Abstract
Resistance of pathogens to antimicrobial therapeutics has become a widespread problem. Resistance can emerge naturally, but it can also be engineered intentionally, which is an important consideration in designing therapeutics for bioterrorism agents. Blocking host receptors used by pathogens represents a powerful strategy to overcome this problem, because extensive alterations to the pathogen may be required to enable it to switch to a new receptor that can still support pathogenesis. Here, we demonstrate a facile method for producing potent receptor-directed antitoxins. We used phage display to identify a peptide that binds both anthrax-toxin receptors and attached this peptide to a synthetic scaffold. Polyvalency increased the potency of these peptides by >50,000-fold in vitro and enabled the neutralization of anthrax toxin in vivo. This work demonstrates a receptor-directed anthrax-toxin inhibitor and represents a promising strategy to combat a variety of viral and bacterial diseases.
Keywords: antimicrobial resistance, phage display, therapeutics
Pathogens can develop resistance to drugs directed against microbial targets by modifying the drug, by lowering the concentration of drug that reaches the target, or by mutating the target (1, 2). There is also an increasing concern that therapeutics developed for bioterrorism agents may be rendered ineffective if the microbial target is altered intentionally. This problem could be overcome, however, by designing inhibitors that block host proteins used by the pathogen or its toxins to cause disease.
Microbial pathogens and their products interact with host structures to facilitate colonization or to promote cellular uptake. Many of these interactions are polyvalent, meaning that they involve the simultaneous binding of multiple ligands on one entity to multiple receptors on another (3). The design of synthetic polyvalent (4–8) or oligovalent (9, 10) molecules also represents a promising approach to enhance the potency of inhibitors of microbial pathogens and toxins. Current examples of this approach have involved the design of molecules that bind directly to the pathogen or toxin. Inhibitors that bind host proteins would represent an effective way to attenuate virulence that may be less susceptible to resistance mechanisms, and the use of polyvalency could provide a significant enhancement in the potency of these inhibitors.
ANTXR1 and ANTXR2 are host receptors that bind and internalize anthrax toxin (11, 12). These proteins are likely important for anthrax pathogenesis because the toxin impairs the immune response and is responsible for the major symptoms and death associated with anthrax. Thus, blocking these receptors could represent a promising approach to anthrax therapy.
ANTXR1 and ANTXR2 are widely expressed type I membrane proteins that bind components of the extracellular matrix (13). They both contain an extracellular I domain, which binds the protective antigen (PA) component of anthrax toxin. The two proteins are 40% identical overall and share 60% identity within their I domains. The PA–receptor interaction is mediated through an aspartate side chain of PA that completes the coordination of a metal ion bound by a metal ion-dependent adhesion site of the receptor I domain (14, 15). PA is processed by a protease into a 63-kDa fragment, PA63, which oligomerizes into ring-shaped heptamers (16). Heptameric PA63 ([PA63]7) binds the enzymatic components of the toxin, edema factor (EF) and lethal factor (LF), and the resulting complexes are internalized. Internalization depends on a coreceptor, low-density lipoprotein (LDL) receptor-related protein 6, that binds ANTXR1/2 directly (17). Upon reaching an acidic compartment, [PA63]7 dissociates from its receptors and inserts flexible loops into the membrane to form a β-barrel pore (18). Insertion of [PA63]7 that has been internalized by ANTXR1 occurs at a higher pH found in early endosomes compared with ANTXR2-internalized [PA63]7, which inserts only at a lower pH found in late endosomes. This difference likely results from the higher affinity of the PA–ANTXR2 interaction. The pore that forms as a result of [PA63]7 insertion allows EF and LF to translocate across the endosomal membrane (19). EF is an adenylate cyclase, and LF is a protease that cleaves mitogen-activated protein kinase kinases. The enzymatic activities of these proteins contribute to disease progression in several ways and at different stages of infection (16).
Several anthrax-toxin inhibitors have been described that interfere with different steps in this intoxication pathway (8, 11, 12, 16, 20–26), but none has targeted the host receptors. Here, we describe the development of a polyvalent receptor-directed anthrax-toxin inhibitor that binds both receptors and protects animals from toxin challenge.
