Abstract
In Xenopus extract meiotic spindles, microtubules slide continuously towards their minus ends, a process called poleward flux. This article discusses recent progress in determining the mechanism of poleward flux, and its functions in spindle organization and generating force on chromosomes. Bipolar organization is required for flux and inhibition of the mitotic kinesin Eg5 inhibits flux, suggesting the sliding force for flux is generated by Eg5 pushing anti-parallel microtubules apart. An important function of flux in spindle organization may be to transport minus ends nucleated at chromatin towards the pole. By pulling microtubules through attachment sites at kinetochores, flux may generate poleward force on metaphase chromosomes.
Keywords: spindle, microtubule, dynamics, chromosome, force
1. Introduction
At metaphase, mitotic and meiotic spindles are poised to segregate their chromosomes. The spindle is bipolar, and chromosomes are aligned with their kinetochores attached to opposite poles, already experiencing the poleward force that will later separate sister chromatids. Thus, understanding the dynamic organization of metaphase spindles will tell us a great deal about both spindle assembly mechanisms and how forces on chromosomes are generated. Although spindles achieve a steady state in mass and length at metaphase, the microtubules within them are highly dynamic, and these dynamics are intimately related to assembly and force producing mechanisms. In this article, I will discuss recent progress in understanding dynamic organization and force production in Xenopus extract spindles. With respect to spindle assembly mechanisms, this system recapitulates meiosis II in unfertilized Xenopus eggs. Spindle assembly can occur without centrosomes, by nucleation of microtubules near chromatin followed by self-organization of bipolarity through a combination of microtubule dynamics and motor protein activity (Walczak et al. 1998, Karsenti & Nedelec 2004). Precisely how microtubules are nucleated, bipolarity self-organized and chromosomes segregated, is not known.
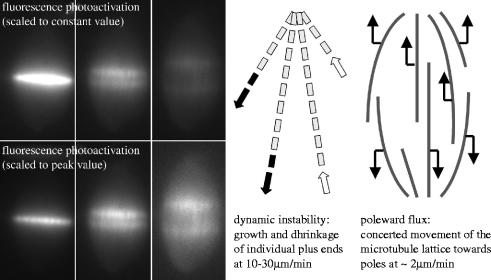
Two techniques have been most useful in probing microtubule dynamics in Xenopus extract spindles: fluorescence photoactivation (figure 1; Sawin & Mitchison 1991) and fluorescence speckle microscopy (Maddox et al. 2003). The former measures both turnover and sliding, the latter mainly sliding. Using these methods, two superimposed dynamic processes were inferred (figure 1). The majority of spindle microtubules turn over with approximately exponential kinetics and a half time of approximately 100 s. Turnover is thought to be driven primarily by dynamic instability of the plus ends, a process in which individual ends switch stochastically between polymerization and depolymerization driven by GTP hydrolysis by beta-tubulin. Microtubules with plus ends attached to kinetochores are presumably protected from turnover by dynamic instability. Simultaneously, spindle microtubules slide towards their minus ends at a rate of approximately 2 μm min−1. This sliding, which must be balanced by polymerization and depolymerization at steady state, is called ‘poleward flux’. To the limit of current detection methods, all spindle microtubules undergo poleward flux (Miyamoto et al. 2004), though its rate is heterogeneous (Vallotton et al. 2003), and may be slower by approximately 10%, on average, in microtubules attached to kinetochores(kMTs) than in neighbouring non-kinetochore microtubules (nkMTs) (Maddox et al. 2003). Until recently, both the molecular mechanism and biological function of flux were enigmatic. Speckle microscopy combined with physical and chemical perturbation experiments have now allowed significant progress in both areas, which are discussed below. Many of these experiments were conducted by the Woods Hole Marine Biological Lab Cell Division Group (MBL-CDG), a consortium of investigators primarily from the laboratories of Ted Salmon and Tim Mitchison, who have gathered in Woods Hole for the last six summers to investigate dynamic organization and force production in Xenopus extract spindles.
Figure 1.
