Abstract
In meiosis, a physical attachment, or cohesion, between the centromeres of the sister chromatids is retained until their separation at anaphase II. This cohesion is essential for ensuring accurate segregation of the sister chromatids in meiosis II and avoiding aneuploidy, a condition that can lead to prenatal lethality or birth defects. The Drosophila MEI-S332 protein localizes to centromeres when sister chromatids are attached in mitosis and meiosis, and it is required to maintain cohesion at the centromeres after cohesion along the sister chromatid arms is lost at the metaphase I/anaphase I transition. MEI-S332 is the founding member of a family of proteins that protect centromeric cohesion but whose members also affect kinetochore behaviour and spindle microtubule dynamics. We compare the Drosophila MEI-S332 family members, evaluate the role of MEI-S332 in mitosis and meiosis I, and discuss the regulation of localization of MEI-S332 to the centromere and its dissociation at anaphase. We analyse the relationship between MEI-S332 and cohesin, a protein complex that is also necessary for sister-chromatid cohesion in mitosis and meiosis. In mitosis, centromere localization of MEI-S332 is not dependent upon the cohesin complex, and cohesin retains its association with mitotic chromosomes even in the absence of MEI-S332.
Keywords: meiosis, sister chromatids, centromere, kinetochore, chromosome segregation
1. Introduction: the MEI-S332 cohesion protein family
Cohesion, or physical attachment, of the sister chromatids is essential in order that they attach stably to microtubules emanating from opposite spindle poles and, thus, segregate properly to daughter cells (for reviews see Lee & Orr-Weaver 2001; Nasmyth 2001). In mitosis, the sister chromatids are attached along their lengths and cohesion is released at the metaphase/anaphase transition, permitting anaphase segregation. In meiosis, cohesion along the sister-chromatid arms plays an additional role in linking homologous chromosomes by stabilizing chiasmata, the relics of reciprocal exchanges that serve to attach homologues in the first meiotic division. Thus, arm cohesion is crucial for bipolar attachment of the homologues in meiosis I and arm cohesion is released at the metaphase I/anaphase I transition. Centromere cohesion is retained until the metaphase II/anaphase II transition, and these attachments are necessary for bipolar attachment and accurate segregation of the sister chromatids in meiosis II.
A conserved complex of proteins called cohesin plays an instrumental and direct role in sister-chromatid cohesion via its association with the chromosomes (for review see Uhlmann 2003). The cohesin complex is composed of four subunits and can form a ring structure. Cleavage of one of the subunits, Scc1/Mcd1/Rad21, by the separase protease opens the ring, causing dissociation of cohesin and release of cohesion. In the prometaphase in metazoans, there is an additional mechanism leading to release of cohesin from the arms that is independent of separase activity but possibly promoted by phosphorylation of the cohesin Rad21 by the Polo mitotic kinase. In meiosis, in many organisms, one or more cohesin subunits are replaced by meiotic counterparts. In yeast, the pool of cohesin at the centromere containing the Rec8 meiotic cognate of Scc1/Mcd1/Rad21 is resistant to separase cleavage at metaphase I/anaphase I, permitting retention of cohesion at the centromere until meiosis II (Buonomo et al. 2000). In fission yeast Schizosaccharomyces pombe, another cohesin subunit has two meiotic forms that show differential localization on the arms and centromere, with Rec11 being present along the chromosome and necessary for recombination and Psc3 at the centromere (Kitajima et al. 2003). The resistance of cohesin at the centromeres to cleavage by separase pointed to a mechanism of protection of this pool of cohesin, active in meiosis I but inactive in meiosis II, when centromere cohesion is released.
