Abstract
Phototropins, plant blue light receptors, mediate stomatal opening through the activation of the plasma membrane H+-ATPase by unknown mechanisms. Here we report that type 1 protein phosphatase (PP1) positively regulates the blue light signaling between phototropins and the H+-ATPase in guard cells of Vicia faba. We cloned the four catalytic subunits of PP1 (PP1c) from guard cells and determined the expression of the isoforms in various tissues. Transformation of Vicia guard cells with PP1c isoforms that had lost enzymatic activity by one amino acid mutation, or with human inhibitor-2, a specific inhibitor protein of PP1c, suppressed blue light-induced stomatal opening. Addition of fusicoccin, an activator of the plasma membrane H+-ATPase, to these transformed guard cells induced normal stomatal opening, suggesting that the transformations did not affect the basic mechanisms for stomatal opening. Tautomycin, an inhibitor of PP1, inhibited blue light-induced H+ pumping, phosphorylation of the plasma membrane H+-ATPase in guard cell protoplasts, and stomatal opening. However, tautomycin did not inhibit the blue light-dependent phosphorylation of phototropins. We conclude that PP1 functions downstream of phototropins and upstream of the H+-ATPase in the blue light signaling pathway of guard cells.
Keywords: phototropin, H+-ATPase, light signaling, phosphorylation/dephosphorylation
Pairs of guard cells forming stomatal pores regulate gas exchange between plants and the atmosphere, allowing the uptake of CO2 for photosynthetic carbon fixation and the loss of water vapor by transpiration (1–3). Stomata open in response to blue light perceived by phototropins (phot1 and phot2), plant-specific Ser/Thr protein kinases with two LOV (light, oxygen, or voltage) domains (4–6). The light-stimulated phototropins undergo autophosphorylation with a subsequent binding of 14-3-3 proteins (7). Signals from phototropins ultimately activate the plasma membrane H+-ATPase via phosphorylation of the C terminus with a subsequent binding of 14-3-3 protein (8). The activated H+-ATPase creates an inside-negative electrical potential across the plasma membrane, which drives the uptake of K+ through inward-rectifying voltage-gated K+ channels (9–11); this leads to an increase in turgor pressure that results in stomatal opening. However, the mechanisms of signal transduction from phototropins to the plasma membrane H+-ATPase are largely unknown. Several components, including cytosolic Ca2+, RPT2 (ROOT PHOTOTROPISM2), and protein kinase in the plasma membrane, have been suggested to play a role, but the evidence to support these claims is not conclusive (12–14).
A previous pharmacological study showed that type 1 or type 2A protein phosphatases (PP1/PP2A) are involved in blue light-dependent stomatal opening in Vicia faba (15). Furthermore, PP1 appears more likely than PP2A to be responsible for this reaction because blue light-dependent H+ pumping was more strongly inhibited by calyculin A than okadaic acid. However, no conclusive evidence for the functional involvement of PP1 in the stomatal response is available so far.
PP1 is the major class of the PPP family of Ser/Thr protein phosphatases and is ubiquitously distributed throughout higher eukaryotes (16). In animals, PP1 has been shown to regulate a broad range of cellular processes such as glycogen metabolism, protein synthesis, cell cycle progression, muscle contraction, transcription, and neuronal signaling (17). Regulation of these multiple processes is accomplished by a few PP1 catalytic subunits (PP1c) associating with multiple regulatory subunits that specify the catalytic activity, the substrate specificity, and the subcellular localization of the holoenzyme (18, 19). Considering the functional importance of PP1 in animals, it is likely that PP1 also regulates fundamental processes in plant cells; however, very little is known about PP1 function in plants (16).
Recently, several experimental approaches to separate the functions of the PP1 and PP2A families in vivo have been reported. Mutational studies have shown that the expression of a dominant negative-type mutant of the PP2A catalytic subunit causes growth defects and reduction of PP2A activity without affecting its holoenzyme assembly in yeast (20, 21). In Drosophila cells, overexpression of inhibitor-2, which inhibits PP1c activity as a regulatory subunit, specifically reduces PP1 activity without affecting that of PP2A (22).
In the present study, we investigated the function of PP1 in guard cells, a highly developed model system for characterizing signal transduction mechanisms in plants (23). In this system, we can induce the signaling cascade by applying blue light and directly observe specific effects caused by any genetic or pharmacological perturbation. We transformed Vicia guard cells with a dominant negative-form of PP1c as well as with inhibitor-2 and demonstrated that PP1 functions as a positive regulator between phototropins and the H+-ATPase in the blue light signaling pathway.
Results
Cloning and Expression of PP1c Genes in V. faba.