Dynamic organization of Xenopus extract spindles. The left panels illustrate use of fluorescence photoactivation to probe microtubule dynamics. Tubulin labelled with caged fluorescein was incorporated into spindles, and a UV beam is used to uncage fluorescence in a bar near the chromosomes. The images show how this bar evolves over approximately 1.5 min intervals. The fluorescence decays rapidly, and remaining fluorescence splits into two bars that move towards the poles. The images are presented scaled to a constant value to emphasize turnover, and scaled to peak intensity to emphasize movement of remaining microtubules. This image is courtesy of Tarun Kapoor (Rockefeller University); for details see Sawin & Mitchison (1991). The models illustrate how we currently interpret these dynamics. Dynamic instability (centre) causes individual microtubules to turn over, while poleward flux (right) causes microtubules to slide steadily towards their minus ends.
2. Flux requires bipolar organization
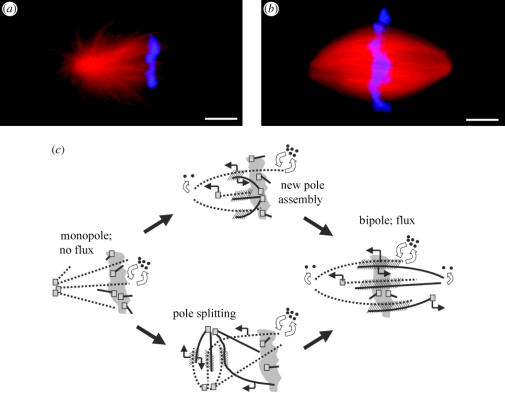
What physical arrangement of microtubules is required for flux? Addressing this basic question is a prerequisite for understanding molecular mechanism. In 1994, Ken Sawin and I used fluorescence photoactivation to conclude that microtubules asters, where all microtubules are oriented in the same direction, displayed flux (Sawin & Mitchison 1994). Re-investigating this question using speckle microscopy, the MBL-CDG came to the opposite conclusion, and accounted for the earlier, incorrect conclusion (Mitchison et al. 2004). In this work, we followed microtubule asters nucleated by sperm centrosomes as they evolved from initially monopolar structures to bipolar spindles (figure 2a,b). Spontaneous bipolarization occurs owing to a combination of two pathways, assembly of a new pole near chromatin and splitting of the old pole (figure 2c). We found that when the asters are truly monopolar, they do not exhibit the coherent microtubule sliding characteristic of flux, and this behaviour starts as they bipolarize. We also found that the inhibition of the Ran pathway with excess importin-alpha in established bipoles leads to the loss of overlap microtubules and cessation of flux. Together, these observations suggest that interactions between anti-parallel overlapping microtubules, shown hatched in figure 2c, are important for both bipolarization and flux. They also suggest that the Ran pathway is continuously required to generate anti-parallel microtubules at steady state. Xenopus extract spindles contain prominent bundles of anti-parallel overlapping microtubules throughout their central region, and limited overlaps extend all the way to the poles. Using electron microscopy (EM), it has been found that the microtubules in overlap bundles are close together, not regularly organized, and embedded in an amorphous substance of unknown composition that may include sliding motors for flux.
Figure 2.
Flux correlates with bipolarity. When sperm is added to mitotic extract, monopolar asters form initially (a). After approximately 30 min, these evolve into bipolar spindles (b). (Red is tubulin, blue DNA Bars, 10 μm). Fluorescence speckle microscopy of these structures allowed us to track the pathways that lead to bipolarization, and revealed that monopoles do not flux and that the onset of flux correlates with onset of bipolarity (Mitchison et al. 2004). (c) A model for bipolarization and flux is shown. Solid and dotted lines are microtubules of opposite polarity, squares are nucleation/capping complexes, open arrows indicate polymerization dynamics and closed arrows indicate sliding. Bipolarization occurs by a combination of new pole assembly near chromatin and splitting of the old pole. The monopolar structures do not flux, and flux starts as they become bipolar. This model suggests that interactions between overlapping, anti-parallel microtubules (shown hatched) are important for driving both bipolarization and flux.