In addition to the conserved cohesin complex, other proteins have been identified that are essential for meiotic cohesion. Moreover, it is possible that subunit composition of the cohesin complex may vary in meiosis in some organisms. For example, a Rec8 subunit has not been identified in Drosophila (Heidmann et al. 2004). Budding yeast Spo13 is a novel protein crucial for retention of centromere cohesion in meiosis and capable of inhibiting release of cohesin when ectopically expressed in mitosis (Lee et al. 2002; Shonn et al. 2002). Spo13 does not localize to the centromere, however, and may act indirectly. The Drosophila ORD protein is required in meiosis for both arm and centromere cohesion, and ORD localizes to centromeres after chromosome condensation in prophase I (Bickel et al. 1997; Balicky et al. 2002). To date, proteins homologous to ORD have not been identified in other organisms. Drosophila MEI-S332 was predicted to protect centromere cohesion because mutations in the mei-S332 gene cause premature separation of the sister chromatids in anaphase I after the release of arm cohesion and segregation of homologues (Davis 1971; Goldstein 1980; Kerrebrock et al. 1992). This results in random segregation in meiosis II in mei-S332 mutants. Furthermore, the MEI-S332 protein localizes to centromeres from prometaphase I until the metaphase II/anaphase II transition, consistent with MEI-S332 regulating cohesion directly (Kerrebrock et al. 1995). Another gene likely to encode a protein crucial to maintain cohesion at the centromeres in meiosis is the pc gene from tomatoes, but its protein product has not yet been identified (Clayberg 1959).
Orthologues to MEI-S332 that share limited sequence homology have now been identified in organisms from yeast to humans. The first family sibling, named shugoshin (sgo1; Japanese for ‘guardian spirit’), was recovered in fission yeast from its similar phenotypes (Kitajima et al. 2004). The sgo1 gene causes lethality in mitotically growing cells when coexpressed with the rec8 gene (Kitajima et al. 2004). Ectopic expression of rec8 in mitosis results in centromere localization of the protein, but it is cleaved at the metaphase/anaphase transition and does not hinder sister separation. Coexpression of Sgo1 protein, however, protects Rec8 protein such that it is retained on the centromere, leading to non-disjunction and lethality. Strains deleted for sgo1 lose sister-chromatid cohesion at the centromere in anaphase I and exhibit premature loss of Rec8 protein, presumably because of separase cleavage (Kitajima et al. 2004; Rabitsch et al. 2004). Normally, sgo1 is expressed specifically in meiosis, localizing to centromeres and dissociating at the metaphase I/anaphase I transition. Similarly, in budding yeast, Sgo1 is needed to prevent premature loss of centromere cohesion and Rec8 localization at anaphase I, thereby averting non-disjunction in meiosis II (Katis et al. 2004; Kitajima et al. 2004; Marston et al. 2004). The S. cerevisiae Sgo1 protein also localizes to centromeres but it persists until the metaphase II/anaphase II transition, and it is also expressed and functions in mitosis. The analyses in the yeasts suggest that Sgo1 functions to protect Rec8 from separase cleavage at metaphase I/anaphase I, possibly via a direct physical interaction (Kitajima et al. 2004).
The identification of S. pombe and S. cerevisiae Sgo1, based on similar roles to MEI-S332 in maintaining centromere cohesion, revealed that the three proteins are members of a conserved family (Kitajima et al. 2004; Rabitsch et al. 2004). The sequence conservation is limited to regions of only 20–45 amino acids at the N and C termini of the proteins, and the extent of homology is small enough that sequence alone does not provide significant criteria for identification of family members. Despite the limited conservation, three of the conserved residues were altered by mutations in mei-S332, and mutation of others in S. pombe affected protein function (Kitajima et al. 2004). In addition, the conserved N and C terminal domains were shown to be essential for localization of the Drosophila protein to centromeres (Lee et al. 2004).
The N and C terminal domains appear to have distinct functions because the Drosophila protein is capable of homotypic interactions, and mutations in the N terminus show intragenic complementation with mutations in the C terminus (Tang et al. 1998). In addition, the N terminal half of the vertebrate Sgo1 protein has been found to bind to and stabilize microtubules (Salic et al. 2004).
The siblings in this rapidly evolving gene family have acquired distinct attributes and functions. They differ in their importance in terms of mitotic chromosome segregation and differ strikingly in the biological processes they affect and their sites of localization. Family members have distinct roles in meiosis in controlling kinetochore behaviour and in mitosis in affecting microtubule dynamics.
