Abstract
The pathogenic species of Yersinia contain the transcriptional regulator RovA. In Yersinia pseudotuberculosis and Yersinia enterocolitica, RovA regulates expression of the invasion factor invasin (inv), which mediates translocation across the intestinal epithelium. A Y. enterocolitica rovA mutant has a significant decrease in virulence by LD50 analysis and an altered rate of dissemination compared with either wild type or an inv mutant, suggesting that RovA regulates multiple virulence factors. Here, we show the involvement of RovA in the virulence of Yersinia pestis, which naturally lacks a functional inv gene. A Y. pestis ΔrovA mutant is attenuated ≈80-fold by LD50 and is defective in dissemination/colonization of spleens and lungs after s.c. inoculation. However, the ΔrovA mutant is only slightly attenuated when given via an intranasal or i.p. route, indicating a more important role for RovA in bubonic plague than pneumonic plague or systemic infection. Microarray analysis was used to define the RovA regulon. The psa locus was among the most highly down-regulated loci in the ΔrovA mutant. A ΔpsaA mutant had a significant dissemination defect after s.c. infection but only slight attenuation by the pneumonic-disease model, closely mimicking the virulence defect seen with the ΔrovA mutant. DNA-binding studies revealed that RovA specifically interacts with the psaE and psaA promoter regions, indicating a direct role for RovA in regulating this locus. Thus, RovA appears to be a global transcription factor in Y. pestis and, through its regulatory influence on genes such as psaEFABC, contributes to the virulence of Y. pestis.
Keywords: CUS-2 phage, IcmF-associated homologous protein, MarR/SlyA, pH 6 antigen
Yersinia pestis is the etiological agent of plague, an acute zoonotic infection that is often a fatal disease in humans. Plague is primarily a disease among rodent populations, and bacteria are usually transmitted to human hosts by the bite of an infected flea (1–3). Y. pestis can rapidly disseminate from a s.c. infection site into the lymphatic system and regional lymph nodes. The swelling of these infected lymph nodes into characteristic buboes is the classical symptom of bubonic plague (4, 5). Infection can progress into the circulatory system, resulting in a systemic disease that can lead to colonization of a variety of tissues including the lungs (6, 7). Development of secondary pneumonic infection poses a significant health risk as Y. pestis becomes highly transmissible during patient coughing. Aerosolized bacteria can be easily inhaled, causing a primary pneumonic infection in a new host, a serious and rapidly fatal disease (8). Both systemic and pneumonic plague result in high mortality rates because of rapid proliferation of bacteria and quick onset of disease pathology.
Y. pestis is a highly virulent pathogen because of its ability to escape the host immune system and rapidly proliferate within host tissues. Major virulence determinants encoded on the pCD virulence plasmid limit killing by host phagocytes, whereas other virulence determinants have evolved for essential nutrient acquisition from the host, such as the siderophore yersiniabactin (9–11). The type of plague pathology observed in a host depends on the mode of transmission, because patients can develop primary bubonic, pneumonic, or gastrointestinal disease. Interestingly, Y. pestis has evolved infectious route-specific virulence factors. Pla protease has been demonstrated to be a plasminogen activator important for establishing a disseminated infection from a s.c. infection site but does not appear to play a role in systemic disease (12, 13). Although Y. pestis spreads rapidly throughout the host, it lacks functional inv and yadA genes, well characterized invasion and adhesion factors of the closely related Yersinia enterocolitica and Yersinia pseudotuberculosis (14, 15). This is one example of the evolutionary divergence of these closely related species that has resulted in altered modes of transmission and virulence of Y. pestis in contrast to the enteropathogens Y. enterocolitica and Y. pseudotuberculosis.
The success of any pathogen depends on the tight regulation and coordinate expression of virulence factors during an infection. Y. pestis must adapt between two very different hosts (flea vs. mammal) and two very different modes of disease within the mammalian host (bubonic vs. pneumonic). Thus, adaptation of regulatory systems to integrate environmental signals and coordinate gene expression as appropriate is likely to be a key aspect of the virulence of this organism. RovA is a transcriptional regulator of the MarR/SlyA family that has been shown to regulate expression of inv in both Y. enterocolitica and Y. pseudotuberculosis (16, 17). Virulence analysis indicates that a rovA mutant is significantly more attenuated than either wild-type Y. enterocolitica or an inv mutant (16, 18), suggesting that RovA regulates other loci, some of which may be virulence determinants. Because rovA is conserved among all three pathogenic species of Yersinia yet Y. pestis lacks a functional inv gene, a key target of RovA, we addressed the role of RovA in plague pathogenesis. The results indicate that rovA is required for virulence in a bubonic, but not pneumonic, model of plague. In addition, we used microarray technology to begin to define the RovA regulon in Y. pestis.
Results
RovA Is Required for Full Virulence in Y. pestis.
To investigate the contribution of RovA in Y. pestis virulence, a null mutant was generated in the fully virulent strain CO92. Because rovA appears to be a single gene locus with flanking genes divergently transcribed from the deleted ORF, this mutation should not have polar effects on nearby genes. The ΔrovA mutant demonstrated growth characteristics similar to wild type when cultivated in Brain–Heart Infusion (BHI) media at both 26°C and 37°C (data not shown). In addition, loss of RovA regulatory function in the mutant strain was demonstrated by using inv and rovA transcriptional reporters (see Supporting Materials and Methods and Fig. 4, which are published as supporting information on the PNAS web site).