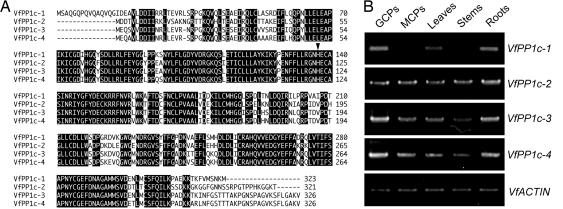
We isolated genes that encode PP1c expressed in guard cells from V. faba. Degenerate primers were designed from the conserved region of PP1c, and four partial cDNA fragments were obtained by RT-PCR using total RNA. We determined the overall cDNA sequences by 5′ and 3′ RACE, and designated the genes V. faba PP1c-1 through 4 (VfPP1c-1 through -4). There were high levels of similarity in the deduced amino acid sequences (95.8–99.7%, Fig. 1A). We confirmed the mRNA expression of all four PP1c genes in guard cells by RT-PCR, and determined their isoform- and tissue-specific expression (Fig. 1B). VfPP1c-1, -3, and -4 were expressed predominantly in guard cells. Actually, VfPP1c-1 was not expressed at all in mesophyll cells or stems. However, VfPP1c-2 was expressed in all tissues examined to the same degree.
Fig. 1.
Amino acid sequences and expression patterns of Vicia PP1c isogenes. (A) Alignment of the deduced amino acid sequences of VfPP1c isoforms. Identical amino acids appear in negative print. Arrowhead indicates the conserved His residue in the catalytic site. The sequences were aligned by using CLUSTAL X. (B) Expression of Vicia PP1c isogenes in guard cell protoplasts (GCPs), mesophyll cell protoplasts (MCPs), leaves, stems, and roots determined by RT-PCR. Actin mRNA was used as an internal standard.
Inhibition of Blue Light-Dependent Stomatal Opening by Transformation of Guard Cells with Mutant Forms of PP1c.
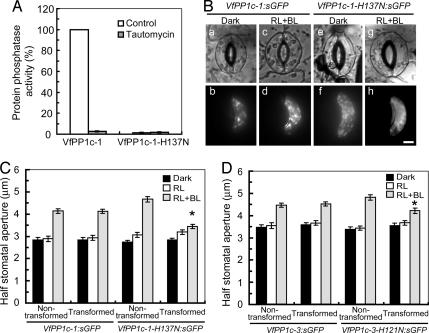
To investigate the function of PP1 in blue light-induced stomatal opening in vivo, we transiently overexpressed mutated forms of PP1c that were impaired in their catalytic activities, by particle bombardment in Vicia guard cells. The His residue in the catalytic site (highlighted in Fig. 1A) was replaced by Asn to abolish PP1c activity (24); we expected that the mutant functioned as a dominant negative form in vivo, similarly as in the case of PP2A (20, 21). We prepared recombinant VfPP1c-1 and VfPP1c-1-H137N and found that the activity was lost completely in the latter. The VfPP1c-1 activity was inhibited by 10 nM tautomycin, a potent inhibitor of PP1c (Fig. 2A).
Fig. 2.
Inhibition of blue light-dependent stomatal opening by expression of mutant forms of PP1c in guard cells. (A) Protein phosphatase activity of VfPP1c-1 and VfPP1c-1-H137N prepared from E. coli cells. Phosphatase activity was measured by using [32P]-labeled myelin basic protein in the absence (control) or presence of 10 nM tautomycin. Each value was expressed as percentage of VfPP1c-1 (55,498 ± 2,914 nmol·min−1·mg−1 protein). Data are means ± SE (n = 3). (B) Images of guard cells in the bright field (a, c, e, and g) and fluorescence microscope (b, d, f, and h). Typical cases of guard cells transformed with VfPP1c-1:sGFP (a–d) or VfPP1c-1-H137N:sGFP (e–h) are shown. Gene constructs were introduced into Vicia guard cells by particle bombardment. Epidermal strips were detached and preincubated for 2.5 h in darkness, and then illuminated by red light (RL, 150 μmol·m−2·s−1) with or without blue light (BL, 10 μmol·m−2·s−1) for 2.5 h. (Scale bar, 10 μm.) (C and D) Half stomatal apertures measured next to transformed and nontransformed guard cells. Results are means ± SE (n = 45, pooled from triplicate experiments) from VfPP1c-1:sGFP or VfPP1c-1-H137N:sGFP (C) and VfPP1c-3:sGFP or VfPP1c-3-H121N:sGFP (D) transformants. Asterisks indicate significant differences between nontransformed and transformed guard cells (P < 0.01).
