has previously been shown that ectopically expressed PipB2 accumulates at the cell periphery at sites of membrane projections in HeLa cells (22, 23). Upon transfection of HeLa cells with a plasmid expressing GFP.PipB2 we saw a clear redistribution of endogenous kinesin-1 toward peripheral GFP.PipB2-positive structures (Fig. 4B), although for unknown reasons this phenotype was observed in only a small percentage of transfected cells. However, a complete colocalization of myc.PipB2 and GFP.C15 was observed upon coexpression in HeLa cells (data not shown). We next assessed the ability of PipB2 to pull down the C15 protein and the TPR domain of the mouse KLC2 (mTPR). Purified GST.PipB2 specifically pulled down both the GFP.C15 and the GFP.mTPR from transfected HeLa cell extracts (Fig. 4C). The direct interaction between human KLC and PipB2 was further confirmed in vitro by using purified GST-PipB2 and His-6.C15 (Fig. 4D). When coexpressed in HeLa cells, GFP.PipB2, but not GFP nor GFP.PipB, coimmunoprecipitated myc.C15 (Fig. 4E). Finally, endogenous kinesin heavy chain (HC) was specifically coimmunoprecipitated with PipB2 expressed in HeLa cells (Fig. 4F). Altogether these results demonstrate the relevance of the interaction between PipB2 and kinesin-1 in eukaryotic cells.
Fig. 4.
PipB2 recruits kinesin-1 by a direct interaction with the KLC. (A) Schematic representation of clone C15 isolated in the yeast two-hybrid screen compared with human KLC (hKLC1) (National Center for Biotechnology Information accession no.AAH08881). The C15 protein has the six TRP motifs (hexagons) and a C-terminal deletion of nine amino acid residues. (B) Endogenous kinesin-1 is relocalized to PipB2-positive structures at the cell periphery. HeLa cells expressing GFP.PipB2 were immunostained with an antibody against kinesin HC. (Scale bar: 20 μm.) (C and D) GST.PipB2 pulls down KLC. Extracts of HeLa cells expressing GFP, GFP.mTPR, or GFP.C15 (C) or His-6.mTPR purified from Escherichia coli (D) were incubated with GST or GST.PipB2 proteins immobilized on beads. Bound proteins were analyzed by Western blotting with an anti-GFP (C) or an anti-His6 (D) antibody. GST.PipB2, but not GST alone, pulled down both murine and human KLC. (E) PipB2 immunoprecipitates KLC. HeLa cells were cotransfected with plasmids encoding for myc.C15 and GFP, GFP.PipB2, GFP.PipB2(Δ341–345), or GFP.PipB. Cell lysates were incubated with a rabbit anti-GFP antibody and protein A beads. Coimmunoprecipitated proteins were analyzed by Western blotting with an anti-myc antibody. (F) PipB2 immunoprecipitates endogenous kinesin-1. Lysates of HeLa cells expressing GFP.PipB2, GFP.PipB2(Δ341–345), or GFP.PipB were incubated with a mouse anti-GFP antibody and protein G beads. Coimmunoprecipitated proteins were analyzed by Western blotting with the rabbit anti-kinesin HC antibody PCP42.
PipB2 has recently been shown to trigger the redistribution of late endocytic compartments to the cell periphery in a microtubule-dependent manner (23). This redistribution has been observed for both ectopically expressed and bacterially delivered PipB2. This activity is abolished by deletion of a C-terminal pentapeptide of PipB2 (residues 341–345) (23). Because this relocalization is suggestive of the action of a plus-end-directed molecular motor such as kinesin-1, we tested whether the C-terminal pentapeptide of PipB2 is also required for kinesin-1 recruitment onto the SCV. HeLa cells were infected with sifA−/pipB2− mutants expressing 2HA-tagged versions of PipB, PipB2, or PipB2(Δ341–345). All three proteins localized to the SCV membranes (Fig. 6A, which is published as supporting information on the PNAS web site). Whereas PipB expression did not restore kinesin-1 recruitment onto the SCV, PipB2(Δ341–345) recruited kinesin-1, although less efficiently than full-length PipB2 (Fig. 6A). Quantitative analysis of confocal images (described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site) demonstrated a linear correlation between the presence of translocated PipB2.2HA on the SCV and the recruitment of kinesin-1 (Fig. 6B). Although a similar correlation was observed with PipB2(Δ341–345).2HA, the threshold of HA signal necessary to detect kinesin-1 enrichment onto the SCV was higher. This finding demonstrates a requirement for the C-terminal pentapeptide motif of PipB2 to efficiently recruit kinesin-1 and is consistent with the reduced amount of myc.C15 coimmunoprecipitated by GFP.PipB2(Δ341–345) compared with full-length GFP.PipB2 (Fig. 4E). Collectively, these results suggest that the ability of PipB2 to redistribute the late endocytic compartments at the cell periphery is a direct consequence of its efficient kinesin-1 binding activity.