One family member, Sgo2, primarily acts to regulate kinetochore behaviour in meiosis I. In meiosis I, the two kinetochores of each sister chromatid pair face the same pole and capture microtubules from the same pole, a process termed mono-orientation. This is essential to ensure that the two sister chromatids migrate together to the same pole in anaphase I. In both meiosis II and mitosis, the sister kinetochores face and capture microtubules from opposite spindle poles. The monopolin protein complex, so far identified only in S. cerevisiae, is essential for mono-orientation of the sister chromatid kinetochores in meiosis I (Toth et al. 2000; Rabitsch et al. 2003). In S. pombe, the cohesin complex plays a crucial role in kinetochore orientation in meiosis I (Watanabe & Nurse 1999). S. pombe Sgo2 is a duplicate family member present in S. pombe and is expressed in both mitosis and meiosis (Kitajima et al. 2004; Rabitsch et al. 2004). Its main role appears to be in ensuring mono-orientation of sister kinetochores in meiosis I (Rabitsch et al. 2004). Although it contributes to accurate mitotic segregation, it is not essential in an otherwise wild-type background (Kitajima et al. 2004; Rabitsch et al. 2004).
The family members also differ in the nature of their activities in mitosis. Drosophila MEI-S332 is expressed in mitotic cells, localizes to mitotic centromeres from prometaphase until anaphase and augments (but is not normally essential) for mitotic centromere cohesion (Moore et al. 1998; LeBlanc et al. 1999). While also not essential for mitosis, S. cerevisiae Sgo1 plays a more important role in mitosis than does MEI-S332 (Katis et al. 2004; Kitajima et al. 2004; Marston et al. 2004). In contrast, ablation of the vertebrate Sgo1 protein leads to premature sister-chromatid separation and chromosome missegregation, indicating an essential mitotic function (Salic et al. 2004). In addition to this more prominent role in mitosis, vertebrate Sgo1 was identified initially by the ability of its N terminus to bind and stabilize microtubules. Loss of vertebrate Sgo1 leads to release of some kinetochore proteins, activation of the spindle assembly checkpoint and a decrease in kinetochore tension, apparently owing to destabilization of microtubules. Thus, it appears that vertebrate Sgo1 plays a more important function in spindle dynamics than do other family members, although it cannot be excluded that the effects on the spindle in vivo arise from loss of sister-chromatid cohesion.
Consistent with their diversity of functions, family members have diverged in terms of cellular localization as well as the timing of their centromere association. In S. pombe, Sgo1 localizes onto the centromeres at metaphase I but is no longer detectable at anaphase I (Kitajima et al. 2004; Rabitsch et al. 2004). The Bub1 spindle checkpoint protein is needed for Sgo1 localization (Kitajima et al. 2004). Sgo1 localization is in the vicinity of the inner centromere but not coincident with the kinetochore (Rabitsch et al. 2004). S. pombe Sgo2 is on telomeres, adjacent to the inner centromere, and along the spindle microtubules in meiosis (Rabitsch et al. 2004). In S. cerevisiae, Sgo1 is at spindle poles in G1-S then localizes throughout the nucleus in mitosis, but can be shown to be kinetochore associated in chromosome spreads (Katis et al. 2004; Marston et al. 2004). In meiosis, it localizes to kinetochores from metaphase I until metaphase II, but is also associated with the spindles in meiosis II (Katis et al. 2004; Marston et al. 2004). Vertebrate Sgo protein is found coating microtubules as well as at the centromere, and it appears to be associated with the inner kinetochore plate (Salic et al. 2004).
The chromosomal position and localization requirements have been defined most precisely for MEI-S332. In both mitosis and meiosis, there are pools of MEI-S332 in the cytoplasm, but it specifically and consistently localizes only onto centromeres in mitosis and meiosis (Moore et al. 1998; Tang et al. 1998). We have not observed it on microtubules, although in some (but not all) male meiosis I cells, it is present at the spindle poles (T. Tang et al. unpublished work).