Results and Discussion
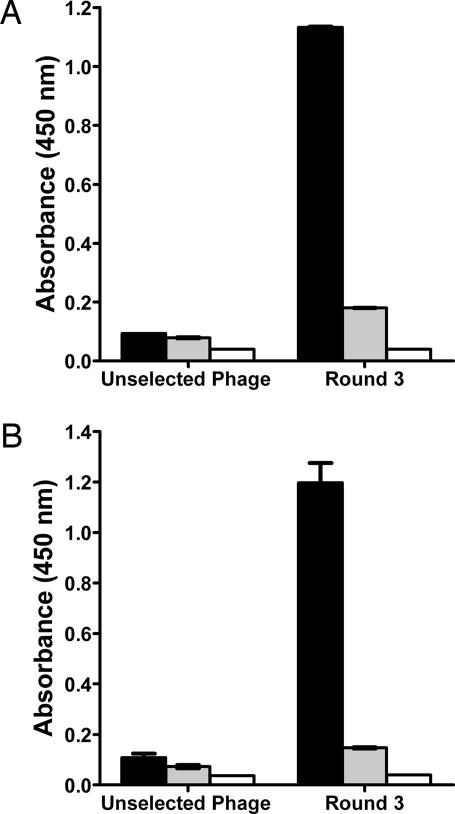
To develop an anthrax-toxin inhibitor, we first used phage display (8, 27) to identify peptides that bind to the cellular receptors ANTXR1 and ANTXR2. The I domains of ANTXR1 and ANTXR2 were purified, and each was immobilized on plastic tubes and then exposed to libraries of M13 phage. Each I domain was exposed to a phage library presenting random 12-residue, 7-residue, or constrained (cyclic) 7-residue peptides. In total, six screens were performed by using the two I domains and the three libraries. After incubation of the phage with the I domain, the tube was washed to remove unbound and weakly bound phage; phage of interest were then eluted by adding PA. This protocol was repeated three times, and the isolated phage were characterized by ELISA. As seen in Fig. 1A and B, the pool of phage selected from the 12-residue library after three rounds of panning bound the bait receptor, and the binding was inhibited by PA. Phage did not bind to the unrelated protein BSA.
Fig. 1.
Identification of peptides that bind the I domains of anthrax-toxin cellular receptors ANTXR1 and ANTXR2. (A and B) Phage from the unselected 12-mer library or pooled phage obtained after three rounds of panning against immobilized ANTXR1 (A) or immobilized ANTXR2 (B) were subjected to ELISA. In each case, the phage pool isolated after three rounds bound specifically to the I domain (black bars) and was inhibited by PA (gray bars). Immobilized BSA was used as a control (white bars).
We isolated a number of phage from the screens and tested each for the ability to bind the I domain that was used in the selection process (data not shown). Two 12-residue peptides that bind the I domain of ANTXR1 share the sequence QLDHS (Table 1). Four peptides that bind ANTXR2 contain the sequence STD and two of these peptides, one from the 12-residue library and the other from the 7-residue library, shared an extended sequence, STDHS. Interestingly, the sequence DHS was shared by some of the peptides identified in screens against ANTXR1 and ANTXR2, indicating that these peptides might bind both receptors.
Table 1.
Sequence of peptides isolated from screening phage display libraries against either ANTXR1 or ANTXR2
| ANTXR1-binding peptides | ANTXR2-binding peptides |
|---|---|
| AWPLSQLDHSYN* | SPHGSTDHSTTA* |
| YHLSSQQLDHSL* | STDHSLY‡ |
| ATWGHPRSSQGM* | STDSGWV‡ |
| CPSSTLFAC† | CTSTDATYC† |
Underlined sequences are common to at least two peptides. The sequence DHS, found in screens against both receptors, is in bold.
*Peptides isolated from 12-mer phage library.
†Peptides isolated from cyclic 7-mer peptide library.
‡Peptides isolated from 7-mer library.
We synthesized four polyvalent inhibitors by attaching multiple copies of each DHS-containing peptide to liposome scaffolds, which are assemblies composed of a phospholipid bilayer and an aqueous core. Liposomes used for these experiments were made from a mixture of 1,2-distearoyl-sn-glycero-3-phosphocholine and a thiol-reactive pyridyldithiopropionate derivative of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine in a molar ratio of 9:1. The liposomes were functionalized with DHS-containing peptides with an additional C-terminal cysteine, which allowed attachment to the liposomes via a reaction with the thiol group. The remaining unreacted thiol-reactive groups on the liposomes were quenched with thioglycerol.