Discussion
In this study we have unequivocally demonstrated that the TTSS-2 effector PipB2 is a kinesin-1 linker that is necessary and sufficient for the recruitment of this molecular motor onto sifA− mutant vacuoles. As SifA down-regulates kinesin-1 recruitment, PipB2 and SifA demonstrate antagonist activities in terms of regulating kinesin-1 recruitment on the SCV. However, the inhibitory activity of SifA appears to be dominant over the action of PipB2 during a Salmonella infection. Indeed, very few SCVs are positive for kinesin-1 during the course of a wild-type infection of tissue culture cells (12), and expression of HA.PipB2 from a transfected plasmid could hardly induce the recruitment of kinesin-1 on wild-type SCV (data not shown). Moreover, controlling kinesin-1 activity seems more important for Salmonella virulence than recruiting this motor, because a pipB2− mutant is only moderately attenuated in mice (13), whereas a sifA− mutant is more severely attenuated (17). Finally, the sifA−/pipB2− mutant is significantly more virulent that the sifA− mutant, thereby indicating that PipB2-dependent kinesin-1 activity plays an important role in the virulence attenuation of the sifA− mutant and that hyperactivity of kinesin-1 associated with the SCV is detrimental for Salmonella replication. Paradoxically, with the exception of kinesin-1 recruitment, other phenotypes associated with the sifA− mutant in tissue culture cells were not significantly affected by the deletion of pipB2−. Perhaps SifA regulates some low level of kinesin-1 recruited to the SCV independent of PipB2 or possesses another unidentified activity, the lack of which is responsible for SCV membrane instability and the atypical intracellular positioning. This activity could possibly be the regulation of another molecular motor such as dynein because inhibition of dynein activity has been shown to stabilize the sifA− SCV to the same extent as does inhibition of kinesin activity (18), and this retrograde molecular motor is recruited onto the SCV by binding to a rab7/RILP complex under certain circumstances (24, 25). SifA contains a pentapeptide motif that has the potential to mimic small GTPases in their active GTP-bound form (26) and thus could potentially modulate the rab7/RILP/dynein cascade (27). Finally, a double sifA−/sseJ− mutant does not lose its vacuole (28), suggesting that SifA and SseJ exert antagonist activities. Whether SseJ controls the activity of a molecular motor is not known. However, because SseJ is predicted to be an acyl transferase it is more likely to modify vacuolar membrane lipids, thereby facilitating membrane exchange leading to membrane disruption (29).
Our data show that TTSS-2 regulates a molecular motor activity by translocating effectors with opposite functions and that these molecules exert antagonistic roles in vivo. This fact raises questions about the temporal regulation of SifA and PipB2 activities. Whether these effectors act sequentially or coordinately is unknown. Kinesin-1 might be required at specific stages of the infection. However, this possibility appears unlikely because it is not possible to detect a window of time in which there is an increased recruitment of kinesin-1 on the SCV. Another possibility is that Salmonella constantly modulates kinesin-1 activity on the SCV to facilitate its needs. This finely tuned regulation of kinesin-1 recruitment could be achieved by regulating the translocation of these two effectors. Alternatively, an eukaryotic-based mechanism might control the intracellular levels of the two antagonistic effectors. Indeed, such a process has been demonstrated for the TTSS-1 effector proteins SopE and SptP, which exert antagonistic effects on Rho GTPase-induced actin cytoskeleton remodeling during the initial invasion process. These effectors are both delivered during the early infection phase, but SopE is rapidly degraded by a proteasome-mediated pathway whereas SptP exhibits a longer half-life (30). It remains to be shown whether either of these mechanisms accounts for the temporal regulation of PipB2 and SifA functions.