Several lines of evidence show that although MEI-S332 localization requires the centromere, its precise site of localization is distinct from the chromosomal regions at which the kinetochore assembles (figure 1). MEI-S332 was localized onto minichromosome derivatives with deletions of centromere regions necessary for chromosome transmission (Lopez et al. 2000). These studies mapped the MEI-S332 localization domain to the centromere region. In addition, translocation chromosomes with blocks of heterochromatin moved distal from the centromere showed that MEI-S332 is not distributed throughout the heterochromatin. Importantly, on minichromosome derivatives that lack adequate centromere activity and are poorly transmitted but nevertheless assemble kinetochores, MEI-S332 is not localized, although kinetochore proteins are detectable (Lopez et al. 2000). Thus, there are distinct requirements for MEI-S332 localization and kinetochore assembly.
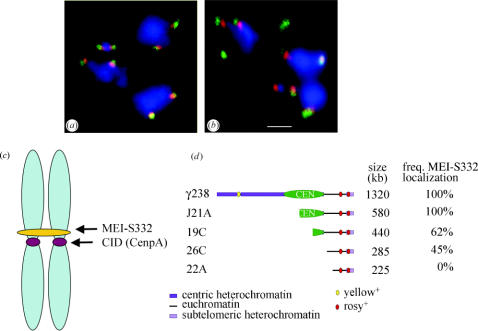
Figure 1.
MEI-S332 localizes adjacent to the kinetochore and binding is dependent on a functional centromere. (a) MEI-S332 (red) localizes adjacent to the outer kinetochore protein ZW10 (green) at the centromeres of bivalents in prometaphase I Drosophila spermatocytes. DNA is stained in blue. (b) MEI-S332 (red) also localizes adjacent to rather than coincident with dynein (green). MEI-S332 is closer to the DNA (blue) than this outer kinetochore protein. Photomicrographs in a and b are courtesy of Jacqueline Lopez; scale bar is 1 μm. (c) In spread chromosomes from mitotic Drosophila S2 culture cells, MEI-S332 is observed localized in a band on one side of the centromere, marked by localization of CID (Drosophila CenpA; Blower & Karpen 2001). (d) Deletion derivatives of a minichromosome were used to map the chromosomal sites needed for MEI-S332 localization. MEI-S332 localization was analysed by immunofluorescence in prometaphase I spermatocytes. The frequency of localization refers to the percentage of prometaphase I spermatocytes in which MEI-S332 could be detected on the minichromosome. The functional centromere region is designated by the green oval. Although the derivatives 19C, 26C and 22A are missing the centromere and are transmitted poorly, they nevertheless organize a kinetochore. The outer kinetochore protein ZW10 is always detectable on these minichromosomes. In contrast, the frequency with which MEI-S332 could be observed diminished in derivatives lacking a fully functional centromere, and MEI-S332 was not detected on the 22A derivative. Panel d is adapted from Lopez et al. (2000).
Deconvolution microscopy confirmed that the site of MEI-S332 localization on mitotic chromosomes is adjacent to, but distinct from, the kinetochore (Blower & Karpen 2001). MEI-S332 localizes as a band across the two sister chromatids next to the kinetochore (figure 1). Unexpectedly, MEI-S332 is present on only one side of the kinetochore, rather than on both chromosomes arms, and the significance of this is not clear. The results of Blower & Karpen (2001) raise the possibility that the localization of MEI-S332 and vertebrate Sgo1 differ in how tightly linked the proteins are to the kinetochore, and more precise localization of vertebrate Sgo1 proteins is warranted. In meiosis I, immunofluorescence localization of MEI-S332 relative to kinetochore proteins shows MEI-S332 to be closer to the chromatin than components of the outer kinetochore plate (figure 1). However, these studies do not specify whether MEI-S332 is present on only one side of the centromere as on mitotic chromosomes.