We tested the ability of these polyvalent liposome-based inhibitors to neutralize anthrax toxin in vitro by incubating RAW264.7 cells with a mixture of PA and LF in the presence of several concentrations of the inhibitor. As seen in Table 2, liposomes presenting AWPLSQLDHSYN, YHLSSQQLDHSL, SPHGSTDHSTTAY, and STDHSLY (2.7% peptide) could inhibit the toxicity of a mixture of PA and LF in RAW264.7 cells. The most potent inhibitor, a liposome presenting multiple copies of AWPLSQLDHSYN, inhibited cytotoxicity with a half-maximal inhibitory concentration (IC50) of 40 nM on a per-peptide basis (Fig. 2A); in contrast, the corresponding monovalent peptide did not inhibit cytotoxicity at concentrations as high as 2 mM. The use of polyvalency therefore enabled a >50,000-fold enhancement in the activity of this peptide. Liposomes presenting only thioglycerol showed no inhibitory activity (Fig. 2A).
Table 2.
Inhibitory activities of polyvalent inhibitors
| Peptide | Activity against toxin-induced cell death in RAW264.7 cells (IC50), nM |
|---|---|
| AWPLSQLDHSYN | 40 |
| YHLSSQQLDHSL | 100 |
| SPHGSTDHSTTAY | 250 |
| STDHSLY | 900 |
Indicated peptides were attached to liposomes, and their inhibitory activity was then measured. Values of IC50 reported are on a per-peptide basis.
Fig. 2.
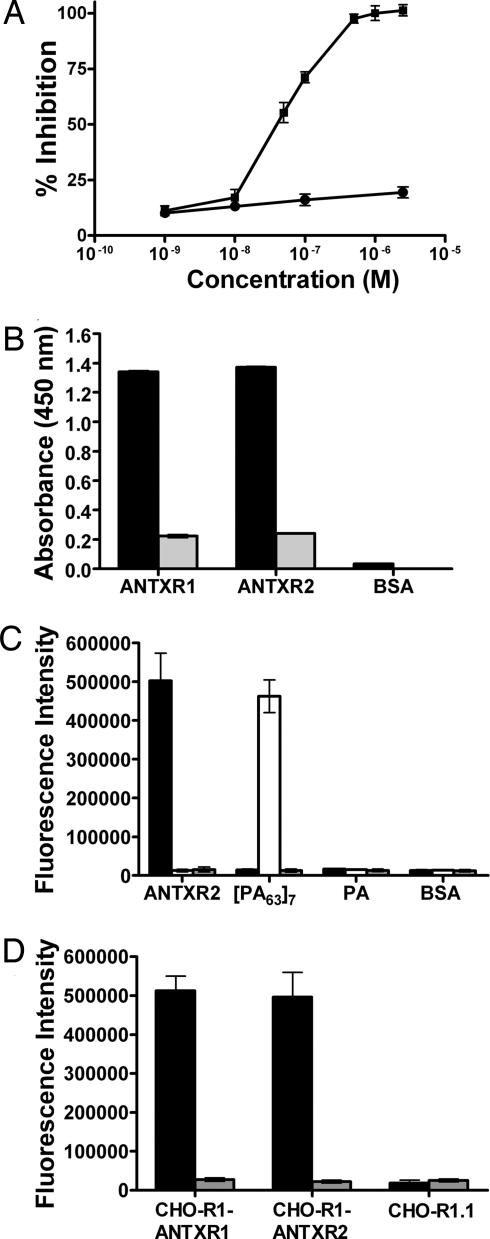
Characterization of AWPLSQLDHSYN-functionalized polyvalent inhibitors in vitro. (A) Inhibition of anthrax toxin-induced RAW264.7 cytotoxicity by polyvalent liposomes presenting AWPLSQLDHSYN (■) or control thioglycerol-functionalized liposomes (●). (B) Phage presenting the sequence AWPLSQLDHSYN, which were isolated by panning against ANTXR1, bind specifically to both ANTXR1 and ANTXR2 (black bars) but do not bind to BSA (white bars). Binding of phage to the I domains was inhibited by PA (gray bars). (C) Binding of fluorescein-containing liposomes presenting the sequence AWPLSQLDHSYN (black bars), the sequence HTSTYWWLDGAP (white bars), and thioglycerol (gray bars) to ANTXR2, [PA63]7, PA, and BSA. (D) Binding of fluorescein-containing liposomes presenting AWPLSQLDHSYN (black bars) and thioglycerol-functionalized control liposomes (gray bars) to CHO-R1-ANTXR1, CHO-R1-ANTXR2, and receptor-deficient CHO-R1.1 cells.