We then transiently expressed constructs in which sGFP was fused to wild-type VfPP1c-1 (VfPP1c-1:sGFP) or VfPP1c-1-H137N (VfPP1c-1-H137N:sGFP). Transformed guard cells were identified by their green fluorescence in isolated epidermal peels. We measured half apertures in stomata formed by one transformed and one nontransformed guard cell (Fig. 2 B and C). When the epidermal peels were irradiated with red light at 150 μmol·m−2·s−1, stomata opened slightly as a result of identical activities of nontransformed and transformed guard cells (Fig. 2C). When the epidermal peels were irrdiated additionally with weak blue light at 10 μmol·m−2·s−1, nontransformed guard cells reacted strongly to increase the aperture (left cells in Fig. 2 B c and g and C), whereas transformed guard cells that overexpressed VfPP1c-1-H137N:sGFP (right cell in Fig. 2 Bg and C) did not. Guard cells expressing VfPP1c-1:sGFP (right cell in Fig. 2 Bc and C) behaved like the nontransformed wild type. Similar inhibition of blue light-dependent stomatal opening was observed when guard cells were transformed with the PP1c mutant VfPP1c-3-H121N:sGFP (Fig. 2D). These results indicated that PP1c mutants expressed in guard cells caused dominant negative effects and inhibited the blue light signaling pathway.
Transformation of Vicia Guard Cells with Human Inhibitor-2 Inhibits Stomatal Opening by Blue Light.
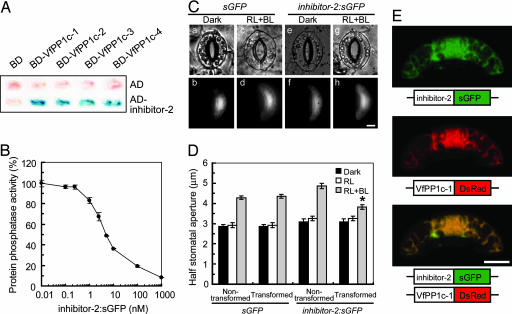
We tested the role of PP1 in blue light-dependent stomatal opening by transiently overexpressing human inhibitor-2, a protein that specifically binds and inhibits PP1c. Human inhibitor-2 was used because no corresponding genes have been identified in plants so far. We examined the interactions of human inhibitor-2 with all VfPP1cs-1 to -4 in a yeast two-hybrid assay, and observed β-galactosidase activities only when inhibitor-2 and PP1cs were coexpressed in yeast (Fig. 3A). As a control, inhibitor-2 did not interact with the catalytic subunit of PP2A under the same conditions (data not shown), suggesting that the interaction between inhibitor-2 and PP1c from Vicia was specific. Moreover, inhibitor-2:sGFP inhibited the activity of VfPP1c-1 in vitro with the half-inhibitory concentration (IC50) ≈ 5 nM (Fig. 3B).
Fig. 3.
Inhibition of blue light-dependent stomatal opening by transformation of guard cells with inhibitor-2. (A) Interaction of Vicia PP1cs with inhibitor-2 determined by yeast two-hybrid assay. β-Galactosidase activity was measured by filter assay using X-gal as a substrate. (B) Inhibition of VfPP1c-1 activity by inhibitor-2:sGFP. Experimental conditions were the same as in Fig. 2A. Phosphatase activities are expressed as percentages of that measured without inhibitor-2:sGFP (65,760 ± 2,418 nmol min−1·mg−1 protein). Data are means ± SE (n = 3). (C) Images of guard cells in the bright field (a, c, e, and g) and fluorescence microscope (b, d, f, and h). Typical guard cells transformed with sGFP (a–d) and inhibitor-2:sGFP (e–h) are shown. Other experimental conditions are the same as in Fig. 2B. (Scale bar, 10 μm.) (D) Determination of half stomatal apertures of guard cells transformed with sGFP and inhibitor-2:sGFP as well as of nontransformed neighboring guard cells. Data represent means ± SE (n = 45, pooled from triplicate experiments). Asterisk indicates significant difference between nontransformed and transformed guard cells (P < 0.01). (E) Subcellular localization of inhibitor-2 and VfPP1c-1. Gene constructs encoding inhibitor-2:sGFP and VfPP1c-1:DsRed were cotransformed into Vicia guard cells by particle bombardment, and fluorescent images were obtained by confocal laser-scanning microscopy. (Scale bar, 10 μm.)
We next examined the in vivo effects of inhibitor-2 on blue light-dependent stomatal opening. Guard cells were transformed with inhibitor-2:sGFP by particle bombardment. When the epidermal peels of nontransformed and transformed guard cells were irradiated with red light, stomata opened slightly. When the peels were irradiated with additional weak blue light, nontransformed cells and cells transformed with sGFP alone responded by increasing the aperture of their stoma (Fig. 3 Cc and D). However, stomatal opening was suppressed in guard cells carrying inhibitor-2:sGFP (Fig. 3 Cg and D).
