Abstract
Chromosome cohesion and condensation are essential prerequisites of proper segregation of genomes during mitosis and meiosis, and are supported by two structurally related protein complexes, cohesin and condensin, respectively. At the core of the two complexes lie members of the structural maintenance of chromosomes (SMC) family of ATPases. SMC proteins are also found in most bacterial and archaeal species, implicating the existence of an evolutionarily conserved theme of higher-order chromosome organization and dynamics. SMC dimers adopt a two-armed structure with an ATP-binding cassette (ABC)-like domain at the distal end of each arm. This article reviews recent work on the bacterial and eukaryotic SMC protein complexes, and discusses current understanding of how these uniquely designed protein machines may work at a mechanistic level. It seems most likely that the action of SMC proteins is highly dynamic and plastic, possibly involving a diverse array of intramolecular and intermolecular protein–protein interactions.
Keywords: SMC proteins, condensin, cohesin, chromosome segregation, ABC ATPases
1. Introduction
The duplication and segregation of genomes are two of the most fundamental events for cell reproduction. Historically, the molecular mechanism underlying genome duplication was first studied in bacterial cells and was later explored in eukaryotic cells. Despite apparent differences in the complexity and variations, it appears that the basic theme of DNA replication is shared between bacteria and humans. In retrospect, this concept was intuitively apparent when the complementary nature of the DNA double helix was first discovered. In contrast, the double helix structure did not immediately tell us how the duplicated DNA molecules might be segregated from each other. Therefore, it was possible that bacteria and eukaryotic cells use completely different ‘molecular logic’ of chromosome segregation. In fact, eukaryotic cells assemble a highly elaborate structure called the mitotic spindle to segregate chromosomes to opposite poles of the cell, whereas bacterial cells apparently lack such a structure (figure 1). However, recent technical advancements in cell imaging, combined with powerful bacterial genetics, have started to uncover a number of similarities in the machineries and mechanisms of chromosome segregation between bacteria and eukaryotes (Sherratt 2003). In particular, the discovery of structural maintenance of chromosomes (SMC) proteins as major chromosome organizers raises the intriguing possibility that most (if not all) organisms may share a common theme of chromosome organization at the most fundamental level (Nasmyth 2001; Hirano 2002). This paper reviews recent progress in our understanding of the structure and function of SMC proteins with a major focus on their mechanistic actions. It will attempt to deduce the ‘basics’ of SMC actions that regulate a diverse range of higher-order chromosome dynamics from bacteria to humans.
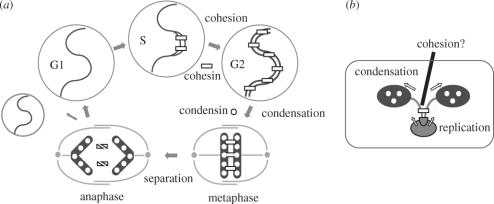
Figure 1.
Chromosome segregation in eukaryotes and bacteria. (a) In eukaryotic cells, the linkage between duplicated DNA molecules (cohesion) is established during S phase and maintained throughout G2 phase. The cohesin complex (indicated by rectangles) plays a central role in this process. At the onset of mitosis, most cohesin dissociates from chromosomes and is replaced with the condensin complex (indicated by circles), leading to the formation of metaphase chromosomes (condensation). A small amount of cohesin left on the chromosome is sufficient to hold the sister chromatids together. Proteolytic cleavage of cohesin subunits promotes final separation of sister chromatids at anaphase, allowing them to be pulled apart to opposite poles of the cell. (b) In bacterial cells, duplication and segregation of chromosomes occur simultaneously. The SMC protein may promote segregation by pulling and condensing duplicated chromosomes at opposite poles of the cell. It remains controversial whether there may be a process corresponding to cohesion in the bacterial chromosome cycle.