Because the polyvalent inhibitor that displayed AWPLSQLDHSYN was more potent than the others we tested, we decided to characterize this inhibitor further. This peptide was isolated from a screen for its ability to bind ANTXR1, but the protection of RAW264.7 cells by AWPLSQLDHSYN-functionalized liposomes suggested that this peptide also binds ANTXR2, because ANTXR2 is expressed in RAW264.7 cells (28). To test this possibility, we examined the binding of phage presenting the peptide AWPLSQLDHSYN to ANTXR1 and ANTXR2 by ELISA. As seen in Fig. 2B, phage presenting this peptide bound to both receptors and did not bind to the unrelated protein BSA.
As a further test of the ability of the AWPLSQLDHSYN peptide to bind the ANTXR2 I domain, we determined whether AWPLSQLDHSYN-functionalized liposomes bound ANTXR2 in an ELISA. We attached the AWPLSQLDHSYN peptide or a control [PA63]7-binding peptide, HTSTYWWLDGAP (8), to liposomes containing fluorescein (29). Binding of fluorescein-containing liposomes to proteins adsorbed to plastic wells was detected by using a fluorescence plate reader. Liposomes presenting AWPLSQLDHSYN bound to ANTXR2 but did not bind to [PA63]7, PA, or BSA (Fig. 2C). In contrast, liposomes presenting HTSTYWWLDGAP bound to [PA63]7 and not to the other three proteins (Fig. 2C). Liposomes presenting only thioglycerol did not bind to any of the proteins tested.
We next tested whether liposomes presenting AWPLSQLDHSYN bind to full-length ANTXR1 and ANTXR2 in the physiologically relevant context of the cell surface (Fig. 2D). This assay was carried out by using the receptor-deficient CHO-R1.1 (11) cell line and two cell lines derived from it: CHO-R1-ANTXR1 cells that express only ANTXR1 and CHO-R1-ANTXR2 cells that express only ANTXR2. Fluorescein-containing liposomes functionalized with AWPLSQLDHSYN bound to CHO-R1-ANTXR1 and CHO-R1-ANTXR2 cells but not to CHO-R1.1 cells, whereas thioglycerol-functionalized fluorescein-containing liposomes did not bind to any of the three cell lines (Fig. 2D).
To probe the mechanism of inhibition of cytotoxicity, we tested the ability of AWPLSQLDHSYN-functionalized liposomes to inhibit the binding of PA to cells (Fig. 3A). CHO-R1.1 or CHO-R1-ANTXR1 cells were treated with a mixture of 2-deoxyglucose, sodium azide, and bafilomycin A1 (dGAB) to inhibit receptor internalization. The cells were treated for 2 h with PA and liposomes presenting AWPLSQLDHSYN, HTSTYWWLDGAP, or thioglycerol; the cells were then washed with PBS. Cellular lysates were made and probed for PA by Western blotting. PA bound CHO-R1-ANTXR1 cells but not CHO-R1.1 cells. Control liposomes presenting either HTSTYWWLDGAP or thioglycerol did not inhibit the association of PA with CHO-R1-ANTXR1 cells and, surprisingly, neither did liposomes presenting the receptor-binding peptide, AWPLSQLDHSYN. Furthermore, the AWPLSQLDHSYN-functionalized liposomes did not inhibit the binding of PA to CHO-R1-ANTXR2 or RAW264.7 cells (data not shown). These results suggest that the peptide-binding site on ANTXR1/2 does not substantially overlap with the PA-binding site. The interaction between the receptor and the phage presenting AWPLSQLDHSYN may have been inhibited by PA (Fig. 2B) as a result of steric clashes between PA and the phage.
Fig. 3.
The AWPLSQLDHSYN-functionalized liposomes prevent LFN binding to oligomeric PA63. (A) CHO-R1.1 cells (lane 1) or CHO-R1-ANTXR1 cells (lanes 2–5) were treated for 2 h with PA alone (lanes 1 and 2); PA and AWPLSQLDHSYN-functionalized liposomes (lane 3), PA and HTSTYWWLDGAP-functionalized liposomes (lane 4), or PA and thioglycerol-functionalized liposomes (lane 5). Cells were washed with PBS and lysed, and the resulting lysates were subjected to Western blotting using anti-PA antibody. (B) The association of [35S]LFN with cells in the presence of PA and liposomes functionalized with either AWPLSQLDHSYN (white bars), HTSTYWWLDGAP (black bars), or thioglycerol (gray bars) was measured by scintillation counting.