LD50 analysis was conducted with wild-type and ΔrovA Y. pestis strains by intranasal (i.n.), i.p., and s.c. routes. No difference in the LD50 between wild type or ΔrovA mutant was observed after i.n. or i.p. inoculation, although there was a slight delay in the mean day to death for the i.p. route. However, s.c. inoculation resulted in the ΔrovA mutant having an 80-fold higher LD50 and delay in mean day of death compared with wild type (Supporting Materials and Methods).
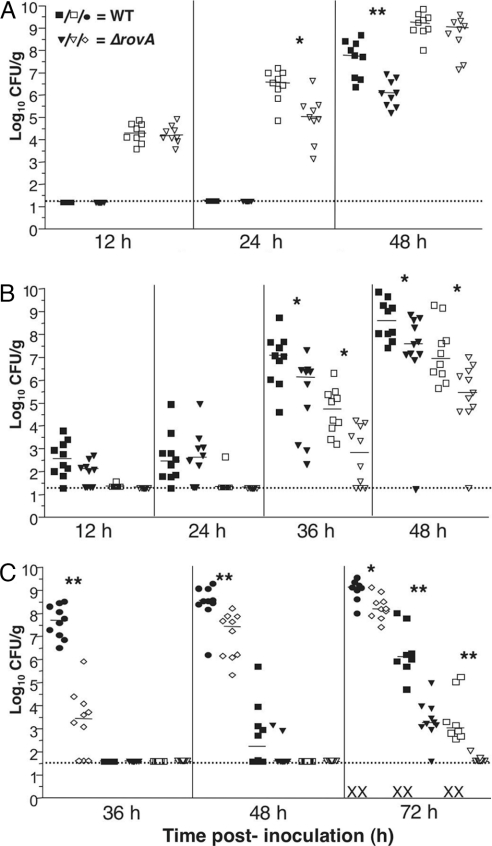
To further assess the role of rovA in Y. pestis virulence, the progression of infection was analyzed for each route. During i.n. infection, no major difference in the ability of the mutant to establish infection within the lungs was detected at 12 h (Fig. 1A). However, a modest decrease in colony forming units (cfu) in the lung for the ΔrovA mutant was seen by 24 h after inoculation, and, at 48 h, the dissemination to the spleen appeared to be slightly delayed. The ΔrovA mutant was more attenuated by the i.p. route, with ≈1–2 logs fewer mutant bacteria recovered from both tissues at later time points (Fig. 1B). The greatest defect again was observed during s.c. inoculation (Fig. 1C). At 36 h after inoculation, cfu recovered from cervical lymph nodes of mice infected with the ΔrovA mutant was ≈4 logs lower than from wild-type-infected mice. Significantly fewer cfu were also observed within the cervical lymph nodes of ΔrovA mutant-infected animals at 48 and 72 h after inoculation. At 48 h, a more disseminated disease is observed, with cfu recovered from the spleens of infected animals. Mice infected with the ΔrovA mutant had ≈3 logs fewer bacteria recovered from their spleens compared with wild-type-infected mice at 72 h after inoculation. In addition, only one ΔrovA mutant-infected mouse had detectable cfu within its lungs, whereas cfu were recovered from all wild-type-infected mice at 72 h after inoculation. Furthermore, by 72 h, some wild-type-infected mice had died, but all ΔrovA mutant-infected mice were viable. Complementation of rovA at another chromosomal site downstream of glmS restores wild-type virulence of the ΔrovA strain, indicating virulence attenuation is due to the lack of the regulator rather than a consequence of a secondary mutation within the mutant strain (Fig. 5A, which is published as supporting information on the PNAS web site). Thus, RovA appears to be a virulence factor for Y. pestis. Particularly, RovA seems be more important for infection from an s.c. site of inoculation, indicating a stronger role for RovA in bubonic rather than pneumonic plague or in the later systemic phases of infection.
Fig. 1.
Virulence analysis of the ΔrovA mutant by different routes of infection. (A–C) Dissemination analysis of CO92 and the ΔrovA mutant. Ten mice per strain for each time point analyzed were infected s.c. (102 cfu) (C), i.n. (104 cfu) (A), or i.p. (102 cfu) (B) with CO92 or ΔrovA. Infection was allowed to progress for the indicated time, and mice were then killed to harvest superficial cervical lymph nodes (filled circle, wild type; open diamonds, ΔrovA mutant), spleens (filled squares, wild type; filled triangles, ΔrovA mutant), and lungs (open squares, wild type; open triangles, ΔrovA mutant). Colonization of bacteria was quantified by enumerating viable cfu in each organ. Each symbol represents a single mouse, and these results are the composite of two independent experiments. The limit of detection is indicated by the dotted line, and symbols in the dotted line indicate cfu below the limit of detection. The X symbol on the x axis denotes dead mice at the designated time point. Significance was assayed by using Student’s t test with a two-tailed nonparametric analysis. For statistically significant comparisons, single asterisks indicate P ≤ 0.05, and double asterisks indicate P ≤ 0.005.
Characterization of the RovA Regulon in Y. pestis.