2. SMC proteins and their conserved roles in chromosome segregation
Eukaryotic cells have at least six different SMC proteins in individual species (Cobbe & Heck 2003). Each of them has a specific binding partner, thereby creating three different heterodimers. SMC2 and SMC4 act as the core of the condensin complex that plays a central role in chromosome assembly and segregation (figure 1a; Hirano & Mitchison 1994; Saka et al. 1994; Hirano et al. 1997; Sutani et al. 1999), whereas SMC1 and SMC3 function as the core of the cohesin complex essential for sister chromatid cohesion (figure 1a; Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998). SMC5 and SMC6 form another heterodimer implicated in DNA repair and checkpoint responses (Fousteri & Lehmann 2000; Taylor et al. 2001). Most archaea and bacteria have a single SMC protein in individual species, which forms a homodimer (Hirano & Hirano 1998; Melby et al. 1998). In a subclass of Gram-negative bacteria including Escherichia coli, SMC is replaced with its distant relative called MukB (Hiraga 2000). In bacterial cells, SMC/MukB may contribute to chromosome segregation by ‘pulling’ duplicated DNA strands to opposite poles of the cell (figure 1b; Britton et al. 1998; Moriya et al. 1998; Lindow et al. 2002; Volkov et al. 2003). Although it remains controversial whether there may be a cohesion process in the bacterial chromosome cycle, there is evidence that MukB may also be involved in holding duplicated origin-proximal regions together in E. coli (Sunako et al. 2001). Therefore, at present, it is not safe to assume that the bacterial SMC/MukB protein is the functional counterpart of the eukaryotic condensin complex. Conceivably, the bacterial protein may be regarded as the common ancestor of the eukaryotic SMC protein complexes, and that condensin and cohesin acquired their specialized biochemical and cellular functions during evolution.
3. Conserved architecture of SMC protein complexes
Substantial efforts have been made to address the mechanistic actions of the eukaryotic condensin and cohesin complexes, and to compare and contrast their biochemical activities in vitro. As judged by electron microscopy, condensin and cohesin share the V-shaped, two-armed structure characteristic of SMC proteins; however, their conformations are drastically different (Anderson et al. 2002). For example, the hinge of condensin is closed and the coiled-coil arms are placed close together. In contrast, the hinge of cohesin is largely open and the coiled coils are spread from each other. Consistent with their conformational differences, the two complexes display distinct sets of biochemical activities in vitro (Losada & Hirano 2001; Sakai et al. 2003). Although these findings are crucial to our understanding of the specialized functions of condensin and cohesin, the apparent differences between the two complexes preclude us from deducing the ‘basic’ mechanism of action of SMC proteins. For this reason, we started structural and functional characterization of the homodimeric SMC protein from the Gram-positive bacterium Bacillus subtilis (Hirano & Hirano 1998, 2002; Hirano et al. 2001). We anticipated that the bacterial SMC protein would provide us with an excellent opportunity to explore fundamental properties that may be shared by condensin and cohesin. We assumed that any information obtained from the analysis of this bacterial protein would be applicable to our understanding of the more sophisticated eukaryotic protein complexes.
Recent studies in genetic and bioinformatics studies identified two proteins called ScpA and ScpB that might function together with SMC in B. subtilis (Mascarenhas et al. 2002; Soppa et al. 2002). We first attempted to reconstitute this putative complex from highly purified individual components (Hirano & Hirano 2004). As judged by sucrose gradient centrifugation, purified ScpA and ScpB are present as monomers (approximately 2.6S) and dimers (approximately 3.5S), respectively. SMC forms a V-shaped dimer with a sedimentation coefficient of approximately 6.5S. When the three subunits are mixed together, they form a stable complex with a sedimentation coefficient of approximately 8.6S. It is most likely that a single complex contains two copies of each subunit. Co-immunoprecipitation experiments show that ScpA binds directly to SMC and bridges the interaction between ScpB and SMC. Additional work using a panel of depletion mutants shows that the major binding site of ScpA lies in the head domain of SMC. A similar conclusion was obtained independently from a study using yeast two-hybrid interaction assays (Dervyn et al. 2004).
The molecular architecture of the bacterial SMC complex is very similar to that of the eukaryotic SMC protein complexes (figure 2). ScpA belongs to the kleisin superfamily of proteins, which also contains the Scc1 subunit of cohesin and the CAP-H subunit of condensin (Schleiffer et al. 2003). In the case of cohesin, Scc1 binds to the SMC head domains and mediates the interaction between Scc3 and SMC (Haering et al. 2002). Less is known about the order of subunit interactions in the condensin complex except that the non-SMC subcomplex binds to the SMC heads (Anderson et al. 2002; Yoshimura et al. 2002). It is of great interest to consider how the SMC protein complexes evolved from the symmetric structure to the asymmetric complexes and acquired more sophisticated and elaborated functions. In fact, such evolution from symmetric to asymmetric structures is often found in many DNA transaction proteins including DNA mismatch repair proteins (Schofield & Hsieh 2003).