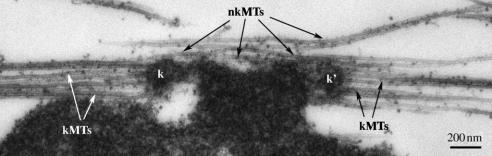
If anti-parallel organization is required for flux, how do kMTs, with their uniform polarity, flux? We believe this occurs either because they are cross-linked to overlapping microtubules that flux, and/or because they interact directly with microtubules of the opposite polarity. By EM, it is found that nkMTs pass close to every sister kinetochores pair, often appearing to interact laterally with chromatin in the kinetochore–centromere region (e.g. figure 3; see Ohi et al. (2003) for more examples and discussion). These microtubules are of mixed polarity and co-bundle with kMTs, leading to close contact between kMTs and microtubules of the opposite polarity. This overlap may allow engagement of the flux engine with kMTs. Microtubules that make lateral interactions with the centromere and co-bundle with kMTs have been observed near every sister kinetochore pair in other types of spindle (Jensen 1982; McDonald et al. 1992). I believe these laterally associated microtubules play an important, and under-appreciated, role in chromosome segregation. As well as driving flux in kMTs, they may be important for congression movements, by providing a substrate for plus-end-directed kinesins attached to kinetochores. If unregulated, laterally associated microtubules may tend to promote merotelic attachments to kinetochores (attachment of a single kinetochore to both poles). Because of their risk and importance, we suspect laterally associated microtubules are subject to positive and negative regulation by a system that includes KinI kinesins and Aurora B kinase (Ohi et al. 2003).
Figure 3.
Sister kinetochore pair in a Xenopus extract spindle by thin section EM. The two kinetochores are designated k and k′. Note kMTs with their plus ends terminating at kinetochores, and nkMTs that interact laterally with the kinetochore–centromere region of the chromosome and co-bundle with kinetochore microtubules. These nkMTs are of mixed polarity, and presumably make anti-parallel interaction with kinetochore microtubules. By speckle microscopy, all the microtubules in this image undergo polewards flux, though kMTs may slide by approximately 10% slower, on average, than nearby nkMTs (Maddox et al. 2003). For more examples, see Ohi et al. (2003).
3. Eg5 drives microtubule sliding
What drives the microtubules sliding component of poleward flux? Two possibilities have been discussed: force from microtubule polymerization dynamics (Margolis & Wilson 1981) and force from plus-end-directed molecular motor proteins (Mitchison & Sawin 1990). The MBL-CDG tested the role of polymerization dynamics by perturbation experiments (Mitchison et al. In press). Hexylene glycol, which blocks all depolymerization, did not inhibit sliding, instead converting it into spindle expansion. Inhibition of the major catastrophe factor in extracts, MCAK (a KinI kinesin), caused displacement of growing plus ends out of the spindle by unbounded growth, and severe disorganization of poles. Again, sliding was not inhibited. We concluded that neither microtubule depolymerization nor polymerization within the spindle are required for the sliding component of flux, implicating a motor protein as the force generator by default.
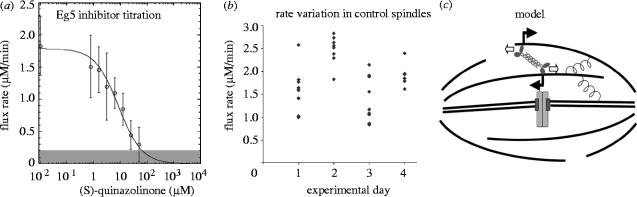
The obvious candidate motor to drive the sliding component of flux is Eg5, a plus-end-directed kinesin. This motor is required for bipolarity in extract spindles (Sawin et al. 1992); it is present in regions of microtubule overlap, though not enriched there (Sawin et al. 1992), and its bipolar, tetrameric organization suggests it might push overlapping, anti-parallel microtubules apart (Kashina et al. 1996). An older study suggested that Eg5 might not be required for flux (Sawin & Mitchison 1994), but a group of students in the Mitchison laboratory readdressed this issue, armed with speckle microscopy, a new method for quantifying sliding by autocorrelation of sequential images and Eg5 inhibiting drugs (Miyamoto et al. 2004). They discovered that pharmacological inhibition of Eg5 causes a dose-dependent reduction in the velocity of microtubule sliding in metaphase spindles (figure 4a). The dose–response fits well to a simple hyperbolic binding isotherm, consistent with flux rate being proportional to the number of active Eg5 heads. Furthermore, immunodepletion of Eg5 largely blocked sliding, and re-addition of expressed Eg5 partially rescued it. Together, these data strongly suggest that Eg5 is the major sliding motor for poleward flux. They do not rule out more minor contributions from other plus-end-directed kinesins in the spindle.