As a transcriptional regulator, RovA likely contributes to the virulence of Y. pestis through regulation of virulence genes. To identify these potential virulence factors, as well as to begin to define the RovA regulon, we compared transcriptional profiles from wild type and ΔrovA mutant by using CO92 whole-genome microarrays. Total cellular RNA was extracted from four biological replicate cultures grown to late log/early stationary phase, where high RovA expression and peak rovA-dependent inv expression was observed in vitro by using an inv-gfp reporter (data not shown). The expression of 73 genes was significantly altered by RovA (≥2-fold, P ≤ 0.05) (Table 1; and see Table 2, which is published as supporting information on the PNAS web site). Of these genes, 51 were down-regulated in the absence of RovA, and 22 genes were up-regulated. Most of the disregulated genes are characterized as hypothetical genes or putative genes related to cellular processes such as extracellular transport, energy metabolism, and biosynthesis. Thirteen genes in multiple phage operons were identified, most strongly repressed by RovA. Expression of the inv pseudogene was decreased in the ΔrovA strain; this finding is consistent with data obtained by reporter fusions and serves as an internal control for the microarray (Fig. 4A). Other interesting loci with altered regulation include pst encoded on the pPCP virulence plasmid, >90% of genes from one of the five IcmF-associated homologous proteins (IAHP) loci, and four genes of the psaEFABC locus. Subsequent analysis by quantitative (q)RT-PCR also demonstrated that psaC was similarly disregulated (data not shown). To confirm the microarray data, 25 genes were chosen, and transcript levels were measured by qRT-PCR. More than 90% of these genes correlated with the disregulation observed by microarray analysis (Tables 1 and 2). In addition, eight random genes not shown to be RovA-regulated were also tested by qRT-PCR, and there was no observed difference in regulation of these loci between the wild-type and ΔrovA mutant samples (data not shown).
Table 1.
Microarray analysis of RovA regulon in Y. pestis
| Identification | ORF description | Fold change |
|
|---|---|---|---|
| Array* | qRT-PCR† | ||
| YPO0499‡ | Hypothetical protein | −4.6 | −4.4 |
| YPO0506‡ | clpB/htpM putative Clp ATPase | −5.5 | −3.1 |
| YPO0510‡ | Hypothetical protein | −4.6 | −5.3 |
| YPO0514‡ | Putative OmpA-family protein | −4.4 | −3.6 |
| YPO1301 | psaE, putative regulatory protein | −5.7 | −9.8 |
| YPO1302 | psaF, putative membrane protein | −4.7 | −11.3 |
| YPO1303 | psaA, pH 6 antigen | −25.9 | −133 |
| YPO1304 | psaB, chaperone protein | −8.7 | −66.5 |
| YPO2277§ | Phage hypothetical protein | 23.2 | 66.7 |
Shown is a partial list of genes from the microarray analysis that were ≥2-fold different with a P value of ≤0.05, as described in Materials and Methods. A complete list of regulated genes from the array can be found in the supporting information.
*Fold difference in gene transcripts between wild type and the ΔrovA mutant as determined by microarray analysis.
†Fold difference in gene transcripts between wild type and the ΔrovA mutant as determined by qRT-PCR. The value listed is the average fold difference from multiple repeats of qRT-PCR performed on different biological replicates, as described in Materials and Methods.
§Cus-2 phage locus (34).
RovA Binds to the Promoters of Disregulated Loci.
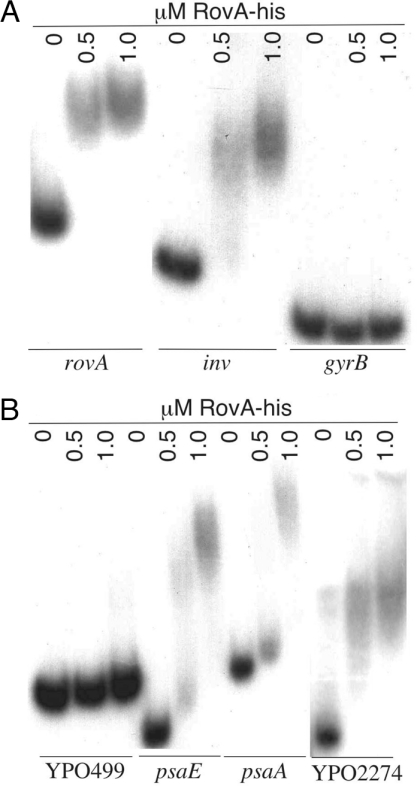
Because the array results indicate only transcriptional differences between wild type and the ΔrovA mutant, we wanted to determine whether RovA directly or indirectly regulates these loci. EMSAs were performed on DNA fragments upstream of selected affected genes to determine whether RovA directly binds these promoters. Previous data has shown that RovA binds to regions within both the inv and rovA promoters of Y. enterocolitica and Y. pseudotuberculosis (17, 19, 20). To ascertain the binding ability of Y. pestis RovA, both 32P-labeled inv or rovA promoter fragments from Y. pestis were incubated with purified RovA-His. For each promoter, 0.5 μM RovA-His is sufficient to observe complete shifting of radiolabeled DNA (Fig. 2A). In contrast, we did not observe any binding to the control gyrB promoter region, even with 1.0 μM RovA-His, indicating a specific interaction of RovA to the inv and rovA promoters. In Y. pseudotuberculosis, the rovA promoter has been shown to have two distinct binding regions (representing high- and low-affinity binding sites) (17, 19). These regions are conserved in Y. pestis, and similar binding-affinity patterns were observed when similar rovA promoter fragments were tested (data not shown).
Fig. 2.
EMSA of RovA with Y. pestis promoters. Randomly 32P-labeled rovA [+1 kb to −20 bp from start codon (SC)], inv (+500 to −20 bp from SC), gyrB (+500 to −20 bp from SC), psaA (+500 to −20 bp from SC), and psaE (+400 to −20 bp from SC) were incubated with the indicated concentrations of purified RovA in the presence of competitor DNA (1 μg of salmon sperm) as described in Materials and Methods.
Because 30% of the disregulated genes fall in the IHAP locus, we investigated a putative promoter region for RovA binding. Although no promoters of this locus have been defined, we examined the region upstream of the first gene of the locus, YPO499. No binding was observed with as much as 1 μM RovA-His, suggesting that there is no direct interaction, at least within the first 500 bp upstream of the ATG start codon (Fig. 2B). However, this same putative promoter region showed RovA-dependent regulation by using a reporter fusion expressed in wild type versus a ΔrovA mutant background (data not shown). Thus, RovA probably contributes indirectly to the regulation of this locus.