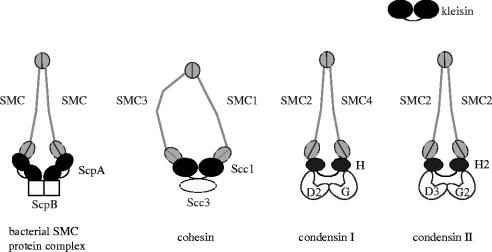
Figure 2.
SMC protein complexes in eukaryotes and bacteria. The B. subtilis SMC protein complex is composed of an SMC homodimer and two regulatory subunits (ScpA and ScpB). The eukaryotic cohesin complex contains a heterodimeric pair of SMC1 and SMC3 and two non-SMC subunits (Scc1 and Scc3). Vertebrate cells possess two different condensin complexes, known as condensin I and condensin II. The two condensin complexes share the same SMC core subunits (SMC2 and SMC4) but differ by their unique sets of non-SMC subunits (CAP-D2, -G, -H for condensin I; CAP-D3, -G2, -H2 for condensin II). Among the non-SMC subunits of these protein complexes, ScpA, Scc1, CAP-H and CAP-H2 are distantly related with each other, belonging to the kleisin superfamily of proteins.
4. ATP-driven, head–head engagement modulates SMC–DNA interactions
The ATP-binding head domain of SMC is composed of the N-terminal and C-terminal sequences and is structurally related to the nucleotide-binding domain (NBD) of ATP-binding cassette (ABC) transporters (Saitoh et al. 1994; Lowe et al. 2001). Accumulating lines of evidence suggest that, like ABC transporters, two ATP molecules are sandwiched between two SMC head domains and that ATP binding and hydrolysis modulate the cycle of engagement (association) and disengagement (dissociation; Hirano et al. 2001; Arumugam et al. 2003; Weitzer et al. 2003; Hirano & Hirano 2004). In solution there is no evidence that SMC dimers may oligomerize regardless of the presence or absence of ATP. Therefore, ATP binding would induce intramolecular engagement of the two head domains and close the arms, whereas ATP hydrolysis would trigger their disengagement and lead to opening of the arms. In contrast, a significant body of evidence suggests that engagement of two head domains from different SMC dimers (intermolecular engagement) might occur in the presence of DNA (Hirano et al. 2001; Hirano & Hirano 2004). Characterization of SMC–SMC interactions on DNA is likely to be the key to our understanding of the dynamic and diverse actions of this class of proteins (see below).
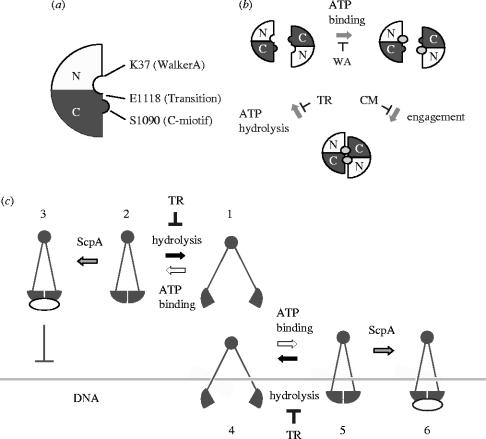
To dissect the mechanochemical cycle of SMC proteins, we have introduced mutations into the B. subtilis SMC protein that block its ATPase cycle at three different stages (figure 3a,b). A mutation in the Walker A motif (K37I) blocks ATP binding. A mutation in the C-motif (S1090R) allows ATP binding but blocks ATP-driven engagement (Hirano et al. 2001). Finally, a so-called transition-state mutation (E1118Q) stabilizes the engaged state by slowing down ATP hydrolysis (Hirano & Hirano 2004). As judged by a simple gel-shift assay, ATP has little impact on double-stranded DNA (dsDNA) binding by wild-type SMC, and the Walker A- and C-motif mutants behave similarly. In contrast, however, the transition-state mutant protein displays a striking ATP-stimulated dsDNA-binding activity. Thus, the transition-state mutation makes the SMC head domains ‘sticky’ in an ATP-dependent manner, and this stickiness has great impacts on the interaction of SMC with dsDNA.