If the effects observed in vivo were actually caused by the interaction between inhibitor-2 and PP1c, the subcellular localization of these binding partners should overlap, at least partly. We coexpressed inhibitor-2:sGFP and VfPP1c-1 fused to red fluorescent protein from Discosoma sp. (VfPP1c-1:DsRed) in guard cells, and inspected the subcellular localization of these two fusion proteins by confocal laser-scanning microscopy. As shown in Fig. 3E, both inhibitor-2:sGFP and VfPP1c-1:DsRed were distributed uniformly in the cytosol and nucleus. Merged images revealed that green fluorescence overlaid exactly with red fluorescence.
Stomata Consisting of Guard Cells Transformed with Mutated PP1c and Inhibitor-2 Open in Response to Fusicoccin, an H+-ATPase Activator.
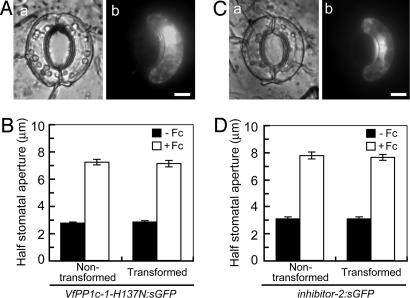
The inhibition of blue light-dependent stomatal opening by mutated PP1c or inhibitor-2 suggested that PP1 mediated the physiological response. However, it is possible that the transformation of the guard cells impaired downstream mechanisms necessary for stomatal opening, including voltage-gated K+ uptake channels, anion transporters, the production of organic anions, or ATP production. To test this possibility, we administered fusicoccin, a fungal toxin that activates the plasma membrane H+-ATPase, to transformed epidermal peels and measured stomatal opening. As shown in Fig. 4, fusicoccin acted on transformed guard cells (fluorescent cells on the right) similarly as on nontransformed guard cells (nonfluorescent cells on the left), suggesting that downstream components responsible for stomatal opening had remained operational in the transformants.
Fig. 4.
Response to fusicoccin of guard cells transformed with PP1c mutant and inhibitor-2. (A and C) Images of guard cells under bright field (a) and fluorescence conditions (b). Typical guard cells transformed with VfPP1c-1-H137N:sGFP (A) and inhibitor-2:sGFP (C) are shown. Epidermal strips were preincubated for 2.5 h in darkness before fusicoccin (10 μM) was added; images were taken after further incubation for 2.5 h in darkness. (Scale bars, 10 μm.) (B and D) Determination of half stomatal apertures of guard cells transformed with VfPP1c-1-H137N:sGFP (B) or inhibitor-2:sGFP (D). Data are means ± SE (n = 45, pooled from triplicate experiments).
Tautomycin Inhibits Phosphorylation of the Plasma Membrane H+-ATPase Without Affecting Phosphorylation of Phototropins in Response to Blue Light.
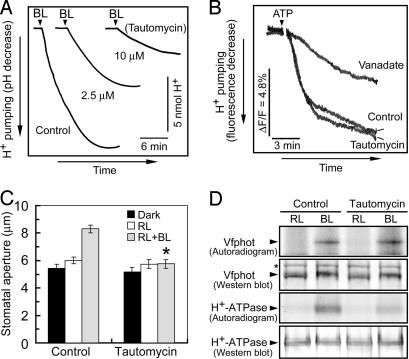
PP1 is most likely to mediate signaling between phototropin and the plasma membrane H+-ATPase (15). To test this, we used tautomycin, a membrane-permeable inhibitor of PP1, which is used frequently to investigate the role of PP1 in various cellular events; because it inhibits PP2A with 10-fold lower sensitivity in vitro (25–28), it can help to distinguish between responses mediated by the two classes of enzymes. We expected that blue light-dependent H+ pumping and stomatal opening were inhibited by this reagent. In fact, blue light-dependent H+ pumping in guard cell protoplasts was decreased by tautomycin in a concentration-dependent manner (Fig. 5A). The degree of inhibition increased with the time of exposure of guard cell protoplasts to tautomycin. Tautomycin is unlikely to affect the plasma membrane H+-ATPase itself, because tautomycin had no effect on ATP-dependent and vanadate-sensitive H+ pumping in microsomal membranes isolated from guard cell protoplasts (Fig. 5B). Furthermore, blue light-dependent stomatal opening in epidermal strips was abolished by tautomycin (Fig. 5C). Complete inhibition of the opening is probably due to the long incubation time of the epidermis with tautomycin (see Materials and Methods), which allows sufficient permeation of tautomycin into guard cells in the epidermis. Tautomycin did not inhibit stomatal opening in the epidermis by red light, suggesting that effect of the reagent was specific to blue light signaling.
Fig. 5.