Our data demonstrate that tight control of kinesin-1 activity is important for establishment and maintenance of the Salmonella replicative niche. It is becoming increasingly clear that TTSS-2 effectors are involved in the regulation of other molecular motors (11, 31). A deeper understanding of how Salmonella regulates and utilizes these molecular motors to survive inside host cells will emerge from identifying the function and host cell targets of the entire set of TTSS-2 effector proteins.
Materials and Methods
Antibodies and Reagents.
The rabbit anti-kinesin HC (KIF5B) antibody PCP42 [generously provided by R. Vale (University of California, San Francisco, CA)] was absorbed on Salmonella acetone powder to remove contaminating antienterobacteria antibodies and used at a dilution of 10−3. The mouse monoclonal antibody against Lamp-1, H4A3 [developed by J. T. August and J. E. K. Hildreth (Johns Hopkins University School of Medicine, Baltimore, MD), obtained from the Developmental Studies Hybridoma Bank (Iowa City, IA), developed under the auspices of the National Institute of Child Health and Human Development, and maintained by the University of Iowa (Iowa City, IA)] was used at a dilution of 10−3. The mouse anti-myc 9E10, anti-HA (clone 16B12; Covance, Richmond, CA), and anti-GFP (clone JL-8; Clontech, Mountain View, CA) antibodies and the rat anti-HA (clone 3F10; Roche Molecular Biochemicals, Indianapolis, IN) antibodies were used at a dilution of 10−3. The rabbit anti-GFP was obtained from Molecular Probes (Eugene, OR). Goat anti-mouse and anti-rabbit coupled to peroxidase (Sigma, St. Louis, MO) were used at 10−4. Secondary antibodies [donkey anti-rabbit, anti-rat, or anti-mouse IgG conjugated to FITC, Texas red, or cyanine 5 (Cy5)] were purchased from Jackson ImmunoResearch (West Grove, PA) and used at a dilution of 5 × 10−3. The goat anti-rabbit or anti-mouse IgG conjugated to Alexa Fluor 350 were from Molecular Probes and were used at a dilution of 5 × 10−3.
Bacterial Strains, Plasmids, and Culture Conditions.
The S. enterica serovar Typhimurium strains and plasmids used in this study and their relevant characteristics are listed in Tables 1 and 2, which are published as supporting information on the PNAS web site. Strains were cultured routinely in LB broth (Difco, San Jose, CA) at 37°C. Ampicillin (50 μg·ml−1), kanamycin (50 μg·ml−1), tetracycline (15 μg·ml−1), and chloramphenicol (30 μg·ml−1) were added when required.
Bacterial Mutagenesis.
Mutagenesis was carried out by using the gene disruption method described by Datsenko and Wanner (32), except that 10 mM arabinose was used to induce expression of the red recombinase. Oligonucleotide primers used to amplify pKD4 kanamycin or pKD3 chloramphenicol resistance genes are listed in Table 3, which is published as supporting information on the PNAS web site. All mutagenesis was performed in the 12023 wild-type strain. Double mutants were created by transduction by using phage P22 HT105 int−. Phage-free transductants were selected for analysis. Gene deletions and transductions were checked by PCR.
Plasmid Construction.
Plasmids were constructed by using Gateway Technology. PipB2 ORF was amplified by PCR from Salmonella 12023 wild-type genomic DNA with the pipB2GWFW and pipB2GWREV oligonucleotides. The sequence of the TPR domain of the mouse KLC2 was amplified from pGFP.KLC2-mTPR with the oligonucleotides O301 and O302. PCR products were cloned by recombination into pDONRzeo vector (Gateway; Invitrogen, Carlsbad, CA). The entry clones were then transferred into Gateway destination vectors for yeast two-hybrid (pDBa), prokaryotic expression (pDEST15 and pDEST17), or eukaryotic expression (pCMVmyc-GW). The pCMVmyc-GW was constructed by using the Gateway Vector conversion system (Invitrogen) by EcoRI digestion of the pCMVmyc (Clontech), fill-in of cohesive ends, and blunt-end cloning of the Gateway reading frame cassette A.
Yeast Two-Hybrid Screening.