Figure 3.
The mechano-chemical cycle of SMC proteins. (a) The ATP-binding head domain of SMC proteins is composed of the N-terminal (N) and C-terminal (C) sequences. The positions of three key residues in the B. subtilis SMC protein are shown. (b) ATP binding and hydrolysis trigger engagement and disengagement of two SMC head domains, respectively. The Walker A mutation K37I (WA) blocks the ATP-binding step. The C-motif mutation S1090R (CM) allows ATP binding but interferes with the engagement step. The transition-state mutation E1118Q (TR) stabilizes the engaged state by slowing down hydrolysis of the bound ATP molecules. (c) A model of how ATP and ScpA might modulate SMC–DNA interactions both negatively and positively. ATP binding induces intramolecular head–head engagement (stage 1–2). The transition-state mutation (TR) stabilizes the engaged state by slowing down the rate of ATP hydrolysis. ScpA binds to the head domains and reduces the residual level of SMC's ATPase activity (stage 3). This ‘closed’ SMC protein binds poorly to dsDNA. In contrast, when SMC is first allowed to interact with dsDNA (stage 4), the combination of the transition state mutation and ScpA results in stable DNA binding (stages 5–6). Note that only the simplest mode of SMC–DNA interactions is depicted here. The same idea is applicable to the more complex modes of SMC–DNA interactions shown in figure 4.
The dsDNA-binding activity of wild-type SMC is hardly affected by ScpA or ScpB alone, but the simultaneous addition of ScpA and ScpB severely interferes with this activity. Virtually no difference is found in the presence or absence of ATP, and the Walker A- and C-motif mutant proteins respond to ScpA and ScpB in a manner indistinguishable from wild-type SMC. In contrast, we find that ScpA greatly suppresses the ATP-stimulated DNA-binding activity of the transition-state mutant (Hirano & Hirano 2004). Interestingly, when the transition-state SMC mutant is first incubated with DNA and then ScpA is added into the reaction, ScpA no longer suppresses dsDNA binding and instead produces nucleoprotein complexes of a larger size. We also find that ScpA suppresses SMC's ATPase activity in a dose-dependent manner. The combination of ScpA and ScpB further enhances this inhibitory effect, although ScpB alone displays no inhibition. The inhibition is observed in the presence or absence of DNA, indicating that ScpA and ScpB affect both DNA-independent and DNA-stimulated ATPase activities of SMC.
Taking these results together, we propose that ScpA and ScpB regulate SMC–DNA interactions both negatively and positively and they do so in an ATP-dependent manner (Hirano & Hirano 2004). The transition-state mutation slows down ATP hydrolysis (figure 3c, stages 1 and 2) and ScpA further reduces the residual level of ATP hydrolysis triggered by this mutant protein (figure 3c, stage 3). In this way, the combination of ScpA and the transition-state mutation effectively stabilizes the engaged state and this ‘closed’ SMC displays poor DNA binding. On the other hand, when SMC is first allowed to interact with DNA (figure 3c, stage 4), the combination of ScpA and the transition-state mutation stabilizes the engaged state on the DNA, leading to stable DNA binding (figure 3c, stages 5 and 6). The action of ScpA on wild-type SMC is not easily detectable presumably because it hydrolyses ATP very quickly. Moreover, the C-motif mutant does not respond to ScpA, suggesting that ATP binding is not sufficient to see the effect of ScpA and that ATP-driven head–head engagement is essential. Thus, the transition-state mutation provides us with a powerful tool for studying how the mechanical cycle of an SMC protein is related to its catalytic cycle and is modulated by its accessory proteins.