2. Conservation in the Drosophila MEI-S332 family
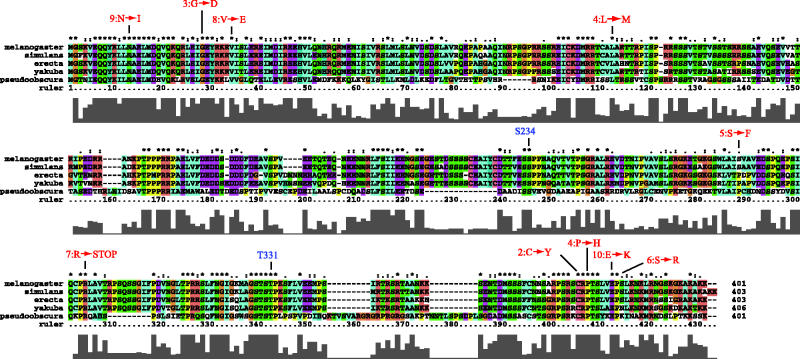
The homologies between family members are so limited that it is difficult to define functional amino acid residues by conservation. Thus, we examined genomic sequences of Drosophila species for MEI-S332 homologues to test which domains of the protein are conserved and likely to play crucial roles in cohesion (figure 2). Drosophila melanogaster, D. erecta, D. simulans and D. yakuba are all in the melanogaster species subgroup, estimated to have diverged from each other in only the last 5 Myr. The predicted coding regions for these MEI-S332 homologues are nearly identical, with only three non-conservative changes and two amino acid additions between D. melanogaster and D. simulans across the entire coding sequence. Although D. erecta is used as an outgroup to the melanogaster subgroup, it is very homologous to D. melanogaster, having only 21 non-conservative changes, allowing for six spacing changes. D. yakuba also is highly homologous, with 22 non-conservative changes plus five spacing changes (figure 2). The extent of identity within the melanogaster subgroup precludes conclusions about functional residues, with the exception of two amino acids changed by mutations in mei-S332 that weakly affect chromosome segregation. The mei-S3325 mutation causes low levels of non-disjunction in males and females, and changes serine 277 to phenyalanine (Tang et al. 1998). In D. erecta and D. yakuba, this residue is a proline. Mei-S3326 affects female meiosis but has only weak effects on male meiosis (Kerrebrock et al. 1992). This mutation hinders the ability of the protein to localize to centromeres (Tang et al. 1998). The mei-S3326 mutation converts serine 384 to arginine (Kerrebrock et al. 1995). Surprisingly, in D. yakuba, this residue is an asparagine.
Figure 2.
Homology alignment of predicted Drosophila MEI-S332 homologues. The sequence alignment of Drosophila melanogaster, D. simulans, D. erecta, D. yakuba, and D. pseudoobscura is shown. The position of mutations in the D. melanogaster mei-S332 are indicated (in red), as are the putative POLO binding sites (in blue).
D. pseudoobscura is estimated to have diverged from the melanogaster group 25 Myr ago, and this divergence is reflected in sequence changes in the MEI-S332 homologue (figure 2). The D. pseudoobscura homologue is 36% identical across the protein sequence with an additional 19% of amino acids that are similar and, thus, it is clearly identifiable as the homologous gene. Comparison of the sequences of the MEI-S332 homologues between D. melanogaster and D. pseudoobscura yields several conclusions. All of the amino acids affected by mutations in mei-S332 are identical except serine 277, which also is a proline in D. pseudoobscura, and serine 384, which, surprisingly, is a lysine. Thus, it is unclear why the change of this residue to arginine in D. melanogaster affects centromere localization and segregation in females so profoundly. Although the largest continuous blocks of homology lie at the N and C termini, there are conserved stretches in the internal region. These may lie in domains that are important for the function of the protein in promoting centromere cohesion. In the melanogaster subgroup, an acidic domain from asparate 173 to glutamate 198 is particularly striking because 14 out of 26 residues are aspartate or glutamate. Interestingly, the acidic nature of this region is not conserved in D. pseudoobscura, even though the block of proline and arginine that resides immediately N-terminal is present, as is a homologous block C terminal (LFSILEEXXSE). The third notable feature concerns the regulation of MEI-S332 by POLO. We have found that D. melanogaster MEI-S332 binds to, and is phosphorylated by, POLO, a step necessary for dissociation of the protein from centromeres in anaphase of mitosis and anaphase II of meiosis (Clarke et al. 2005). A consensus POLO binding site of Ser-pSer/pThr-Pro/X has been delineated, and this is present at two positions in D. melanogaster MEI-S332 at serine 234 and threonine 331 (Elia et al. 2003). Although both of these affect POLO binding, threonine 331 is more essential for phosphorylation and centromere dissociation of MEI-S332 (Clarke et al. 2005). It is interesting that in D. pseudoobscura, the proline at the first site is changed to valine, whereas threonine 331 retains the consensus site. The sequences surrounding Thr331 also are more conserved than those around Ser234.