Competent MaV203 yeast cells were successively transformed with pDBa-PipB2 and 100 μg of human fetal brain cDNA library in pEXP-AD502 vector, and ≈5 × 105 transformants were plated onto minimal selective medium lacking tryptophane, leucine, and histidine, were supplemented with 20 mM 3-amino-1,2,4-triazole, and were incubated at 30°C for 5 days. Positive clones were then tested for two other two-hybrid reporters, i.e., uracil and β-galactosidase, as described (33). Positive colonies were PCR-amplified, and phenotypes were retested by reintroducing PCR products and pEXP-AD502 vector linearized into fresh yeast cells containing the bait (34). Colonies giving rise to more than one PCR product were eliminated. pEXP-AD502 inserts of positive clones after the second round of two-hybrid screening were sequenced and identified. The two clones with ORF sequences in-frame with the GAL4AD coding sequence were considered for further investigation.
GST Pull-Down.
GST, GST.SKIP (1–310), and GST.PipB2 were expressed in Escherichia coli BL21(DE3), purified from bacterial lysates on glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech, Piscataway, NJ) following the manufacturer’s instructions, and stored at −80°C in 20% glycerol in PBS. Transfected HeLa cells were resuspended in lysis buffer (PBS, 1% Nonidet P-40, and EDTA-free protease inhibitor mixture; Roche) for 30 min at 4°C and centrifuged at 20,000 × g for 10 min. For pull-down experiments 250–300 μg of protein of HeLa cell extracts were incubated with 2 μg of purified protein bound to 5 μl of stacked glutathione-Sepharose 4B beads and incubated for 2 h at 10°C. Beads were washed four times with lysis buffer supplemented with 0.5 M NaCl. Ten percent lysates and pulled-down proteins were analyzed by SDS/PAGE and Western blotting by using the anti-GFP JL-8 antibody.
Coimmunoprecipitation Assays.
Transfected HeLa cells were resuspended in immunoprecipitation buffer (PBS/0.5% Nonidet P-40/1 mM EGTA/1.5 mM MgCl2/10% glycerol/EDTA-free protease inhibitor mixture; Roche) for 30 min at 4°C and centrifuged at 20,000 × g for 10 min. Ten percent of the lysate was boiled in SDS/PAGE sample buffer, and the remainder was used in immunoprecipitations. A total of 250–300 μg of protein of HeLa cell extracts were incubated with 2 μl of rabbit anti-GFP antibody for 3 h at 4°C. Ten microliters of a 50% slurry of protein A beads were added and further incubated for 1 h at 4°C. Beads were washed four times in immunoprecipitation buffer supplemented with 0.5 M NaCl and boiled in 20 μl of SDS/PAGE sample buffer. Two percent lysates and the immunoprecipitates were analyzed by SDS/PAGE and Western blotting by using mouse anti-myc or anti-GFP antibodies.
Eukaryotic Cells and Culture Conditions.
RAW-264.7 and HeLa cell lines were grown routinely in DMEM (GibcoBRL, Gaithersburg, MD) supplemented with 10% FCS (Life Technologies, Rockville, MD), 2 mM nonessential amino acids, and glutamine (GibcoBRL) at 37°C in 5% CO2. Bone marrow cells were isolated from femurs of 6- to 10-week-old C57BL/6 female mice and differentiated into macrophages as previously described (35).
Bacterial Infection and Replication Assays.
HeLa cells were seeded onto glass coverslips (12-mm diameter) in 10-cm dishes at a density of 106 cells per dish 24 h before infection. Bacteria were incubated overnight at 37°C with shaking, diluted 1:33 in fresh LB broth, and incubated in the same conditions for 3.5 h. The cultures were diluted in Earle’s buffered salt solution (pH 7.4) and added to the cells at a multiplicity of infection of ≈100:1. The infection was allowed to proceed for 10 min at 37°C in 5% CO2. Macrophages were seeded at a density of 2 × 105 cells per well in 24-well tissue culture plates 24 h before use. Bacteria were cultured overnight at 37°C with shaking and were opsonized in DMEM containing FCS and 10% normal mouse serum for 20 min. Bacteria were added to the cells at a multiplicity of infection of ≈100:1 and incubated for 20 min at 37°C in 5% CO2. Cells were washed three times with growth medium containing 100 μg·ml−1 gentamicin and incubated in this medium for 1 h, after which the gentamicin concentration was decreased to 10 μg·ml−1 for the remainder of the experiment. Where indicated, HeLa cells were transfected with FuGENE 6 (Roche Molecular Biochemicals) following the manufacturer’s instructions at 2 h after invasion, and cells were further incubated for 14 h.