Figure 4.
Eg5 drives the sliding component of flux. (a) Small molecule inhibitors of Eg5 cause a dose-dependent inhibition of flux. The sliding component of flux was measured using cross-correlation between sequential images. (b) Variation of flux rate in control spindles. Note large spindle-to-spindle and preparation-to-preparation variation. Flux rate was approximately constant in individual spindles over a period of minutes. (c) Model showing how the plus-end-directed mitotic kinesin Eg5 may drive the sliding component of flux by pushing anti-parallel microtubules apart. Open arrows indicate the direction that Eg5 walks on microtubules, driven by ATP hydrolysis. The closed arrows indicate the resulting microtubule sliding if the Eg5 molecule is static. The springs are drawn to emphasize that the flux motors are probably working against a mechanical load that limits sliding rate. The molecular nature of this load is unknown. See Miyamoto et al. (2004) for details.
Precise measurement of flux by cross-correlation revealed that flux in control spindles was surprisingly variable, both from spindle to spindle and between extract preparations (figure 4b). For a single spindle, flux rate was constant over many minutes. This variability between spindles implies that flux rate is not simply determined by the gliding velocity of Eg5. The system behaves as if the Eg5 motors are acting against a load, and either the load or the number of active Eg5 molecules varies between spindles. The idea of a load is cartooned as springs in figure 4c. Its real molecular nature, and the reason it varies between spindles, is unclear. Addressing this issue might tell us much about spindle mechanics. One potential source of mechanical load is kinetochores that may generate friction opposing the flux motor (discussed below). It will be interesting to test if spindles lacking kinetochores, such as DNA bead spindles, show faster or more uniform flux rates.
The bipolarity requirement, Eg5 inhibition experiments and variability of rate suggest a simple model for the molecular mechanism of the sliding component of flux (figure 4c). While this model rationalizes the data discussed here, it does not explain all previous observations, and the flux engine is certainly more complicated in reality. Future models will need to explain why Eg5 is enriched at poles rather than overlaps, why Eg5 needs to be phosphorylated on a conserved cdk1 site to target to spindles (Blangy et al. 1995) and why poly(ADP-ribose) is required in addition to Eg5 to stabilize bipolarity in extract spindles (Chang et al. 2004). Understanding the precise role of Eg5 in dynamic organization may also have practical implications, since Eg5 inhibitors related to the one used in figure 4a are currently being tested for therapeutic utility in cancer.