As one of the most strongly down-regulated loci in the ΔrovA mutant, the psaEFABC locus was examined by EMSA. This locus is known to encode proteins involved in the regulation (psaEF) and biosynthesis (psaBC) of a fimbrial structure (psaA) on the surface of the bacterial cell (21, 22). This locus has been shown to have at least two distinct promoter regions in front of psaE and psaA, respectively (23, 24). RovA-His bound both promoter fragments, but higher concentrations of recombinant protein were required to achieve the full shift observed with the inv and rovA promoters (Fig. 2B) (17, 19, 20), suggesting that RovA directly influences the expression of psa genes.
Among the most repressed loci identified by microarray analysis, the CUS-2 filamentous phage locus (YPO2274–YPO2279) was assessed for interaction with RovA. A putative promoter region of the YPO2274 ORF was shown to be completely shifted by 0.5 μM RovA. The phage locus had a similar affinity for RovA as was observed for the inv and rovA promoters, indicating a possible direct role of RovA in repression for this phage operon. The ability of RovA to repress gene transcription has been observed; previous work in Y. pseudotuberculosis demonstrated that elevated levels of RovA leads to repression of rovA, implicating an autoregulatory mechanism to maintain appropriate regulator levels within the bacteria (19).
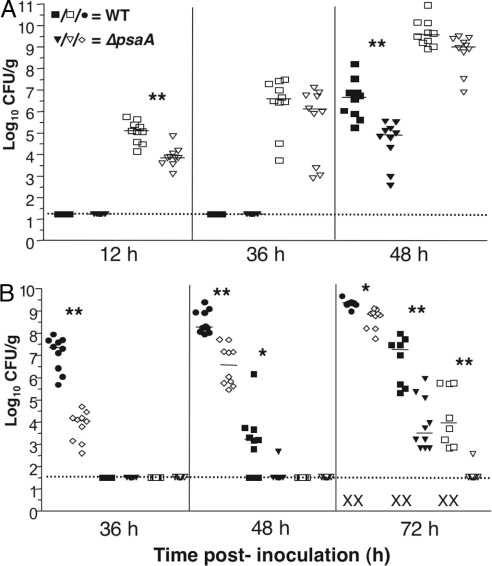
psaA Is Required for Full Virulence in Y. pestis.
pH 6 antigen (PsaA) previously has been implicated in Y. pestis virulence. A psaA mutant of a KIM5(pgm−) strain showed a 100-fold increase in LD50 by the retroorbital route (22). Because transcription of psaA is significantly decreased in the ΔrovA mutant, we wanted to determine whether the attenuation observed in the ΔrovA mutant was due to the decreased psaA expression. To this end, an in-frame deletion of psaA was generated in CO92 and tested in our mouse model. After i.n. inoculation, fewer cfu were recovered from the lungs of the ΔpsaA mutant than wild type at 12 h after inoculation (Fig. 3A), but no differences in lung colonization were observed after 36 or 48 h. In the spleen, no cfu were detected from wild-type- or ΔpsaA mutant-infected mice until 48 h. At this time, fewer cfu were recovered from ΔpsaA mutant- than wild-type-infected mice. Thus, the ΔpsaA mutant exhibited a slight lag in colonization of both the lung and spleen compared with wild-type.
Fig. 3.
Virulence analysis of the Δpsa mutant by different routes of infection. (A and B) Mice were infected i.n. (104 cfu) (A) or s.c. (102 cfu) (B) with either CO92 or the psaA mutant. Infection was allowed to progress for the indicated time, and mice were then killed to harvest superficial cervical lymph nodes (filled circles, wild type; open diamonds, ΔrovA mutant), spleens (filled squares, wild type; filled triangles, ΔrovA mutant), and lungs (open squares, wild type; open triangles, ΔrovA mutant). Bacterial load and significance were determined as in Fig. 1. Results are the composite of two independent experiments.
After s.c. inoculation, significantly fewer cfu were obtained in the cervical lymph nodes of the ΔpsaA mutant-infected mice throughout the course of infection, with the ΔpsaA mutant colonizing at ≈2–3 logs lower compared with wild type at earlier time points (Fig. 3B). At 48 h after infection, most wild-type-infected animals demonstrated a disseminated disease with spleen colonization but only one ΔpsaA mutant-infected mouse had detectable cfu. This reduction in bacterial colonization is again observed, with ≈4 logs less bacteria recovered in the spleens of ΔpsaA mutant-infected animals at 72 h after inoculation. In addition, all wild-type-infected mouse lungs were highly colonized at 72 h after inoculation, but only one mouse infected with the ΔpsaA mutant had detectable cfu within this tissue. Again, we observe death of wild-type-, but not ΔpsaA mutant-infected animals, at 72 h after inoculation, with significant difference in colonization of all organ sites assessed. Overall, the virulence phenotype of the ΔpsaA mutant closely mimicked that of the ΔrovA mutant, indicating that down-regulation of the psa locus in the ΔrovA mutant could account in large part for the observed attenuation of the regulator mutant. Again, chromosomal complementation of psa was used to determine that the virulence defect in the ΔpsaA strain was due to the deletion of this gene (Supporting Materials and Methods).