For enumeration of intracellular bacteria, cells were washed three times with PBS and lysed with 0.1% Triton X-100 for 10 min, and a dilution series was plated onto LB agar plates. Plates were incubated overnight at 37°C, and colonies were counted. Each time point was performed in triplicate, and each experiment was performed three times or more.
Immunofluorescence.
Cells grown on coverslips were fixed with 3% paraformaldehyde (pH 7.4) in PBS at room temperature for 10 min. Fixed cells were washed three times in PBS and permeabilized with 0.1% saponin in PBS. Primary and secondary antibodies were diluted in PBS containing 0.1% saponin and 5% horse serum. Coverslips were incubated with primary antibodies for 30–60 min at room temperature, washed in PBS containing 0.1% saponin, and then incubated with appropriate secondary antibodies. Coverslips were mounted onto glass slides using Mowiol (Aldrich, Lyon, France). Cells were observed with an epifluorescence microscope (Leica, Deerfield, IL) or a LSM510 confocal laser scanning microscope (Zeiss, Thornwood, NY).
Scoring of Phenotypes by Epifluorescence Microscopy.
SCVs were labeled by using antibodies against the lysosomal glycoprotein Lamp-1. Infected cells were observed by epifluorescence, and the percentage of GFP-expressing bacteria present in a vacuole was determined by counting the total number of bacteria and the number of bacteria totally or partially encircled by the Lamp-1 marker. GFP-expressing bacteria enclosed in a Lamp-1-positive compartment were scored as juxtanuclear when the SCV was at less than one bacterial length from the nucleus, visualized by DAPI staining. The percentage of SCVs positive for kinesin-1 was determined by visualizing GFP-expressing bacteria in the green channel, Lamp-1 in the UV channel (using Alexa Fluor 350 secondary antibodies), and kinesin HC in the red channel.
Mouse Mixed Infections.
Six- to 8-week-old BALB/c mice were inoculated i.p. with equal amounts of two bacterial strains for a total of 105 bacteria per mouse. The spleens were harvested 48 h after inoculation and homogenized. Bacteria were recovered and enumerated after plating a dilution series onto LB agar and LB agar with the appropriate antibiotics. CIs were determined for each mouse, and a minimum of three mice were infected (19). The CI is defined as the ratio between the mutant and wild-type strains within the output (bacteria recovered from the mouse after infection) divided by their ratios within the input (initial inoculum). Unpaired t test analysis was performed to compare two CIs, and a one-sample t test comparing the log of the CI to 0 was used to determine whether the CI was significantly different from 1. All statistical analyses were performed by using Prism (GraphPad, San Diego, CA). The two-tailed P value was calculated.
Supplementary Material
Acknowledgments
We thank Suzana Salcedo and David Weiss for critical reading of the manuscript and Stéphanie Balor for help with bone marrow-derived macrophages. T.H., E.B., and A.D. were recipients of a fellowship from the French Ministry of Research and Association pour la Recherche sur le Cancer. N.S. was the recipient of a fellowship of the Microban European Union Network (MRTN-CT-2003-504227). This work was supported by the Intramural Research Program of the National Institutes of Health–National Institute of Allergy and Infectious Diseases; the Agence Nationale de la Recherche (Grant 05-BLAN-0028-01); and institutional grants from the Centre National de la Recherche Scientifique and Institut National de la Santé et de la Recherche Médicale.