Next, we tested the ability of the AWPLSQLDHSYN-functionalized liposomes to prevent the binding of radiolabeled LFN, the N-terminal fragment of LF, to oligomeric PA63 on the cell surface. Cells were treated with dGAB and proteolytically activated PA (nPA) for 1 h. The cells were washed and then treated with [35S]LFN and peptide-functionalized liposomes. As seen in Fig. 3B, liposomes presenting AWPLSQLDHSYN inhibited the association of LFN to cells that had been treated with nPA, as did the positive-control HTSTYWWLDGAP-functionalized liposomes that bind [PA63]7 directly. Thioglycerol-functionalized liposomes did not inhibit the association of [35S]LFN with [PA63]7. These results suggest that, although the AWPLSQLDHSYN peptide binds anthrax-toxin receptors, the functionalized liposomes inhibit intoxication by sterically blocking the binding of LF to [PA63]7.
Finally, we tested the ability of these receptor-directed polyvalent inhibitors to neutralize anthrax toxin in vivo, in Fisher 344 rats. Five of six rats that were injected intravenously with toxin (a mixture of 30 μg of PA and 8 μg of LF), and all six rats that were coinjected with toxin and unfunctionalized liposomes became moribund (Table 3). Coinjection of the AWPLSQLDHSYN-functionalized lipsomes with the toxin, however, prevented all six animals from becoming moribund. Six rats injected with the liposome-based inhibitors alone showed no adverse side effects.
Table 3.
Liposome inhibitors protect rats from LeTx challenge
| Treatment | Moribund rats/total |
|---|---|
| LeTx | 5/6 |
| LeTx + 400 nmol inhibitor | 0/6 |
| LeTx + thioglycerol-functionalized liposomes | 6/6 |
Rats were injected intravenously with only LeTx (30 μg of PA and 8 μg of LF), LeTx with thioglycerol-functionalized liposomes or LeTx with liposome inhibitors. The amount of inhibitor is listed in terms of amount of peptide. Results are combined from two independent experiments and are statistically significant by the log-rank test.
We have provided a demonstration of the in vivo efficacy of a receptor-targeted anthrax antitoxin. These anthrax toxin inhibitors might serve as valuable adjuncts to antibiotic therapy. Although Bacillus anthracis can be eradicated from a host by treatment with antibiotics, such treatment is often insufficient to save the patient once symptoms have developed because of the continuing action of the secreted toxin. The administration of a receptor-directed polyvalent inhibitor could neutralize the toxin and help reduce the high mortality rates associated with inhalational anthrax.
Our approach to designing anthrax-toxin receptor-directed inhibitors may be broadly applicable to receptors used by other pathogens and toxins (3, 30–33). Membrane proteins are currently the most highly represented class of drug targets for noninfectious diseases. Given the rapid emergence of antimicrobial drug resistance, targeting membrane receptors also represents a promising approach to design novel anti-infective agents. Phage-display technology is inexpensive and allows for the rapid identification of peptides that can be used as the basis for polyvalent receptor-targeted inhibitors; synthesis of polyvalent inhibitors, such as the ones described here and elsewhere, is simple, inexpensive, and scalable and provides significant enhancements in potency. Although broad-spectrum therapeutics have clear advantages, antimicrobials tailored to specific pathogens, especially those directed against host structures, may be an effective strategy to overcome the problem of resistance.
Materials and Methods
Phage-Display Selection.
M13 phage libraries (New England Biolabs, Beverly, MA) displaying random 12-mer, 7-mer, and cyclic 7-mer peptides were used for panning. The I domains from ANTXR1 and ANTXR2 were purified as described (15, 34). Proteins were allowed to adsorb on Maxisorp tubes (Nunc, Roskilde, Denmark) from a 2 μg/ml solution overnight at 4°C. The tubes were blocked with 2% BSA in PBS for 2 h and washed with PBS buffer. An M13 phage library (1.5 × 1011 pfu in PBS) was added to the tubes and incubated for 1 h at room temperature. The tubes were then washed 10 times with 0.1% Tween 20 in PBS. The remaining bound phages were eluted by adding PA (20 μg/ml in PBS and 1 h incubation at room temperature). The amplified phage pool was panned again as described above, except that the phages were added to the protein-coated tubes and incubated for 30 min in the second round and for 5 min in the third round, and the elution of bound phages was carried out by overnight incubation with PA (20 μg/ml in PBS) in rounds two and three.