Discussion
RovA is an established virulence factor in Y. enterocolitica, due in part to its positive regulation of inv. The observation that a Y. enterocolitica rovA mutant is more attenuated in mice than either wild-type or inv mutant strains suggested that RovA influences the regulation of other virulence genes (16, 18). In this work, we investigated the role of RovA in Y. pestis virulence and showed that its relative importance as a virulence factor depends on the route of infection. The increased LD50 and delayed dissemination of the ΔrovA mutant indicate that RovA may regulate genes important to establish infection during bubonic plague. A more modest degree of attenuation was observed in our pneumonic and systemic infection models, indicating that RovA-regulated genes are either less critical at these sites or at these stages of the disease process. Furthermore, this observation highlights the point that mutant strains are not universally attenuated for virulence, and route of infection is an important consideration when investigating the phenotype of a mutant. This result has been observed for another Y. pestis gene, pla. Virulence studies have shown Pla is essential for establishing a systemic disease after s.c. infection, but a pla mutant does not seem to affect virulence if administered by i.v. inoculation (13, 25).
These findings correlate well with data from Y. enterocolitica, in which the rovA mutant is less attenuated via i.p. inoculation compared with an oral inoculation (18). Pathogenic Yersinia species are lymphotropic pathogens. Y. enterocolitica and Y. pseudotuberculosis rapidly colonize Peyer’s Patches within the small intestine and then disseminate to the mesenteric lymph nodes shortly thereafter, whereas Y. pestis disseminates to and colonizes the lymph node proximal to the site of s.c. inoculation (4, 26, 27). Because a rovA mutant seems to be most attenuated in these species by routes of infection that lead first to colonization of a lymph node, it is tempting to speculate that the RovA regulon might be important for efficient trafficking to or colonization of lymphoid tissues, survival within these tissues, or dissemination from these areas to other organ sites in the infected animals. In this study, we observed a significant decrease in cfu recovered from the cervical lymph nodes of mice infected with the ΔrovA mutant. However, further experiments are needed to better understand this virulence defect and define the role of RovA in colonization of lymph tissues. Additionally, this may indicate that the RovA-regulon contributes to the transmission phase between the flea and host in a natural infection.
DNA microarrays were used to identify genes affected by RovA in an effort to identify potential virulence genes as well as define the RovA regulon in Y. pestis. Although most disregulated genes encode hypothetical proteins with unknown function, several loci are of particular interest. More than half of the positively regulated genes were part of an IAHP locus (YPO0499–YPO0516). IAHP gene clusters have been identified in a number of Gram-negative species, with multiple clusters found in Y. pestis alone (28, 29). IAHP loci have been implicated in virulence of some organisms and recently were shown to encode a new secretion system (30–33). It is interesting that RovA appears to regulate only one of these closely related IAHP loci, perhaps indicating that other IAHPs within Y. pestis are differentially regulated and therefore play other roles during the Y. pestis life cycle.
Regulatory proteins can function to directly modify gene expression by binding to DNA or interacting with other transcriptional elements within the promoter region or indirectly through effects on a secondary factor(s) that influences gene expression. RovA has been characterized as a DNA-binding protein that is capable of directly interacting with specific regions of the inv and rovA promoters of Y. enterocolitica and Y. pseudotuberculosis (17, 19, 20). In this study, we showed that RovA is capable of binding the same DNA regions with similar affinity to inv and rovA in Y. pestis. In addition, we observed that RovA binds directly to the psaE and psaA promoters but not a promoter region from the IAHP locus. Because all of these loci are disregulated in the ΔrovA mutant, this suggests RovA modulates gene expression through direct and indirect mechanisms. Furthermore, studies with other MarR/SlyA family members have shown that these regulatory proteins can function as both activators and repressors of gene expression. Microarray experiments showed that loss of rovA results in increased expression of multiple loci. The CUS-2 filamentous phage locus (YPO2274–2279) contains genes that are among the most RovA-repressed. Our work also has demonstrated that RovA can bind a putative promoter region within the locus, indicating a direct role for the regulator in transcriptional repression. Several of the genes within this locus have been characterized as virulence determinants of Vibrio cholerae and during extraintestinal Escherichia coli infections (34, 35). Interestingly, all Y. pestis biovar orientalis strains tested are positive by PCR for the presence of this locus, whereas biovar mediavalis is negative, and biovar antiqua gave mixed results (36).
Among the most down-regulated genes in the ΔrovA mutant were genes psaEFAB of the pH 6 antigen locus. These results were unexpected, because psaA was shown to be expressed only at 37°C at pH 6. The cultures for microarray analysis were grown at 26°C, conditions that are reported to have low psaA expression (21, 22). Interestingly, expression of Y. enterocolitica inv is low at 37°C in unbuffered media, but, when grown at 37°C in pH 5.5 media, expression is increased to levels similar to that observed at 26°C. Additionally, it has been demonstrated that inv is expressed in vivo in a rovA-dependent manner (16, 37). In a nonpigmented Y. pestis strain, a psaA mutant was shown to be attenuated by the retroorbital route of infection (22). In this report, we demonstrate that the loss of psaA in a fully virulent strain of Y. pestis results in a significant virulence defect in bubonic, and a slight virulence defect in pneumonic, models of disease. Furthermore, it has been observed that genes of the psa locus are down-regulated in the lungs compared with expression in vitro, indicating that transcription of this locus may not be important for the virulence of Y. pestis after i.n. inoculation (38). By examining the progression of infection, this attenuation closely mimics that observed with the ΔrovA mutant, suggesting that a large portion of the virulence defect of the ΔrovA mutant may be attributed to decreased expression of the psa locus. One possible model for the role of PsaA and regulation by RovA in Y. pestis pathogenesis is as follows. RovA could allow expression of the psa locus by Y. pestis while in the flea (low temperature) before transmission. If PsaA is an antiphagocytic factor, as has been proposed (39), this would offer some protection from phagocytic cells immediately after transmission before expression of the antiphagocytic F1 capsule and the Yops, which are expressed only at 37°C. PsaA would not be required in pneumonic plague models, because F1 capsule and Yops would already be expressed.