By homology searches, including the use of solely the N and C terminal of most conserved domains, we were unable to identify a gene homologous to MEI-S332 from the genome sequence of the mosquito Anopheles. This suggests that the protein family is diverging rapidly even within the Diptera.
3. Role of MEI-S332 in mitosis
Although the MEI-S332 protein localizes onto mitotic centromeres in prometaphase and dissociates as the sister chromatids lose their cohesion at the metaphase/anaphase transition, null mutants of mei-S332 are fully viable, even when maternal pools of protein are mutant (Moore et al. 1998; LeBlanc et al. 1999). Overexpression of MEI-S332 can lead to organism lethality and cell death. MEI-S332 appears to contribute to centromeric cohesion in mitosis because the ratio of anaphase to metaphase figures is increased in mei-S332 mutants, and this ratio is decreased when MEI-S332 is overexpressed. Both of these observations are consistent with dosage effects of MEI-S332 contributing to cohesion at the centromere (LeBlanc et al. 1999). In addition, when arm cohesion is released by hypotonic treatment, the frequency of premature separation of the centromeres is increased in mei-S332 mutants and decreased in overexpression lines. Although we did not observe phenotypes consistent with mitotic failures in mei-S332 mutants, the mitotic tissues in Drosophila are extremely plastic during development and can tolerate large amounts of cell death. Proliferation of neighbouring cells or cell growth can compensate for cell loss from mitotic failure or cell death.
As a further test of the function of mei-S332 in mitosis, we set up a situation in which we could measure the division rate of mei-S332 mutant cells in direct comparison with wild-type cells in the eye imaginal disk (figure 3; Xu & Harrison 1994). Using a heterozygous mei-S332 strain for the null allele mei-S3327, we induced mitotic recombination at a specific FRT site with the FLPase recombinase, thus generating homozygous mei-S332 mutant cells and a twin spot of wild-type cells in the adult eyes. The chromosome with the wild-type copy of the mei-S332 gene carried the dominant w+ eye colour marker, and the intensity of red eye pigment is proportional to dosage of the w+ gene. Thus, homozygous wild-type cells can be distinguished from those heterozygous for mei-S332 and w+ by their darker red eye colour. Homozygous mutant mei-S332 cells give rise to a white eye clone. If mei-S332 mutant cells did not proliferate as well as their wild-type sibling cells, then the clone size (measured by counting the number of white ommatidia in the eye) would be smaller than the red clones. We observed that following induction of recombination the size of mei-S332 mutant clones was 75% that of wild-type. This is consistent with a role for mei-S332 in mitosis, although it is not essential for organismal viability.
Figure 3.