5. Are protein–protein interactions important for the action of SMC proteins?
As discussed above, the ATP-modulated engagement/disengagement cycle of the head domains plays a critical role in modulating SMC–DNA interactions. In principle, two different modes of head–head engagement are possible. First, it would occur intramolecularly within an SMC dimer to encircle DNA strands by an ‘embrace’ mechanism (Haering et al. 2002). Alternatively, the ATP-driven head–head engagement could take place intermolecularly between different SMC dimers by a ‘hand-in-hand’ mechanism. Previous studies provide evidence that intermolecular engagement may indeed occur in the presence of DNA (Hirano et al. 2001; Hirano & Hirano 2004). For example, ATP stimulates protein–protein cross-linking of the B. subtilis SMC protein on DNA, and induces the formation of large nucleoprotein complexes that can be precipitated by low-speed centrifugation. Moreover, mutant proteins defective in ATP-binding or ATP-hydrolysis have dominant negative effects on the ATPase activity of wild-type SMC only in the presence (but not the absence) of DNA. Perhaps the most striking evidence for an ATP-dependent protein–protein interaction comes from a recent biophysical study of the E. coli MukBEF protein complex (Case et al. 2004). This study used an optical tweezers to detect real-time interactions of MukBEF with a single DNA molecule and provided evidence that MukBEF may form a robust filamentous structure on the DNA in an ATP-binding-dependent manner. Interestingly, a stretching/relaxation experiment reveals both tension-sensitive and tension-insensitive protein–protein interactions within the nucleoprotein filament. The authors postulate that the former is mediated by stalk–stalk interactions whereas the latter is supported by ATP-dependent, intermolecular head–head interactions. In fact, several biochemical studies suggest that ATP-independent protein–protein interactions may play an important role in cooperative interactions of SMC proteins with DNA (Sakai et al. 2003; Stray & Lindsley 2003; Hirano & Hirano 2004). These results strongly suggest that SMC proteins have the potential to display two different types of protein–protein interaction on DNA; namely, ATP-dependent intermolecular head–head engagement and ATP-independent stalk–stalk interaction. It will be important in the future to set up an experimental system or condition in which the two types of SMC–SMC interactions can clearly be separated from each other.
6. Characterization of the eukaryotic SMC protein complexes
What is known about the actions of the eukaryotic SMC protein complexes? On the basis of their cellular functions, it has been proposed that condensin may act as an intramolecular DNA crosslinker to fold and compact a single DNA molecule, whereas cohesin may act as an intermolecular DNA crosslinker to hold duplicated DNA molecules together (Hirano 1999; Nasmyth 2001). The dramatic differences in arm conformation and biochemical activities between the two complexes further support the idea that condensin and cohesin are structurally and functionally differentiated from each other (Losada & Hirano 2001; Anderson et al. 2002). In the case of cohesin, an attractive ‘ring’ hypothesis has been proposed in which a single cohesin complex embraces two DNA duplexes within its coiled-coil arms (Haering et al. 2002). This model explains how proteolytic cleavage of the Scc1 subunit of the complex might open the ring and thereby trigger sister chromatid separation at anaphase onset (Uhlmann et al. 1999). Despite supporting data largely from genetic studies (Gruber et al. 2003), biochemical evidence for this hypothesis remains scarce. It is possible that, besides the postulated topological linkage between the cohesin ring and DNA, protein–protein interactions also play a crucial role in stabilizing the linkage between two sister chromatids.
In the case of condensin, two in vitro assays have been devised to demonstrate that this complex is able to induce superhelical tension into DNA in an ATP-hydrolysis-dependent manner. In the first, condensin introduces positive supercoils into closed circular DNA in the presence of type I topoisomerase (Kimura & Hirano 1997; Hagstrom et al. 2002). In the second, the complex converts nicked circular DNA into specific type of knots (i.e. three-noded knots) in the presence of type II topoisomerase (Kimura et al. 1999). Both activities are under the control of cdk1-dependent, mitosis-specific phosphorylation (Kimura et al. 1998, 2001), supporting the idea that they are physiologically relevant activities that may initiate and drive mitotic chromosome assembly in vivo. Most recently, a single-DNA-molecule manipulation technique using a magnetic tweezers has shown that condensin is able to physically compact DNA in an ATP-hydrolysis-dependent manner (Strick et al. 2004). It remains to be determined whether a single condensin complex is capable of mediating the compaction reaction, as has been implicated by electron spectroscopic imaging (Bazett-Jones et al. 2002), or whether cooperative interactions of multiple condensins may be required as has been suggested from the analysis of the E. coli MukBEF complex (Case et al. 2004). In the future, it will also be of great interest to critically compare the activities of the canonical condensin complex (now referred to as condensin I) with those of a new condensin complex (condensin II) recently discovered from vertebrate cells (Ono et al. 2003; Yeong et al. 2003).