Effects of tautomycin on H+ pumping, stomatal opening, and the phosphorylation of Vfphots and the plasma membrane H+-ATPase in response to blue light. (A) Inhibition of blue light-dependent H+ pumping in Vicia guard cell protoplasts by tautomycin. Guard cell protoplasts were preincubated for 30 min under the red light (RL, 600 μmol·m−2·s−1), and then were exposed to 10 μM tautomycin for 2 h. Afterward, a pulse of blue light (BL, 100 μmol·m−2·s−1, 30 s) was applied at the times indicated by vertical arrowheads. (B) Effect of tautomycin on ATP-dependent H+ pumping in microsomal vesicles from guard cell protoplasts. The basal reaction mixture (250 μl) contained membrane vesicles (10 μg of protein), 10 mM Mops-KOH (pH 7.0), 0.25 M mannitol, 5 mM MgCl2, 1 mM EGTA, 50 mM KNO3, 5 μg·ml−1 oligomycin, and 1 μM quinacrine. Vanadate at 100 μM and tautomycin at 10 μM were added. ΔF/F, change in fluorescence divided by the initial fluorescence. (C) Inhibition of blue light-dependent stomatal opening by tautomycin. Epidermal strips were preincubated for 2.5 h in darkness with 10 μM tautomycin, and then illuminated by red light (RL, 150 μmol·m−2·s−1) with or without blue light (BL, 10 μmol·m−2·s−1) for 2.5 h. Data represent means ± SE (n = 65, pooled from triplicate experiments). Asterisk indicates significant difference between control and tautomycin treatments (P < 0.01). (D) Levels of phosphorylation and the amounts of Vfphot and the H+-ATPase. Guard cell protoplasts were treated as described for Fig. 5A with [32P]orthophosphate. Autoradiography was carried out on Vfphots and H+-ATPase, which were isolated by immunoprecipitation from 200 and 100 μg of guard cell proteins, respectively. Western blotting of Vfphot and the H+-ATPase was performed by using polyclonal antibodies raised against to individual protein. Asterisk indicates a nonspecific protein. Experiments repeated on two occasions gave similar results.
PP1 is most likely to function upstream of the H+-ATPase and transmits the light signal from phototropins to the H+-ATPase for its activation. If so, inhibition of PP1 would suppress blue light-dependent phosphorylation of the H+-ATPase without affecting phosphorylation of the phototropins. We determined the phosphorylation levels of both phototropins and the H+-ATPase in response to blue light. Vfphots (V. faba phototropins) and H+-ATPase were immediately isolated by immunoprecipitation at the indicated times after the light pulse (7), and separated on SDS/PAGE. Autoradiograms revealed that Vfphots and the H+-ATPase were phosphorylated in response to blue light (Fig. 5D). In the presence of tautomycin, the phosphorylation of Vfphots was not affected, but that of the H+-ATPase was strongly inhibited. Western blot analysis revealed that there was no change in the levels of Vfphots and the H+-ATPase. Mobility shift of Vfphot to the upper site by blue light strengthened the occurrence of the phosphorylation in the presence of tautomycin. These results indicated that PP1 functions downstream of phototropins and upstream of the H+-ATPase in the blue light signaling pathway of guard cells.
Discussion
In this study, we demonstrated that PP1 mediates the signal transduction from phototropins to the plasma membrane H+-ATPase in guard cells. Despite the extensive progress in our understanding of the functions of PP1 in animal cells, functional analysis of plant PP1 has been difficult (16, 29). However, the physiological roles of several members of the PPP family of protein phosphatases in plants have been recently identified by using Arabidopsis transgenic plants with loss- or gain-of-function mutants. For example, PP5 and PP6 have been demonstrated to modulate phytochrome-mediated signaling by dephosphorylating the Pfr form in the phosphorylated state (30, 31). PP7 has been shown to act as a positive regulator in blue light signaling mediated by cryptochromes (32). In these analyses, Arabidopsis T-DNA insertional lines and antisense lines were excellent tools to dissect the role of individual enzymes because these catalytic subunits of protein phosphatases are encoded by one or two genes (33, 34). In sharp contrast to these particular catalytic subunits of protein phosphatases such as PP5, PP6, and PP7, there are at least nine isogenes encoding PP1c isoforms (33–35), and our preliminary results indicated that all of them were expressed in Arabidopsis guard cells (data not shown). Because these isoforms show very high amino acid sequence similarities (90.9–99.7%), these isogenes likely function redundantly in plant cells, making it difficult to characterize the function of PP1 in plant cells by genetic methods. Very high amino acid sequence similarities were obtained in PP1c of Vicia guard cells (Fig. 1A). In addition, it was difficult to obtain the Arabidopsis transformants with antisense constructs encoding PP1c (T. Aoyama, Kyoto University, personal communication), possibly because excessive reductions of PP1c might be lethal. In this study, we thus transiently transformed Vicia guard cells with dominant negative forms of PP1c, and successfully demonstrated the function of PP1 in the stomatal blue light response. However, it should be noted that we could not determine at this stage whether any particular PP1c isoform is involved in the response.