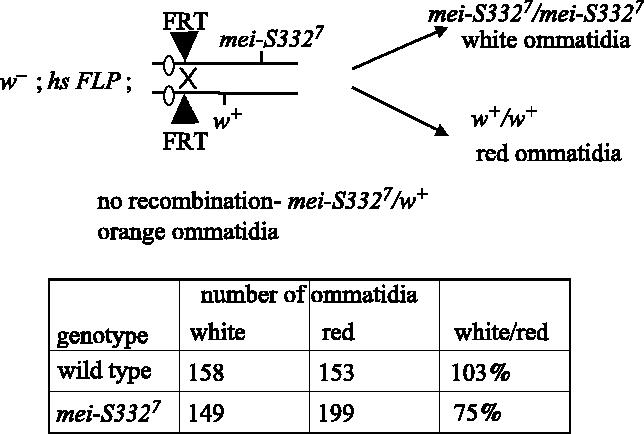
Direct comparison of proliferation capabilities of mei-S332 mutant cells. Twin spot clones were induced in mitotic tissues of Drosophila heterozygous for the mei-S3327 null mutation by site-specific recombination at FRT sites at the base of the right arm of chromosome 2 following induction of the FLPase recombinase. Homozygous mutant mei-S3327 cells gave rise to white ommatidia in the eye, distinguishable from the sibling wild-type red ommatidia. The number of ommatidia in about 10 mutant and 10 wild-type clones were scored and summed.
4. Relationship between MEI-S332 and the cohesin complex
In budding and fission yeast meiosis, the Sgo1 proteins are necessary to prevent cleavage of centromeric Rec8 at the metaphase I/anaphase I transition, ensuring retention of the cohesin complex at the centromeres until anaphase II. We showed that centromere localization of MEI-S332 did not depend on the cohesin complex because if the Rad21 subunit was removed by RNAi in S2 tissue culture cells, then MEI-S332 was retained at the centromere (Lee et al. 2004). Furthermore, MEI-S332 is capable of localizing onto the centromeres of unreplicated sister chromatids, thus it does not require that cohesion be established for centromere binding (Lee et al. 2004). The available antibodies against Drosophila Rad21 permit detection of the protein localized onto chromosomes in mitotic S2 cells (Warren et al. 2000) and, therefore, we could ask the reciprocal question of whether MEI-S332 was necessary for localization of the cohesin complex. In Drosophila, as in vertebrate cells, the bulk of the cohesin complex along the chromosome arms is lost in prometaphase, but cohesin is retained in the mitotic centromere regions until the metaphase/anaphase transition (Warren et al. 2000). We used RNAi to ablate MEI-S332 in order to evaluate whether MEI-S332 was required for retention of cohesin at the centromeres of mitotic chromosomes.
For RNAi against MEI-S332, double-stranded RNA was synthesized. Two 600 bp fragments of mei-S332 cDNA were amplified by PCR for use as templates in dsRNA production. The hybridizing sequences of the primers were fragment 1: 5′-CTGCAAAACCATCGCCAG-3′ and 5′-GGGCGTGACGGTGACTTG-3′ and fragment 2: 5′-AACAATCGTCTGTTTAGC-3′ and 5′-CTTGGCCTTGCCTTTCGA-3′. In vitro transcription was performed with a T7 Megascript kit (Ambion) and dsRNA was ethanol precipitated.
RNAi was performed as described in Lee et al. (2004). For each time point, cells were removed for protein extracts and cytological analysis. Guinea pig anti-MEI-S332 (Tang et al. 1998) was used at 1 per 20 000 in Western blotting to confirm protein depletion. Cells were diluted to 5×105 cells ml−1 and 100 μl were cytospun against a slide in a Thermo Shandon cytocentrifuge at 1000 r.p.m. for 2 min. Slides were fixed in 3.7% formaldehyde/0.5% Triton X-100/PBS for 10 min. After PBS washes, slides were blocked and stained with rabbit anti-DRAD21 (Warren et al. 2000) and anti-MEI-S332, as described in Lee et al. (2004). Microscopy was performed on a Zeiss Axiophot microscope with Zeiss PlanNeofluar 40× and 100× oil objectives. Images were collected with a Spot RT CCD camera and software and processed in Adobe Photoshop.