It is interesting to note that inv, the only identified RovA-regulated virulence gene in Y. enterocolitica, is a pseudogene in Y. pestis. Similar microarray studies in Y. enterocolitica have indicated the homolog to the psa locus, the myf genes, are RovA regulated (J.S.C. and V.L.M., unpublished data); however, it is not clear what contribution the myf locus has to the virulence of this organism. Although the psaA locus is among the most obvious virulence determinates identified by the microarray analysis, the animal data in this study does not exclude the possibility that other RovA-regulated genes contribute to the virulence of Y. pestis. However, this study has demonstrated that RovA maintains an important role in the pathogenesis of Y. pestis and may contribute to the virulence of this organism largely through the regulation of the psa locus.
Materials and Methods
Bacterial Strains, Growth Conditions, and Mice.
Y. pestis strain CO92 (naturally polymyxin B-resistant) is a clinical isolate from a pneumonic infection, provided by the U.S. Army, Fort Dietrich, MD (40). Strains were cultivated at 26°C on BHI (Difco, Sparks, MD) plates for 60 h. Liquid cultures of CO92 were grown at 26°C for 16 h with aeration in BHI or at 37°C in BHI supplemented with 2.5 mM CaCl2. E. coli strains DH5α or S17–1λpir were grown at 37°C for 16 h with aeration in LB broth. Antibiotics, when needed, were used at the following concentrations (unless otherwise noted): kanamycin (100 μg/ml), chloramphenicol (12.5 μg/ml), polymyxin B (25 μg/ml), or ampicillin (50 μg/ml).
All animal experiments were approved by the Washington University Animal Studies Committee and performed as outlined in protocols 20020257 and 20050189. Six- to 8-week-old female C57BL/6 mice from The Jackson Laboratory (Bar Harbor, ME) were maintained in the barrier facility at Washington University and allowed free access to sterilized food and water. Before infection, all mice were anesthetized with a mixture of ketamine HCl (100 mg/ml) and xylazine HCl (20 mg/ml) mixed 1:1 and delivered at a dose of 0.5 ml/kg of body weight by i.p. injection. Mice were euthanized by i.p. injection of Nembutal sodium (pentobarbital sodium) at a dose of 75 mg/kg of body weight.
Virulence Analysis in Animal Model.
Bacteria for i.p. and s.c. infections were cultured at 26°C in BHI for 16–18 h, and, for i.n. infections, bacteria were cultured at 37°C in BHI supplemented with 2.5 mM CaCl2. To determine LD50, five groups of six mice were infected with serial 10-fold dilutions of the bacterial suspension (104 to 1 cfu for i.p. and s.c. or 105 to 10 cfu for i.n. infections). Mice were monitored twice daily for 7 days, and the LD50 was calculated as described (41) (Fig. 6, which is published as supporting information on the PNAS web site).
For colonization/dissemination analysis, 10 mice were infected for each time point examined with the wild type, ΔrovA, or ΔpsaA by i.p. (102 cfu), s.c. in front neck (102 cfu), or i.n. (104 cfu) routes. At various times after infection, mice were euthanized, and lungs, spleens, and superficial cervical lymph nodes were removed. The bacterial load for each organ was determined by plating dilutions of the macerated tissues onto BHI plates and reported as cfu per gram of tissue. Infections were repeated in at least two independent experiments.
Plasmid Construction and Mutagenesis.
The plasmid for generating a null rovA mutant (YPO2374) was constructed as follows. Primers rovA-delA and rovA-delB were used to amplify an ≈500-bp sequence upstream of rovA (all primer sequences are listed in Table 3, which is published as supporting information on the PNAS web site). This product was digested with SalI and BamHI and cloned into those sites of pSR47S (42). Primers rovA-delC and rovA-delD were used to amplify an ≈500-bp fragment downstream of rovA that was subsequently digested with BamHI and NotI. This fragment was then cloned into the same sites of pSR47S next to the upstream fragment, generating plasmid pJC101. This plasmid was conjugated into Y. pestis, and integrants were purified on polymyxin and kanamycin plates. Transconjugants were restreaked onto plates with polymyxin and 10% sucrose to select for plasmid resolution. Deletion candidates were screened by Southern blot and PCR to confirm deletion of rovA. PCR was used to confirm the presence of all three virulence plasmids and the pgm locus on the selected CO92 ΔrovA mutants. One such strain (YP9) was used for further analysis.
To purify RovA, a recombinant form of RovA with a histidine tag was constructed. rovA was amplified by PCR using rovA-his1 and rovA-his2, which included the native ribosome-binding site and initiation codon. The PCR product was digested with XhoI and EcoRI and ligated into pET-24 (+) (Novagen, San Diego, CA) to generate provA-his. rovA was sequenced to confirm that no mutations were present, and the plasmid was moved into E. coli BL-21 (DE3) for protein expression and purification. This provA-his was demonstrated to produce a functional regulator by complementation of a rovA mutant strain (data not shown).