Abbreviations
- HC
heavy chain
- KLC
kinesin light chain
- SCV
Salmonella-containing vacuole
- TPR
tetratricopeptide repeat
- TTSS-2
second type III secretion system
- CI
competitive index.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Garcia-del Portillo F., Finlay B. B. J. Cell Biol. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 3.Shea J. E., Hensel M., Gleeson C., Holden D. W. Proc. Natl. Acad. Sci. USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirillo D. M., Valdivia R. H., Monack D. M., Falkow S. Mol. Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 5.Hensel M., Shea J. E., Waterman S. R., Mundy R., Nikolaus T., Banks G., Vazquez-Torres A., Gleeson C., Fang F. C., Holden D. W. Mol. Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 6.Ochman H., Soncini F. C., Solomon F., Groisman E. A. Proc. Natl. Acad. Sci. USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catron D. M., Sylvester M. D., Lange Y., Kadekoppala M., Jones B. D., Monack D. M., Falkow S., Haldar K. Cell. Microbiol. 2002;4:315–328. doi: 10.1046/j.1462-5822.2002.00198.x. [DOI] [PubMed] [Google Scholar]
- 8.Beuzon C. R., Méresse S., Unsworth K. E., Ruiz-Albert J., Garvis S., Waterman S. R., Ryder T. A., Boucrot E., Holden D. W. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez-Torres A., Xu Y., Jones-Carson J., Holden D. W., Lucia S. M., Dinauer M. C., Mastroeni P., Fang F. C. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 10.Meresse S., Unsworth K. E., Habermann A., Griffiths G., Fang F., Martinez-Lorenzo M. J., Waterman S. R., Gorvel J. P., Holden D. W. Cell. Microbiol. 2001;3:567–577. doi: 10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- 11.Henry T., Gorvel J. P., Méresse S. Cell. Microbiol. 2006;8:23–32. doi: 10.1111/j.1462-5822.2005.00649.x. [DOI] [PubMed] [Google Scholar]
- 12.Boucrot E., Henry T., Borg J. P., Gorvel J. P., Méresse S. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 13.Knodler L. A., Vallance B. A., Hensel M., Jackel D., Finlay B. B., Steele-Mortimer O. Mol. Microbiol. 2003;49:685–704. doi: 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 14.Richter-Dahlfors A., Buchan A. M. J., Finlay B. B. J. Exp. Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salcedo S. P., Noursadeghi M., Cohen J., Holden D. W. Cell. Microbiol. 2001;3:587–597. doi: 10.1046/j.1462-5822.2001.00137.x. [DOI] [PubMed] [Google Scholar]
- 16.Brumell J. H., Rosenberger C. M., Gotto G. T., Marcus S. L., Finlay B. B. Cell. Microbiol. 2001;3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 17.Stein M. A., Leung K. Y., Zwick M., Garcia-del Portillo F., Finlay B. B. Mol. Microbiol. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 18.Guignot J., Caron E., Beuzon C., Bucci C., Kagan J., Roy C., Holden D. W. J. Cell Sci. 2004;117:1033–1045. doi: 10.1242/jcs.00949. [DOI] [PubMed] [Google Scholar]
- 19.Beuzon C. R., Holden D. W. Microbes Infect. 2001;3:1345–1352. doi: 10.1016/s1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 20.Salcedo S. P., Holden D. W. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCart A. E., Mahony D., Rothnagel J. A. Traffic. 2003;4:576–580. doi: 10.1034/j.1600-0854.2003.00113.x. [DOI] [PubMed] [Google Scholar]
- 22.Knodler L. A., Bestor A., Ma C., Hansen-Wester I., Hensel M., Vallance B. A., Steele-Mortimer O. Infect. Immun. 2005;73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knodler L. A., Steele-Mortimer O. Mol. Biol. Cell. 2005;16:4108–4123. doi: 10.1091/mbc.E05-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison R. E., Brumell J. H., Khandani A., Bucci C., Scott C. C., Jiang X., Finlay B. B., Grinstein S. Mol. Biol. Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsman M., Jordens I., Kuijl C., Janssen L., Neefjes J. Mol. Biol. Cell. 2004;15:2954–2964. doi: 10.1091/mbc.E03-08-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., Mattoo S., McMahon S. A., Ghosh P., Hughes T. R., Boone C., Dixon J. E. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 27.Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., Neefjes J. Curr. Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Albert J., Yu X. J., Beuzon C. R., Blakey A. N., Galyov E. E., Holden D. W. Mol. Microbiol. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 29.Waterman S. R., Holden D. W. Cell. Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 30.Kubori T., Galan J. E. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 31.Abraham G. L., Hensel M. Cell. Microbiol. 2006;8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 32.Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walhout A. J., Vidal M. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 34.Orr-Weaver T. L., Szostak J. W. Proc. Natl. Acad. Sci. USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Chastellier C., Frehel C., Offredo C., Skamene E. Infect. Immun. 1993;61:3775–3784. doi: 10.1128/iai.61.9.3775-3784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.