Our results indicate that PP1 transfers the signal from phototropins to the plasma membrane H+-ATPase. To substantiate this finding, we used tautomycin, a cell-permeable and preferential inhibitor of PP1, and investigated the effect of tautomycin on phosphorylation of both phototropins and the plasma membrane H+-ATPase in guard cell protoplasts. When guard cell protoplasts were stimulated with a pulse of blue light in the presence of 10 μM tautomycin, phototropins underwent phosphorylation but the plasma membrane H+-ATPase did not. Thus, PP1 action appeared to be located between phototropins and the H+-ATPase in the blue light-signaling cascade. In accord with this result, H+ pumping in guard cell protoplasts and stomatal opening in epidermal peels in response to blue light were inhibited by tautomycin at 10 μM. The high concentration of tautomycin required for inhibition may be due to low permeability of tautomycin across the plasma membrane (26). Taken together, the results demonstrated that PP1 transmits the signal from phototropins to the plasma membrane H+-ATPase in guard cells.
Because phototropins are protein kinases associated with the plasma membrane (5, 36), one might assume that phototropins can phosphorylate the H+-ATPase directly (37–39). The present results argue against this hypothesis and instead indicate the presence of another protein kinase that directly phosphorylates the H+-ATPase.
In Arabidopsis guard cells, phot1 and phot2 induce the activation of the plasma membrane H+-ATPase by blue light and mediate stomatal opening in a redundant manner (6). Although the signals from phot1 and phot2 are ultimately transduced to the H+-ATPase, it is unclear where such separate signals are converged on the signaling pathways. Recent investigations have shown that both NPH3 (NONPHOTOTROPIC HYPOCOTYL3) and RPT2 interact directly with phot1, but not with phot2, and that these elements are required for phot1-dependent phototropism (14, 40, 41). Therefore, it is likely that phot1 and phot2 transduce separate signals in the initial steps of signaling and that PP1 may function in the common pathway after the convergence point in guard cells. In support of this notion, tautomycin completely inhibited stomatal opening by blue light (Fig. 5C).
Overexpression of PP1c mutants and inhibitor-2 in guard cells suppressed blue light-dependent stomatal opening (Fig. 2 and 3) and inhibited the activity of PP1 in guard cells. However, the modes of inhibitory action of these two proteins are quite different. The dominant negative-type mutants likely compete with endogenous PP1cs for substrate and regulatory subunits and interrupt blue light signaling. In contrast, inhibitior-2 seems to compete with an endogenous regulatory subunit by binding to PP1cs, decreasing their catalytic activity and resulting in the inhibition of blue light signaling. Therefore, both the regulatory subunit and the substrate of PP1c appear to be required for blue light signaling in guard cells. Considering the biochemical properties of PP1, we hypothesize that the regulatory subunit of PP1 is modified by upstream components in response to blue light. One possible candidate for this upstream component is phototropin kinase itself (5, 36). We note that activities of some of the PP1 regulatory subunits in animals, including inhibitor-2, neurabin I, and NIPP1, are regulated by reversible phosphorylation (17). Another possibility is that phototropins directly activate PP1c as a regulatory subunit.
Phototropins mediate a wide range of physiological responses that optimize photosynthesis (42, 43), including phototropism (4), chloroplast movement (36, 44, 45), leaf expansion (36, 46), leaf movement (47), and stomatal opening. Further investigations are needed to clarify whether PP1 is a common signaling component in all of these phototropin-mediated processes.
In conclusion, we have identified PP1 as a component of the blue light signaling pathway leading from phototropins to the plasma membrane H+-ATPase in guard cells. To our knowledge, this is the first demonstration of a functional role of PP1 in plant cells (16).
Materials and Methods
Plant Materials and Isolation of Guard Cell Protoplasts.
V. faba (cv. Ryosai Issun) was cultured hydroponically in a green house as described (48). Guard cell protoplasts were obtained enzymatically from 5- to 6-week-old leaves as detailed (8).
Cloning of PP1c Genes from Vicia Guard Cells.
Two degenerate oligonucleotide primer pairs, (5′-TGYGGNGAYATHCAYGG-3′ and 5′-CAYTCRTCRTARAANCCRTA-3′) and (5′-GARYTNGARGCNCCNATHAAR-3′ and 5′-CATRCANAGDATYTTRTCRTC-3′), were designed on the basis of conserved amino acid sequences in PP1c. Partial sequences were obtained by RT-PCR using total RNA (5 μg) prepared from Vicia guard cell protoplasts. Full-length sequences of VfPP1c-1 (AB038648), VfPP1c-2 (AB254850), VfPP1c-3 (AB254851), and VfPP1c-4 (AB254852) were determined by inverse PCR or 5′ and 3′ RACE.