A deletion of the gene encoding PsaA in Y. pestis strain CO92 was constructed by a modified form of lambda red recombination originally described by Datsenko and Wanner (43). Briefly, 500 bp upstream and 500 bp downstream of psaA were independently amplified by PCR with the oligonucleotides psaA 5′-500 and P1 psaA 3′3 for the upstream fragment, and P4 psaA 5′975 and psaA 3′+500 for the downstream region. The resulting products were gel-purified and combined with a neo cassette flanked by FRT sites (previously amplified by PCR from the plasmid pKD13) in a second PCR amplification using psaA 5′-500 and psaA 3′+500. CO92 carrying pKD46 (AmpR) was grown to midlog phase at 26°C in the presence of 10 mM arabinose to induce the recombinase genes and transformed with the gel-purified psaA-FRT-neo-FRT-psaA PCR product. Recombinants were selected on plates containing kanamycin (50 μg/ml). Strains in which pKD46 had been lost were identified after serial passage in BHI broth and subsequent identification of AmpS colonies. The neo cassette introduced in the previous step was resolved by the introduction of pLH29, a plasmid carrying the FLP recombinase gene under the control of the lac promoter (a gift from A. Darwin, New York University Medical Center, New York, NY), and growth overnight at 26°C in the presence of isopropyl β-d-thiogalactoside (1 mM). KanS and CmS recombinants (indicating the loss of pLH29) were identified and confirmed by PCR to generate the ΔpsaA mutant (YP16).
RNA Isolation and Microarray Hybridization and Analysis.
Saturated cultures of CO92 and ΔrovA mutant were subcultured into BHI to an OD600 of 0.2 and grown for ≈8 h to OD600 ≈ 3.5. Twenty milliliters of culture was combined with 40 ml of RNAprotect Bacteria Reagent (Qiagen) to ensure immediate stabilization of RNA. Cells were centrifuged at 8,000 × g for 10 min, supernatant was removed, and pellets were stored at −80°C. RNA was purified by using a RiboPure-Bacteria kit (Ambion, Austin, TX) as specified by the supplier. Samples from four separate cultures of each strain were prepared and used for hybridization.
All microarray analysis was performed at the Genome Sequencing Center of Washington University. Total RNA was reverse-transcribed, labeled, and hybridized to the oligonucleotide arrays as described (38). Slides were scanned on a ScanArray Express HT Scanner (PerkinElmer, Wellesley, MA) to detect Cy3 and Cy5 fluorescence at 543 and 633 nm, respectively. Each spot was defined on a pixel-by-pixel basis by using a modified Mann–Whitney statistical test. The resulting values were imported into GeneSpring 7 software (Agilent Technologies, Palo Alto, CA) for analysis. The mean signal and control intensities of the on-slide duplicate spots were calculated. A Lowess curve was fit to the log-intensity versus log-ratio plot. Twenty percent of the data were used to calculate the Lowess fit at each point. This curve was used to adjust the control value for each measurement. If the control channel was <10 relative fluorescence units, then 10 was used instead. Data were filtered by using the Student’s t test function in GeneSpring. The P value is the result of a one-sample Student’s t test, which was applied to the natural log of the mean of each normalized value against the baseline value of 0. Genes with differences corresponding to P < 0.05 in either the high or the low photomultiplicator scans and that had signal to control or control to signal ratios >2.0 were considered to be significantly regulated.
qRT-PCR.
cDNA synthesis was performed with SuperScript III reverse transcriptase and 0.2 μg of RNA as specified by the supplier (Invitrogen, Carlsbad, CA). Each 25-μl qRT-PCR mixture contained 0.5 μl of cDNA, 12.5 μl of 2× SYBR green master mix (Qiagen), and 900 nM gene-specific primers (Table 4, which is published as supporting information on the PNAS web site). Data were normalized to Y. pestis gyrase B (gyrB) mRNA and relative fold-change calculated by using the ΔΔCT method.
EMSAs.
Purification of RovA-His and EMSA were conducted as described (20). In brief, radiolabeled promoter fragments of putative RovA-regulated genes (≈500 bp) were generated by PCR, which is published as supporting information on the PNAS web site). Three thousand cpm of 32P-labeled product was mixed with varying concentrations of RovA-His in binding buffer (20 mM Tris·HCl, 15% glycerol, 50 mM NaCl, 5 mM MgCl2, and 1 μg of salmon sperm DNA). The 10-μl reactions were incubated at room temperature for 15 min, separated on 3.5% PAGE gels, and visualized by autoradiogram.
Supplementary Material
Acknowledgments
We thank M. B. Lawrenz and K. A. Walker for critical reading of the manuscript and M. Heinz of the Washington University Genome Sequencing Center Microarray Core for the microarray hybridizations. This study was supported by National Institutes of Health (NIH) Grants AI53298 (to V.L.M.) and U54 AI57160 [to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) to W.E.G.]. W.W.L. was supported by the Infectious Diseases Training Grant to Washington University Clinical/Translational Fellowship Program of the MRCE, the W. M. Keck Foundation, and NIH National Research Service Award F32 AI069688. J.S.C. was supported by Lucille P. Markey Special Emphasis Pathway in Human Biology and Training Program in Cellular and Molecular Biology Grant T32-GM07067.