Enzyme-Linked Immunosorbent Assay.
Purified receptor protein (I domain of ANTXR1 or ANTXR2) was coated in the wells of a 96-well plate overnight at 4°C at 1 μg/ml in PBS (pH 7.5). The wells were blocked with 2% BSA in PBS for 2 h, followed by a PBS wash. Phages (108 pfu) were incubated in these protein-coated wells for 1 h in the presence/absence of purified PA protein (4 μg/ml in PBS). Phage binding to protein was quantified by using an anti-phage antibody conjugated to horseradish peroxidase (Amersham Biosciences, Piscataway, NJ) by using 3,3′,5,5′-tetramethylbenzidine (Pierce Biotechnology, Rockford, IL) as a substrate. These experiments were carried out in duplicate and repeated twice.
Synthesis of Liposome-Based Polyvalent Inhibitor.
Liposomes were made from a mixture of 1,2-distearoyl-sn-glycero-3-phosphocholine and the thiol-reactive lipid 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine in a molar ratio of 9:1. Dynamic light scattering confirmed the presence of vesicles (radius, 51 ± 4 nm). Peptides identified by phage display were synthesized by Genemed Synthesis (South San Francisco, CA). These peptides were acetylated at their N termini and amidated at the C termini and had an extra cysteine residue at the C termini to facilitate their attachment to liposomes. Peptide predissolved in DMSO was added to a solution of liposomes in phosphate buffer (pH 8), and the reaction was allowed to proceed overnight. The remaining unreacted thiol-reactive groups were quenched with thioglycerol. Excess unreacted peptide and thioglycerol were removed by dialysis. The peptide-functionalized liposomes were characterized by UV-Vis spectroscopy to determine the concentration of peptide (8) by measuring absorbance at 280 nm. The procedure used to prepare liposomes incorporating fluorescein was similar to that described above, except that the hydration step was carried out with a 10 mM solution of fluorescein in phosphate buffer (25 mM, pH 8). All liposomes used in this study were functionalized with 2.7% peptide. Inhibitor concentrations are on a per-peptide basis.
Characterization of the Binding of Liposomes to Immobilized Proteins.
Purified protein was coated in the wells of a 96-well plate overnight at 4°C from a 10 μg/ml solution in PBS (pH 7.4). The wells were blocked with 100 μl of 2% BSA in PBS for 2 h. The fluorescein-containing liposomes were then incubated in the wells for 1 h at room temperature. The wells were then washed 10 times with PBS to remove any unbound liposomes. Finally, the intensity of fluorescence in the wells was quantified by using a plate reader (PerkinElmer, Wellesley, MA). Excitation was at 495 nm, and emission was read at 535 nm with a measurement time of 1 s. Results reported here are the average of three independent experiments, each carried out in triplicate.
Characterization of the Binding of Liposomes to Cells.
The binding experiment was carried out by using CHO-R1-ANTXR1, CHO-R1-ANTXR2, and receptor-deficient CHO-R1.1 (11) cells. Cells were cultured in F12 medium supplemented with 10% FBS (Invitrogen). The cells were seeded in a 96-well plate at a density of 10,000 cells per well and incubated overnight. After washing with PBS, the wells were blocked by adding 100 μl of a solution of 5% BSA in PBS for 2 h. Unbound BSA was removed by washing the wells with PBS. Next, the cells were incubated with 100 μl of 50 mM 2-deoxy-glucose (Sigma, St. Louis, MO)/10 mM sodium azide (Sigma)/200 nM bafilomycin A1 (Alexis Biochemicals, San Diego, CA) (dGAB) in F12 medium supplemented with 10% FBS for 45 min at 37°C (18). The fluorescein-containing liposomes were then added to the wells and incubated for 1 h at room temperature. After 10 washes with PBS, the values of the fluorescence intensity in the wells were measured by using a plate reader (PerkinElmer). Excitation was at 495 nm, and emission was read at 535 nm with a measurement time of 1 s. Results reported here are the average of three independent experiments, each carried out in triplicate.
Western Blot Assay.