Transient Expression of sGFP Fusion Proteins in Vicia Guard Cells and Measurement of Stomatal Aperture.
Individual cDNA was cloned into CaMV35SΩ-sGFP(S65T)-nos3′ (49) and introduced into Vicia leaves by particle bombardment (PDS-1000/He particle-delivery system; Bio-Rad, Hercules, CA). Stomatal aperture was determined from epidermal strips prepared from the abaxial side of leaves. Detailed conditions are described in Supporting Text, which is published as supporting information on the PNAS web site.
Preparation of Recombinant PP1c in Escherichia coli and Protein Phosphatase Assay.
Full-length VfPP1c-1 and VfPP1c-1-H137N were subcloned into pGEX-2T (Amersham Pharmacia, Tokyo, Japan), and transformed into E. coli strain BL21. The recombinant proteins were produced and purified according to Watanabe et al. (50) and as described in Supporting Text. Ser/Thr phosphatase activity was assayed according to the manufacturer’s protocol (New England Biolabs, Beverly, MA).
Yeast Two-Hybrid Assay.
Full-length cDNA clones of Vicia PP1cs and inhibitor-2 were inserted into pAS2–1 or pACT2 vectors (Clontech, Palo Alto, CA), and transformed into the Saccharomyces cerevisiae strain Y190. β-Galactosidase filter assays were carried out by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as a substrate.
Measurement of Blue Light- and ATP-Dependent H+ Pumping.
Blue light-dependent H+ pumping from guard cell protoplasts was measured as described (48). Isolation of microsomal membrane from guard cell protoplasts and measurement of ATP-dependent H+ pumping was performed as described (51).
Phosphorylation Levels and Amounts of Vfphots and the H+-ATPase.
Levels of phosphorylation and amounts of Vfphots and H+-ATPase were determined as described (7, 8) with slight modification. To determine in vivo phosphorylation, guard cell protoplasts were labeled with [32P]orthophosphate. Amounts of Vfphots and the H+-ATPase were determined immunologically. A detailed description is provided in Supporting Text.
Supplementary Material
Acknowledgments
We thank Xiao Zhang (Department of Biology, Henan University, Henan, China) for help with H+ pumping measurements. This research was partially supported by Japanese Ministry of Education, Science, Sports, and Culture Grants-in-Aid for Scientific Research on Priority Areas 17084005, Scientific Research (A) 16207003 (to K.-i.S.), and Young Scientists (A) 14704003 (to T.K.).
Abbreviations
- phot
phototropin
- PP1
type 1 protein phosphatase
- PP1c
catalytic subunit of PP1
- VfPP1c
V. faba PP1c.
Footnotes
References
- 1.Zeiger E. Annu. Rev. Plant Physiol. 1983;34:441–475. [Google Scholar]
- 2.Assmann S. M., Shimazaki K. Plant Physiol. 1999;119:809–815. doi: 10.1104/pp.119.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder J. I., Allen G. J., Hugouvieux V., Kwak J. M., Waner D. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 4.Huala E., Oeller P. W., Liscum E., Han I. S., Larsen E., Briggs W. R. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 5.Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., Briggs W. R. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M., Shimazaki K. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- 7.Kinoshita T., Emi T., Tominaga M., Sakamoto K., Shigenaga A., Doi M., Shimazaki K. Plant Physiol. 2003;133:1453–1463. doi: 10.1104/pp.103.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita T., Shimazaki K. EMBO J. 1999;18:5548–5558. doi: 10.1093/emboj/18.20.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assmann S. M., Simoncini L., Schroeder J. I. Nature. 1985;318:285–287. [Google Scholar]
- 10.Shimazaki K., Iino M., Zeiger E. Nature. 1986;319:324–326. [Google Scholar]
- 11.Schroeder J. I., Raschke K., Neher E. Proc. Natl. Acad. Sci. USA. 1987;84:4108–4112. doi: 10.1073/pnas.84.12.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimazaki K., Goh C.-H., Kinoshita T. Physiol. Plantarum. 1999;105:554–561. [Google Scholar]
- 13.Svennelid F., Olsson A., Piotrowski M., Rosenquist M., Ottman C., Larsson C., Oecking C., Sommarin M. Plant Cell. 1999;11:2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inada S., Ohgishi M., Mayama T., Okada K., Sakai T. Plant Cell. 2004;16:887–896. doi: 10.1105/tpc.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita T., Shimazaki K. Plant Cell Physiol. 1997;38:1281–1285. [Google Scholar]