4. Function of flux in spindle organization
As a dramatic dynamic behaviour, it has always seemed likely that flux plays some role in spindle assembly, and correlative observations point towards a function in bipolarization (figure 2c). We have no definitive evidence on this point as yet, but a new model for spindle dynamics, under development by Kendra Burbank in the Mitchison laboratory, makes explicit predictions (figure 5). In this model, microtubules are nucleated near chromatin by local activation of the Ran pathway. The resulting plus ends grow out, and later shrink back, by dynamic instability. Once the growing microtubule couples into the flux machinery, it starts to slide, resulting in transport of its minus end towards the pole at the flux rate. A key innovation in the Burbank model is that the minus ends are ‘reflecting’, meaning that when the plus end depolymerizes back to the minus end, the microtubule does not disappear completely, but rather starts growing again. We hypothesize that this reflecting quality is provided by some molecular cap that also nucleates the microtubule. The molecular composition of this cap is unknown, but gamma-tubulin complex, TPX2 and possibly RHAMM are likely to be part of it (Groen et al. 2004). In the model, minus ends are transported steadily back to the pole at the flux rate, while plus ends grow and shrink freely. At some point, the minus end becomes unstable and loses its reflecting property. When this happens, the next plus-end-depolymerization event leads to complete loss of the microtubule. At steady state, it will be replaced by new nucleation at the chromatin. The model is being developed in part to address the issue of what governs spindle length. The position of the most distal minus ends defines the poles and the length of the spindle. Spindle length is thus controlled by the location at which reflecting ends are converted to non-reflecting. How this location is specified and what biochemical event destabilizes minus ends is unclear. An interesting possibility is specification of the location by the position of plus ends of the opposite polarity growing out from chromatin. In that case, dynamic instability of plus ends sets the length of the spindle, acting indirectly via an influence on minus end stability. Consistent with this idea, bounded dynamic instability naturally sets a length-scale (Verde et al. 1992), and steady-state spindle length in the extract system is known to be sensitive to perturbation of plus-end dynamics (reviewed in Karsenti & Vernos 2001; also see Mitchison et al. In press).
Figure 5.
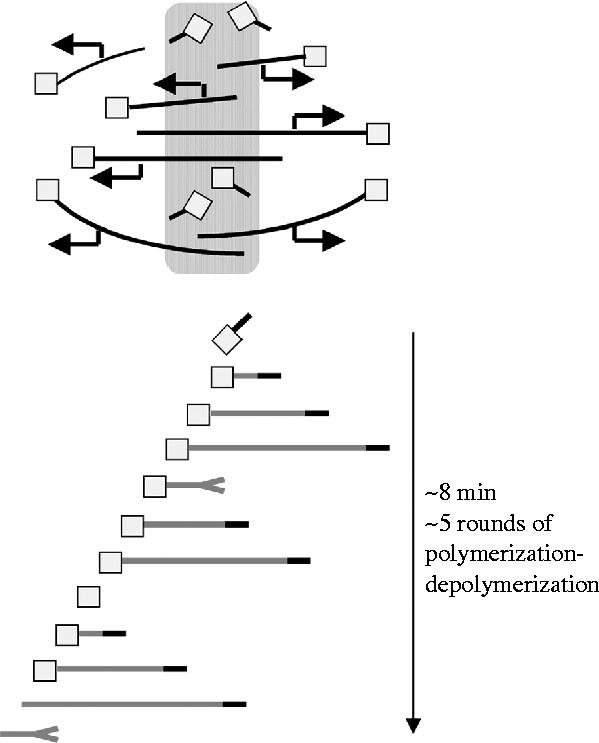
Model for dynamic organization of extract spindles. This model, currently under development by Kendra Burbank, Aaron Groen and Tim Mitchison, seeks a quantitative explanation of metaphase spindle dynamics, emphasizing chromatin-driven microtubule nucleation and dynamic instability of plus ends. The upper panel shows an overview of steady-state organization, the lower panel shows the proposed life history of a single microtubule. Microtubules (grey lines) are nucleated at chromatin by complexes that probably include gamma-tubulin complex and TPX2 (squares). These complexes also cap minus ends. Plus ends grow out (black tip) and shrink back (split tip) by dynamic instability. The capping complex ‘reflects’ shrinking plus ends, causing them to regrow. Minus ends, stabilized in this manner, move towards the pole, driven by the flux engine. At some location the cap is lost, minus ends loose their reflecting property and the next depolymerization event leads to loss of the microtubule.