7. Towards a unified understanding of SMC actions
As discussed above, our current understanding of the mechanistic action of SMC proteins is far from being complete. Nevertheless, recent studies using a number of different approaches including biochemistry, genetics, biophysics and structural biology have started to uncover the highly dynamic behaviours of this class of chromosomal ATPases. An emerging concept is that the action of SMC proteins is highly dynamic and plastic, possibly involving a diverse array of intramolecular and intermolecular protein–protein interactions. Here, a scheme is presented to summarize the potential of these uniquely designed protein machines in which a flexible hinge connects two long coiled-coil arms with sticky ends (figure 4). SMC would be present in a closed form with two ATP molecules sandwiched between the two head domains (figure 4, stage 1). Interaction of SMC with DNA or with an ATPase-activation protein would trigger ATP hydrolysis, resulting in disengagement of the head domains. This would, in turn, allow hinge opening (figure 4, stage 2) or head flipping (figure 4, stage 3), and make it possible for the SMC dimer to associate with DNA by a ‘hooking’ mechanism. Subsequent binding to ATP on the DNA would induce head–head engagement either intramolecularly (figure 4, stages 4 and 6) or intermolecularly (figure 4, stages 5 and 7) and modulate the next level of DNA manipulations. Such manipulations may include ‘topological trapping’ of DNA that contributes either to holding two DNA duplexes together (figure 4, stage 4) or to folding a single DNA molecule into a loop (figure 4, stage 6). Finally, ATP-independent stalk–stalk interactions would lead to the formation of a higher-order structure that reorganizes and stabilizes the initial reactions (figure 4, stages 8–10). Although this scheme is drawn as if the ATP-dependent reactions precede the ATP-independent ones for simplicity, the reverse order of actions is certainly possible. It is tempting to speculate that condensin and cohesin are functionally differentiated in order to support subsets of the specific reactions depicted here. Despite the apparent complexity, this scheme provides a conceptual framework for our understanding of the dynamic and diverse behaviours of SMC proteins. Critical comparisons between the bacterial and eukaryotic SMC protein complexes will continue to be important to draw a comprehensive molecular picture of SMC-mediated chromosome dynamics and to gain further insight into the evolution of chromosome architecture.
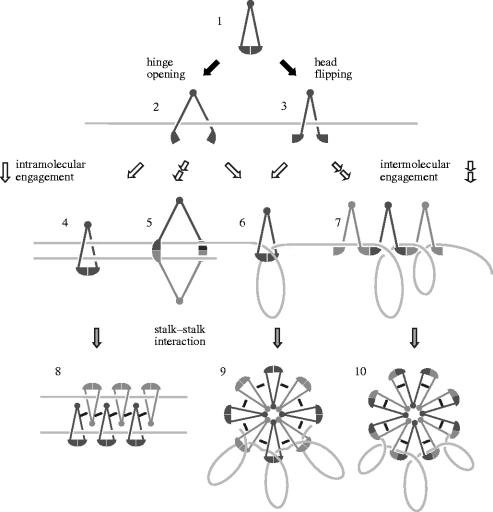
Figure 4.
Dynamic interactions of SMC proteins with DNA. SMC dimers may be present in a closed state in which two ATP molecules are sandwiched between the two head domains (stage 1). Hydrolysis of the bound ATP molecules would trigger disengagement of the head domains, thereby leading to either hinge opening (stage 2) or head flipping (stage 3). These conformational changes would allow the SMC dimer to interact with DNA by a ‘hooking’ mechanism. Subsequent ATP binding to the head domains on the DNA would promote intramolecular engagement (stages 4 and 6) or intermolecular engagement (stages 5 and 7), leading to multiple modes of SMC–DNA interactions that may include ‘topological trapping’ (e.g. stages 4 and 6). Finally, ATP-independent SMC–SMC interactions, possibly mediated by their stalk regions, would contribute to the formation of higher-order nucleoprotein complexes (stages 8–10). These dynamic actions of SMC proteins are likely to be modulated further by non-SMC regulatory subunits or other chromosomal proteins. It may be speculated that eukaryotic SMC protein complexes are functionally differentiated in order to support subsets of the specific reactions depicted here. For example, cohesin may hold two sister chromatids together by one of the mechanisms shown in the left (stages 4, 5 and 8), whereas condensin may organize DNA by one of the mechanisms shown in the right (stages 6, 7, 9 and 10).