CHO-R1.1 or CHO-R1-ANTXR1 cells were seeded in 24-well plates and grown overnight at 37°C. Confluent monolayers were incubated with 50 mM 2-deoxy-glucose, 10 mM sodium azide, and 200 nM bafilomycin A1 (dGAB) in supplemented F12 medium for 45 min at 37°C. Cells were washed with 1 ml of PBS and then treated with 1 nM trypsin-nicked PA in the presence of dGAB and 1 μM peptide-functionalized liposomes (2.7% peptide) for 2 h at 37°C. Cells were washed with PBS before lysis with 60 μl of ECB buffer, which contains 10 mM NaH2PO4, pH 8.0, 1 M NaCl, 10 mM EDTA, 1 mM PMSF, and 1% (vol/vol) Triton X-100. Thirty micrograms of total-protein lysate from each sample was subjected to SDS/PAGE. Proteins were transferred to nitrocellulose, and PA was detected by Western blotting using anti-PA antibody raised in rabbits against full-length PA (a gift from R. J. Collier, Harvard Medical School, Boston, MA).
Toxin-Assembly Assay.
CHO-R1-ANTXR2 cells were seeded in 24-well plates and grown overnight. The cells were incubated on ice with 2 × 10−8 M trypsin-nicked PA for 2 h and washed twice with cold PBS. The cells were then incubated with 35S-labeled LFN in the presence of indicated concentrations of inhibitors for 1 h. After extensive washing with cold PBS, the cells were lysed, and the radioactivity was measured by scintillation counting. Nonspecific binding was measured as the amount of radioactivity associated with the cells in the absence of PA and was subtracted from values obtained in the presence of PA. The percentage inhibition was calculated by dividing the radioactivity of the peptide-containing samples by that of the sample without inhibitor.
Cytotoxicity Assay.
RAW264.7 cells were seeded in 96-well plates and incubated overnight. The cells were treated with 10−9 M PA and 3 × 10−10 M LF in the absence or presence of inhibitors. After an incubation period of 4 h, cell viability was assessed by using the MTS assay according to the manufacturer’s instructions (Promega, Madison, WI).
Rat Intoxication.
A mixture of purified PA (30 μg) and LF (8 μg) mixed with PBS, peptide-functionalized liposomes (2.7% peptide; 400 nmol on a per-peptide basis), or thioglycerol-functionalized liposomes was used for the intoxication assay. Fisher 344 rats (Charles River Laboratories, Wilmington, MA) were injected intravenously in the tail vein. Three rats were used per group, and the appearance of symptoms of intoxication was monitored over 4 h. The rats were killed to avoid unnecessary distress when the symptoms became pronounced.
Acknowledgments
This work was supported by National Institutes of Health Grant R21 AI053506. J.M. holds the Canada Research Chair in Bacterial Pathogenesis.
Abbreviations
- LF
lethal factor
- PA
protective antigen.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levy S. B., Marshall B. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Hogan D., Kolter R. Curr. Opin. Microbiol. 2002;5:472–477. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- 3.Mammen M., Choi S. K., Whitesides G. M. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Mammen M., Dahmann G., Whitesides G. M. J. Med. Chem. 1995;38:4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 5.Mochalova L. V., Tuzikov A. B., Marinina V. P., Gambaryan A. S., Byramova N. E., Bovin N. V., Matrosovich M. N. Antiviral Res. 1994;23:179–190. doi: 10.1016/0166-3542(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 6.Yarema K. J., Bertozzi C. R. Curr. Opin. Chem. Biol. 1998;2:49–61. doi: 10.1016/s1367-5931(98)80035-5. [DOI] [PubMed] [Google Scholar]
- 7.Kamitakahara H., Suzuki T., Nishigori N., Suzuki Y., Kanie O., CH W. Angew. Chem. Int. Ed. 1998;37:1524–1528. doi: 10.1002/(SICI)1521-3773(19980619)37:11<1524::AID-ANIE1524>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 8.Mourez M., Kane R. S., Mogridge J., Metallo S., Deschatelets P., Sellman B. R., Whitesides G. M., Collier R. J. Nat. Biotechnol. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 9.Kitov P. I., Sadowska J. M., Mulvey G., Armstrong G. D., Ling H., Pannu N. S., Read R. R., Bundle D. R. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 10.Fan E., Zhang Z., Minke W. E., Hou Z., Verlinde C. L. M. J., Hol W. G. J. J. Am. Chem. Soc. 2000;122:2663–2664. [Google Scholar]