The model in figure 5 was developed to try and account for experimentally measured distributions of microtubule polarity and plus ends in spindles, given the theory of dynamic instability and the central role of chromatin-driven nucleation in extract spindles. It can be compared with recent qualitative models emphasizing related points (Karsenti & Nedelec 2004). It is very much a work in progress. It does not account for kMTs, or for a contribution from nucleation at poles. We think these simplifications are justified in an initial model, since nkMTs account for approximately 95% of total spindle microtubules, and Xenopus extract spindles assemble to normal length and morphology without kinetochores or centrosomes (reviewed in Karsenti & Vernos 2001). Centrosome-free poles probably recruit nucleating factors, and quantifying the relative contribution of nucleation from chromatin versus poles to steady-state dynamics is an important future goal. Despite its simplifications, the model makes interesting predictions about the function of flux. Firstly, while flux is required for dynamic organization, the rate of flux does not influence spindle length. This is consistent with the observation of relatively normal length spindles when both Eg5 and dynein are partially inhibited (Mitchison et al. In press), and flux rate is much reduced (Miyamoto et al. 2004). Secondly, depolymerization occurs primarily from plus ends, by dynamic instability, rather than from minus ends as in most models. Our model thus requires some factor to uncap minus ends, but it does not require a factor to promote steady depolymerization there (with the possible exception of kMTs). The uncapping factor could work by negatively regulating the cap complex, or by cutting the microtubule.
5. Force on kinetochores
In addition to a role in spindle assembly, flux probably has an important function in generating forces on chromosomes. Kinetochores must experience poleward force in metaphase to congress and silence the spindle checkpoint, and in anaphase to promote physical segregation of the separated sister chromatids. The question of how poleward force is generated is one of the oldest in mitosis research, and despite considerable progress, we have yet to answer it at a molecular or even mechanical level. In recent years, potential force producing mechanisms have been broken into two classes (Inoue & Salmon 1995). In ‘pac-man’ mechanisms, force is generated at the microtubule–kinetochore interface, and microtubules depolymerize at kinetochores. In ‘traction fibre’ mechanisms, force is generated all along kMTs (or at poles). This results in movement of kinetochores by passive attachment to moving microtubules, and depolymerization of kMTs at poles. From microtubule marking experiments we know that kMTs in many systems, including Xenopus extracts, in fact depolymerize at both kinetochores and poles, creating a complex mechanochemical problem (Mitchison & Salmon 2001). The MBL-CDG re-investigated this question in Xenopus extract spindles using fluorescence speckle microscopy to track microtubules, and labelled CenpA to track kinetochores, allowing live imaging of polymerization dynamics at kinetochores for the first time (Maddox et al. 2003). We found continuous polymerization at kinetochores in metaphase, and switching between polymerization and depolymerization in anaphase. We also found that kinetochores are stretched towards the poles in metaphase, indicating strong poleward forces. Integrating those data with the bipolarity and Eg5 requirements, we can begin to build a detailed picture of how forces on chromosomes are generated at metaphase.
In the integrated model (figure 6), kMTs slide polewards in a traction fibre mechanism, pushed by Eg5 acting between anti-parallel microtubules. kMTs co-bundle with microtubules of opposite polarity, allowing the anti-parallel interactions necessary for Eg5 action. Poleward sliding pulls the microtubule lattice through attachment sites at kinetochores. This pulling either drives, or is supported by, polymerization of the attached plus end, creating a reverse pac-man. We hypothesize that the kinetochore attachment sites exert molecular friction that resists this sliding, resulting in poleward force generation, analogous to a slipping clutch on a fishing reel. In this model, the kinetochore is passive with respect to force generation, and harvests poleward force from the flux engine by resisting sliding. Alternatively, it is possible that the kinetochore actively generates poleward force at metaphase, for example by minus end directed motor activity. I currently prefer the passive model. Opposing flux with friction seems to be an easy way to generate force, and I suspect that active generation of poleward force by kinetochores with mature, end-on microtubule attachments requires depolymerization at kinetochores, as proposed in the pac-man model (Inoue & Salmon 1995). Furthermore, inhibition of cytoplasmic dynein, the minus-end-directed motor most often implicated in generating force at kinetochores (Sharp et al. 2000), does not reduce poleward force on metaphase kinetochores in Xenopus extract spindles, as assayed by the distance between sister kinetochores after treatment with p50 dynamitin (MBL-CDG, unpublished observation).
Figure 6.