- 16.Luan S. Annu. Rev. Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 17.Ceulemans H., Bollen M. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 18.Bollen M. Trends Biochem. Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 19.Cohen P. T. W. J. Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 20.Evans D. R. H., Myles T., Hofsteenge J., Hemmings B. A. J. Biol. Chem. 1999;274:24038–24046. doi: 10.1074/jbc.274.34.24038. [DOI] [PubMed] [Google Scholar]
- 21.Lizotte D. L., McManus D. D., Cohen H. R., DeLong A. Gene. 1999;234:35–44. doi: 10.1016/s0378-1119(99)00188-2. [DOI] [PubMed] [Google Scholar]
- 22.Bennett D., Szöor B., Gross S., Vereshchagina N., Alphey L. Genetics. 2003;164:235–245. doi: 10.1093/genetics/164.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan L.-M., Zhao Z., Assmann S. M. Curr. Opin. Plant Biol. 2004;7:537–547. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Egloff M.-P., Cohen P. T. W., Reinemer P., Barford D. J. Mol. Biol. 1995;254:942–959. doi: 10.1006/jmbi.1995.0667. [DOI] [PubMed] [Google Scholar]
- 25.MacKintosh C., Klumpp S. FEBS Lett. 1990;277:137–140. doi: 10.1016/0014-5793(90)80828-7. [DOI] [PubMed] [Google Scholar]
- 26.Favre B., Turowski P., Hemmings B. A. J. Biol. Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- 27.Resjö S., Oknianska A., Zolnierowicz S., Manganiello V., Degerman E. Biochem. J. 1999;341:839–845. [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y., Mumby M. C. J. Biol. Chem. 1999;274:31917–31924. doi: 10.1074/jbc.274.45.31917. [DOI] [PubMed] [Google Scholar]
- 29.Smith R. D., Walker J. C. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:101–125. doi: 10.1146/annurev.arplant.47.1.101. [DOI] [PubMed] [Google Scholar]
- 30.Kim D.-H., Kang J.-G., Yang S.-S., Chung K.-S., Song P.-S., Park C.-M. Plant Cell. 2002;14:3043–3056. doi: 10.1105/tpc.005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu J. S., Kim J.-I., Kunkel T., Kim B. C., Cho D. S., Hong S. H., Kim S.-H., Fernández A. P., Kim Y., Alonso J. M., et al. Cell. 2005;120:395–406. doi: 10.1016/j.cell.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Møller S. G., Kim Y.-S., Kunkel T., Chua N.-H. Plant Cell. 2003;15:1111–1119. doi: 10.1105/tpc.008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arabidopsis Genome Initiative. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 34.Kerk D., Bulgrien J., Smith D. W., Barsam B., Veretnik S., Gribskov M. Plant Physiol. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Q., Li J., Smith R. D., Walker J. C. Plant Mol. Biol. 1998;37:471–481. doi: 10.1023/a:1005912413555. [DOI] [PubMed] [Google Scholar]
- 36.Sakai T., Kagawa T., Kasahara M., Swartz T. E., Christie J. M., Briggs W. R., Wada M., Okada K. Proc. Natl. Acad. Sci. USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin C. Plant Cell. 2002;14(Suppl.):S207–S225. doi: 10.1105/tpc.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liscum E., Hodgson D. W., Campbell T. J. Plant Physiol. 2003;133:1429–1436. doi: 10.1104/pp.103.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M., Chory J., Fankhauser C. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 40.Motchoulski A., Liscum E. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- 41.Sakai T., Wada T., Ishiguro S., Okada K. Plant Cell. 2000;12:225–236. doi: 10.1105/tpc.12.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briggs W. R., Christie J. M. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 43.Takemiya A., Inoue S., Doi M., Kinoshita T., Shimazaki K. Plant Cell. 2005;17:1120–1127. doi: 10.1105/tpc.104.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarillo J. A., Gabrys H., Capel J., Alonso J. M., Ecker J. R., Cashmore A. R. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 45.Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S., Kato T., Tabata S., Okada K., Wada M. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto K., Briggs W. R. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue S., Kinoshita T., Shimazaki K. Plant Physiol. 2005;138:1994–2004. doi: 10.1104/pp.105.062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimazaki K., Kinoshita T., Nishimura M. Plant Physiol. 1992;99:1416–1421. doi: 10.1104/pp.99.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe T., da Cruz e Silva E. F., Huang H.-B., Starkova N., Kwon Y.-G., Horiuchi A., Greengard P., Nairn A. C. Methods Enzymol. 2003;366:321–338. doi: 10.1016/s0076-6879(03)66024-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X., Wang H., Takemiya A., Song C. P., Kinoshita T., Shimazaki K. Plant Physiol. 2004;136:4150–4158. doi: 10.1104/pp.104.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.