- 11.Bradley K. A., Mogridge J., Mourez M., Collier R. J., Young J. A. T. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 12.Scobie H. M., Rainey G. J. A., Bradley K. A., Young J. A. T. Proc. Natl. Acad. Sci. USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell S. E., Mavila A., Salazar R., Bayless K. J., Kanagala S., Maxwell S. A., Davis G. E. J. Cell Sci. 2001;114:2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 14.Santelli E., Bankston L. A., Leppla S. H., Liddington R. C. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 15.Lacy D. B., Wigelsworth D. J., Scobie H. M., Young J. A. T., Collier R. J. Proc. Natl. Acad. Sci. USA. 2004;101:6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier R. J., Young J. A. T. Annu. Rev. Cell Dev. Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 17.Wei W., Lu Q., Chaudry G. J., Leppla S. H., Cohen S. N. Cell. 2006;124:1141–1154. doi: 10.1016/j.cell.2005.12.045. [DOI] [PubMed] [Google Scholar]
- 18.Rainey G. J. A., Wigelsworth D. J., Ryan P. L., Scobie H. M., Collier R. J., Young J. A. T. Proc. Natl. Acad. Sci. USA. 2005;102:13278–13283. doi: 10.1073/pnas.0505865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krantz B. A., Melnyk R. A., Zhang S., Juris S. J., Lacy D. B., Wu Z., Finkelstein A., Collier R. J. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainey G. J. A., Young J. A. T. Nat. Rev. Microbiol. 2004;2:721–726. doi: 10.1038/nrmicro977. [DOI] [PubMed] [Google Scholar]
- 21.Karginov V. A., Nestorovich E. M., Moayeri M., Leppla S. H., Bezrukov S. M. Proc. Natl. Acad. Sci. USA. 2005;102:15075–15080. doi: 10.1073/pnas.0507488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sellman B. R., Mourez M., Collier R. J. Science. 2001;292:695–697. doi: 10.1126/science.109563. [DOI] [PubMed] [Google Scholar]
- 23.Maynard J. A., Maassen C. B. M., Leppla S. H., Brasky K., Patterson J. L., Iverson B. L., Georgiou G. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 24.Numa M. M. D., Lee L. V., Hsu C. C., Bower K. E., Wong C. H. ChemBioChem. 2005;6:1002–1006. doi: 10.1002/cbic.200500009. [DOI] [PubMed] [Google Scholar]
- 25.Panchal R., Hermone A. R., Nguyen T. L., Wong T. Y., Schwarzenbacher R., Schmidt J., Lane D., McGrath C., Turk B., Burnett J., et al. Nat. Struct. Mol. Biol. 2004;11:67–72. doi: 10.1038/nsmb711. [DOI] [PubMed] [Google Scholar]
- 26.Rai P., Padala C., Poon V., Saraph A., Basha S., Kate S., Tao K., Mogridge J., Kane R. S. Nat. Biotechnol. 2006;24:582–586. doi: 10.1038/nbt1204. [DOI] [PubMed] [Google Scholar]
- 27.Zwick M. B., Shen J. Q., Scott J. Curr. Opin. Biotechnol. 1998;9:427–436. doi: 10.1016/s0958-1669(98)80017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banks D. J., Barnajian M., Maldonado-Arocho F. J., Sanchez A. M., Bradley K. A. Cell. Microbiol. 2005;7:1173–1185. doi: 10.1111/j.1462-5822.2005.00545.x. [DOI] [PubMed] [Google Scholar]
- 29.Peer D., Margalit R. Int. J. Cancer. 2004;108:780–789. doi: 10.1002/ijc.11615. [DOI] [PubMed] [Google Scholar]
- 30.Dhiman N., Jacobson R. M., Poland G. A. Rev. Med. Virol. 2004;14:217–229. doi: 10.1002/rmv.430. [DOI] [PubMed] [Google Scholar]
- 31.Cywes C., Stamenkovic I., Weiss M. J. Clin. Invest. 2000;106:995–1002. doi: 10.1172/JCI10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., et al. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger E. A., Murphy P. M., Farber J. M. Annu. Rev. Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 34.Bradley K. A., Mogridge J., Rainey G. J. A., Batty S., Young J. A. T. J. Biol. Chem. 2003;278:49342–49347. doi: 10.1074/jbc.M307900200. [DOI] [PubMed] [Google Scholar]