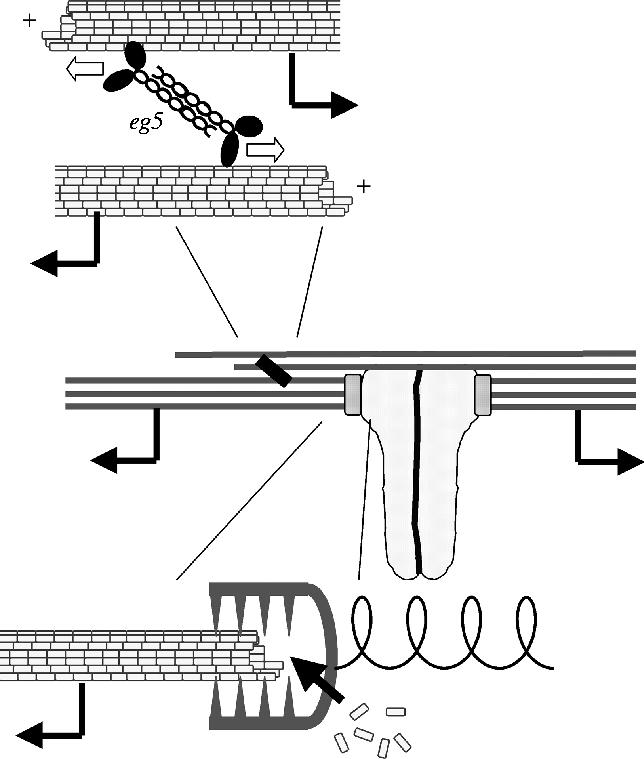
Forces on kinetochores at metaphase. This model, based on discussions in the MBL-CDG, seeks to integrate observations on the role of anti-parallel overlap and Eg5 in driving flux (figures 2–4) with measurements of polymerization dynamics and force production at kinetochores (Maddox et al. 2003). Kinetochore microtubules slide continuously towards their minus ends (middle panel), driven by Eg5 pushing anti-parallel microtubules apart (upper panel) and supported by polymerization of plus ends at kinetochores (lower panel). This sliding pulls the microtubule lattice through kinetochore attachment sites. Transient attachment of these sites to the lattice generates friction at the interface, leading to poleward force on kinetochores (illustrated as stretching a spring). In this model, the metaphase kinetochore is a passive force generator, analogous to the slipping clutch on a fishing reel. In anaphase, kinetochores may switch between passive and active force generation (Maddox et al. 2003).
A key question with respect to the function of flux in chromosome segregation is whether the metaphase mechanisms and molecules diagrammed in figure 6 also function in anaphase. A well-established view is that the only change in spindle mechanics at anaphase is the physical separation of sister chromatids, with force generating mechanisms remaining constant (McNeill & Berns 1981). In that case, the forces operating at metaphase (figure 6) would also drive anaphase A and anaphase B movements. Eg5 targeting, however, requires cdk1 phosphorylation (Blangy et al. 1995), and this presumably declines during anaphase. It is possible that microtubule sliding in anaphase, which is required for both anaphase A and B movements, is driven by different plus-end-directed kinesins. There is no shortage of candidates; CenpE, chromokinesin/kif 4 and MKLP1 are all plus-end-directed kinesins that target to overlap microtubules at anaphase onset (Sharp et al. 2000). Any (or all) of these kinesins might also help drive polewards sliding of kMTs in anaphase, continuing the role that Eg5 occupies in metaphase. It should be possible to address such possibilities in the near future, using speckle microscopy to measure microtubule sliding, and RNAi or immunodepletion to inhibit specific motors.
Finally, it is important to note that spindle dynamics mechanisms defined in the Xenopus extract system will not necessarily translate directly to other systems. It is not yet clear how many systems display poleward flux at metaphase, or whether the mechanism of flux is the same in animal mitosis as it is in egg meiosis. In spindles that do not flux, such as budding and fission yeast, there may still be an important, conserved role for plus-end-directed, bipolar kinesins in pushing poles apart (Sharp et al. 2000). Even this is not universal, however, since nematode blastomere spindles can bipolarize normally in the absence of this type of kinesin (Tony Hyman, personal communication). It is also unclear how important chromatin-driven nucleation of microtubules is outside animal egg meiotic systems. Diversity of mechanical mechanisms is an emerging theme in mitosis research and more comparisons between systems will be required in the future to identify common principles in the mechanics of chromosome segregation.