Abbreviations
- BHI
Brain–Heart Infusion
- i.n.
intranasal
- IAHP
IcmF-associated homologous protein
- qRT-PCR
quantitative RT-PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bacot A. W., Martin C. J. J. Hyg. 1914;13(Plague Suppl. 3):423–439. [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova V. A. Annu. Rev. Entomol. 1977;22:23–32. doi: 10.1146/annurev.en.22.010177.000323. [DOI] [PubMed] [Google Scholar]
- 3.Cavanaugh D. C., Williams J. E. In: International Conference on Fleas, Traub R., Starcke H., editors. Rotterdam, The Netherlands: Balkema; 1980. pp. 245–256. [Google Scholar]
- 4.Jawetz E., Myer K. F. J. Infect. Dis. 1944;74:1–13. [Google Scholar]
- 5.Pollitzer R. W. H. O. Monogr. Ser. 1954;Vol. 22:1–698. [Google Scholar]
- 6.Janssen E., Myer K. F. J. Infect. Dis. 1958;103:183–187. doi: 10.1093/infdis/103.2.183. [DOI] [PubMed] [Google Scholar]
- 7.Une T., Brubaker R. R. Infect. Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poland J. D., Quan T. J., Barnes A. M. Handbook of Zoonoses, Section A Bacteria, Rickettsial, Chlamydial, and Mycotic. Ann Arbor, MI: CRC; 1994. [Google Scholar]
- 9.Viboud G. I., Bliska J. B. Annu. Rev. Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 10.Bearden S. W., Fetherston J. D., Perry R. D. Infect. Immun. 1997;65:1659–1968. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bearden S. W., Perry R. D. Mol. Microbiol. 1999;32:403–4014. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 12.Sodeinde O. A., Sample A. K., Brubaker R. R., Goguen J. D. Infect. Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sodeinde O. A., Subrahmanyam Y. V., Stark K., Quan T., Bao Y., Goguen J. D. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 14.Pierson D. E. In: Molecular Genetics of Bacterial Pathogenesis, Miller V. L., Kaper J. B., Portnoy D. A., Isberg R. R., editors. Washington DC: Am. Soc. Microbiol; 1994. pp. 235–247. [Google Scholar]
- 15.Straley S. C., Starnbach M. N. In: Effects of Microbes on the Immune System, Cunningham M. W., Fujinami R. S., editors. Philadelphia: Lippincott Williams and Wilkins; 2000. pp. 71–92. [Google Scholar]
- 16.Revell P. A., Miller V. L. Mol. Microbiol. 2000;35:677–685. doi: 10.1046/j.1365-2958.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 17.Nagel G., Lahrz A., Dersch P. Mol. Microbiol. 2001;41:1249–1269. doi: 10.1046/j.1365-2958.2001.02522.x. [DOI] [PubMed] [Google Scholar]
- 18.Dube P. H., Handley S. A., Revell P. A., Miller V. L. Infect. Immun. 2003;71:3512–3520. doi: 10.1128/IAI.71.6.3512-3520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heroven A. K., Nagel G., Tran H. J., Parr S., Dersch P. Mol. Microbiol. 2004;53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 20.Ellison D. W., Miller V. L. J. Bacteriol. 2006;188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindler L. E., Tall B. D. Mol. Microbiol. 1993;8:311–324. doi: 10.1111/j.1365-2958.1993.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 22.Lindler L. E., Klempner M. S., Straley S. C. Infect. Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price S. B., Freeman M. D., Yeh K. S. J. Bacteriol. 1995;177:5997–6000. doi: 10.1128/jb.177.20.5997-6000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Isberg R. R. Mol. Microbiol. 1997;24:499–510. doi: 10.1046/j.1365-2958.1997.3511719.x. [DOI] [PubMed] [Google Scholar]
- 25.Sebbane F., Jarrett C., Gardner D., Long D., Hinnebusch B. J. Proc. Natl. Acad. Sci. USA. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cover T. L., Aber R. C. N. Eng. J. Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 27.Sebbane F., Gardner D., Long D., Gowen B. B., Hinnebusch B. J. Am. J. Pathol. 2005;166:1427–1439. doi: 10.1016/S0002-9440(10)62360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S., Chakrabortty A., Banerjee R., Chaudhuri K. Biochem. Biophys. Res. Commun. 2002;295:922–928. doi: 10.1016/s0006-291x(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 29.Parkhill J., Wren B., Thomson N. R., Titball R. W., Holden M. T., Prentice M. B., Sebaihia M., James K. D., Churcher C., Mungall K. L., et al. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 30.Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. Proc. Natl. Acad. Sci. USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons D. A., Heffron F. Infect. Immun. 2005;73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bladergroen M. R., Badelt K., Spaink H. P. Mol. Plant–Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- 33.Rao P. S., Yamada Y., Tan Y. P., Leung K. Y. Mol. Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez M. D., Lichtensteiger C. A., Vimr E. R. FEMS Microbiol. Lett. 2001;198:125–128. doi: 10.1111/j.1574-6968.2001.tb10630.x. [DOI] [PubMed] [Google Scholar]
- 35.Davis B. M., Waldor M. K. Curr. Opin. Microbiol. 2003;6:35–42. doi: 10.1016/s1369-5274(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez M. D., Lichtensteiger C. A., Caughlan R., Vimr E. R. J. Bacteriol. 2002;184:6050–0655. doi: 10.1128/JB.184.21.6050-6055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe J. C., Badger J. L., Miller V. L. Mol. Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 38.Lathem W. W., Crosby S. D., Miller V. L., Goldman W. E. Proc. Natl. Acad. Sci. USA. 2005;102:17786–17791. doi: 10.1073/pnas.0506840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X. Z., Lindler L. E. Infect. Immun. 2004;72:7212–7219. doi: 10.1128/IAI.72.12.7212-7219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doll J. M., Zeitz P. S., Ettestad P., Bucholtz A. L., Davis T., Gage K. Am. J. Trop. Med. Hyg. 1994;51:109–114. doi: 10.4269/ajtmh.1994.51.109. [DOI] [PubMed] [Google Scholar]
- 41.Reed L. J., Muench H. Am. J. Hygiene. 1938;27:493–497. [Google Scholar]
- 42.Walker K. A., Miller V. L. J. Bacteriol. 2004;186:4056–4066. doi: 10.1128/JB.186.13.4056-4066.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.