Acknowledgments
I thank former and current members of the Hirano laboratory for their contribution to this research. I am also grateful to colleagues working in the SMC field who are always willing to share new ideas and to have stimulating discussions. The work from the author's laboratory was supported by grants from the National Institutes of Health.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Chromosome segregation’.
References
- Anderson D.E, Losada A, Erickson H.P, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering C.H, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D.P, Kimura K, Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell. 2002;9:1183–1190. doi: 10.1016/s1097-2765(02)00546-4. [DOI] [PubMed] [Google Scholar]
- Britton R.A, Lin D.C.-H, Grossman A.D. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R.B, Chang Y.P, Smith S.B, Gore J, Cozzarelli N.R, Bustamante C. The bacterial condensin MukBEF compacts DNA into a repetitive, stable structure. Science. 2004;305:222–227. doi: 10.1126/science.1098225. [DOI] [PubMed] [Google Scholar]
- Cobbe N, Heck M.M.S. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 2003;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- Dervyn E, Noirot-Gros M.-F, Mervelet P, McGovern S, Ehrlich S.D, Polard P, Noirot P. The bacterial condensin/cohesin-like protein complex acts in DNA repair and regulation of gene expression. Mol. Microbiol. 2004;51:1629–1640. doi: 10.1111/j.1365-2958.2003.03951.x. [DOI] [PubMed] [Google Scholar]
- Fousteri M.I, Lehmann A.R. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering C.H, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Hagstrom K.A, Holmes V.F, Cozzarelli N.R, Meyer B.J. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S. Dynamic localization of bacterial and plasmid chromosomes. A. Rev. Genet. 2000;34:21–59. doi: 10.1146/annurev.genet.34.1.21. [DOI] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion and repair. Genes Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Positive and negative regulation of SMC–DNA interactions by ATP and accessory proteins. EMBO J. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison T.J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hirano M, Anderson D.E, Erickson H.P, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13 S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13 S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K, Rybenkov V.V, Crisona N.J, Hirano T, Cozzarelli N.R. 13 S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Kimura K, Cuvier O, Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- Lindow J.C, Kuwano M, Moriya S, Grossman A.D. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano T. Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Curr. Biol. 2001;11:268–272. doi: 10.1016/s0960-9822(01)00066-5. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Cordell S.C, van den Ent F. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled coil inserted. J. Mol. Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov A.V, Graumann P.L. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T.E.G, Ciampaglio C.N, Briscoe G, Erickson H.P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moriya S, Tsujikawa E, Hassan A.K, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers M.P, Neuwald A.F, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I.G, Wood E.R, Earnshaw W.C. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Hizume K, Sutani T, Takeyasu K, Yanagida M. Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein–protein assembly. EMBO J. 2003;22:2764–2775. doi: 10.1093/emboj/cdg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Schofield M.J, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. A. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Sherratt D.J. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros M.-F, Oesterhelt D, Ehrlich S.D, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Stray J.E, Lindsley J.E. Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J. Biol. Chem. 2003;278:26 238–26 248. doi: 10.1074/jbc.M302699200. [DOI] [PubMed] [Google Scholar]
- Strick T.R, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. Curr. Biol. 2004;14:874–880. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Sunako Y, Onogi T, Hiraga S. Sister chromatid cohesion of Escherichia coli. Mol. Microbiol. 2001;42:1233–1241. doi: 10.1046/j.1365-2958.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.M, Moghraby J.S, Lees J.H, Smit B, Moens P.B, Lehmann A.R. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell. 2001;12:1583–1594. doi: 10.1091/mbc.12.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Volkov A, Mascarenhas J, Andrei-Selmer C, Ulrich H.D, Graumann P.L. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol. Cell. Biol. 2003;23:5638–5650. doi: 10.1128/MCB.23.16.5638-5650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Yeong F.M, et al. Identification of a subunit of a novel kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr. Biol. 2003;13:2058–2064. doi: 10.1016/j.cub.2003.10.032. [DOI] [PubMed] [Google Scholar]
- Yoshimura S.H, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]