Glossary
- EM
electron microscopy
- KMT
kinetochore microtubule
- MBL-CDG
Woods Hole Marine Biological Laboratory Cell Division Group
- nkMT
non-kinetochore microtubule
Footnotes
One contribution of 17 to a Discussion Meeting Issue `Chromosome segregation'.
References
- Blangy A, Lane H.A, d'Herin P, Harper M, Kress M, Nigg E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Chang P, Jacobsen M.K, Mitchison T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Groen A.C, Cameron L.A, Coughlin M, Miyamoto D.T, Mitchison T.J, Ohi R.O. XRHAMM functions in ran dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. (in press) [DOI] [PubMed] [Google Scholar]
- Inoue S, Salmon E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen C.G. Dynamics of spindle microtubule organization: kinetochore fiber microtubules of plant endosperm. J. Cell. Biol. 1982;92:540–558. doi: 10.1083/jcb.92.2.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Nedelec F. The mitotic spindle and actin tails. Biol. Cell. 2004;96:237–240. doi: 10.1016/j.biolcel.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kashina A.S, Baskin R.J, Cole D.G, Wedaman K.P, Saxton W.M, Scholey J.M. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Straight A, Coughlin P, Mitchison T.J, Salmon E.D. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell. Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis R.L, Wilson L. Microtubule treadmills—possible molecular machinery. Nature. 1981;293:705–711. doi: 10.1038/293705a0. [DOI] [PubMed] [Google Scholar]
- McDonald K.L, O'Toole E.T, Mastronarde D.N, McIntosh J.R. Kinetochore microtubules in PTK cells. J. Cell. Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill P.A, Berns M.W. Chromosome behavior after laser microirradiation of a single kinetochore in mitotic PtK2 cells. J. Cell. Biol. 1981;88:543–553. doi: 10.1083/jcb.88.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J, Salmon E.D. Mitosis: a history of division. Nat. Cell. Biol. 2001;3:E17–E21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J, Sawin K.E. Tubulin flux in the mitotic spindle: where does it come from, where is it going? Cell. Motil. Cytoskeleton. 1990;16(2):93–98. doi: 10.1002/cm.970160202. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J, Maddox P, Groen A, Cameron L, Perlman Z, Ohi R, Desai A, Salmon E.D, Kapoor T.M. Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol. Biol. Cell. 2004;15:5603–5615. doi: 10.1091/mbc.E04-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T. J., Maddox, P., Gaetz, J., Groen, A., Shirasu, M., Desai, A., Salmon, E. D. & Kapoor, T. M. In Press. Roles of polymerization dynamics, opposed motors and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell [DOI] [PMC free article] [PubMed]
- Miyamoto D.T, Perlman Z.E, Burbank K.S, Groen A.C, Mitchison T.J. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Coughlin M.L, Lane W.S, Mitchison T.J. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell. 2003;5:309–321. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- Sawin K.E, Mitchison T.J. Poleward microtubule flux mitotic spindles assembled in vitro. J. Cell. Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E, Mitchison T.J. Microtubule flux in mitosis is independent of chromosomes, centrosomes, and antiparallel microtubules. Mol. Biol. Cell. 1994;5:217–226. doi: 10.1091/mbc.5.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E, LeGuellec K, Philippe M, Mitchison T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sharp D.J, Rogers G.C, Scholey J.M. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Vallotton P, Ponti A, Waterman-Storer C.M, Salmon E.D, Danuser G. Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescent speckle microscopy study. Biophys. J. 2003;85:1289–1306. doi: 10.1016/S0006-3495(03)74564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell. Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E, Vernos I, Mitchison T.J, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]