Both double-stranded RNA products successfully reduced MEI-S332 protein to undetectable levels after 4 days of treatment of S2 cells (figure 4a). In control, untreated cells both RAD21 and MEI-S332 localized to the centromeres of metaphase chromosomes, although MEI-S332 was more restricted in its localization to punctate focuses corresponding to the centromere proper (figure 4b). Strikingly, in cells depleted for MEI-S332 and lacking detectable protein, RAD21 localized normally at the centromere regions of metaphase chromosomes (figure 4c). Thus, in mitosis, MEI-S332 is not essential for centromere retention of RAD21 at metaphase.
Figure 4.
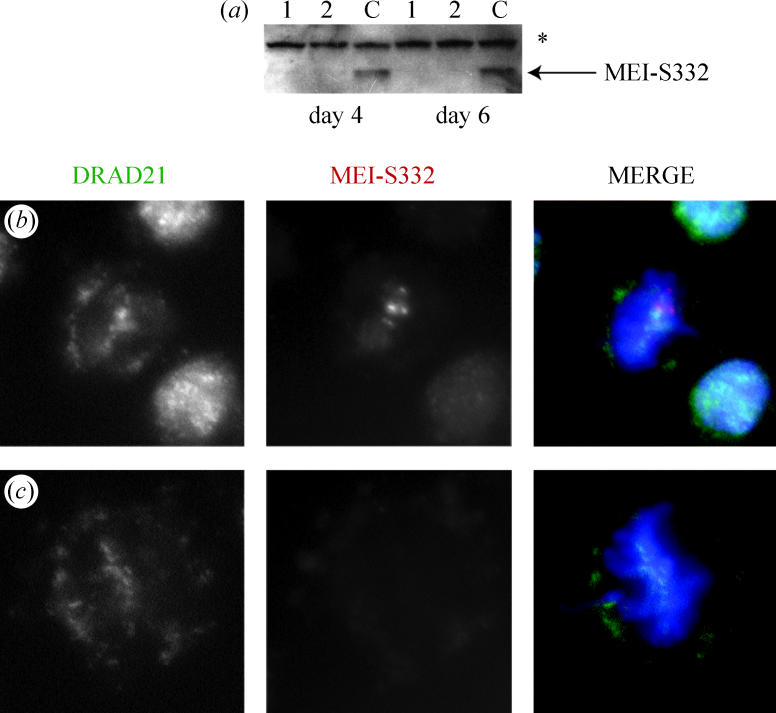
RAD21 localizes independently of MEI-S332 on mitotic metaphase chromosomes. (a) Western blot of mei-S332 RNAi experiment. 15 μg of fragment 1 or fragment 2 dsRNA were added to cells, and samples taken at day 4 and day 6 for Western blotting with a MEI-S332 antibody. No dsRNA was added to control wells (c). Both fragments 1 and 2 effectively depleted the MEI-S332 protein. An asterisk marks a non-specific band and demonstrates equal loading. (b) and (c) cells were cytospun onto slides and stained for DRAD21 (green), MEI-S332 (red) and DNA (blue). DRAD21 localises to metaphase chromosomes in the presence (b) or absence of MEI-S332 (c). (b) Control metaphase cell at 5 days after the start of the experiment. (c) Metaphase cell 5 days after 25 μg of fragment 2 dsRNA was added to cells.
5. Does MEI-S332 have activities in meiosis I?
Drosophila mei-S332 mutants exhibit mostly non-disjunction in meiosis II, although there are low levels of meiosis I non-disjunction (Kerrebrock et al. 1992). This is most detectable in female meiosis I. The characteristics of meiosis I differ in Drosophila between females and males. In females, homologous chromosomes form a synaptonemal complex, undergo recombination and retain linkage via chiasmata. In addition, the spindle is anastral and organized by the chromosomes. In males, the homologues associate via specific pairing sites and do not recombine and the spindle is organized by a centrosome.
MEI-S332 is not essential for kinetochore function in meiosis I or mono-orientation of the two sister chromatid kinetochores because, if so, levels of meiosis I non-disjunction would be more pronounced in the null alleles. It is possible that kinetochore activity in meiosis II is affected in mei-S332 mutants because lagging chromosomes are observed (Kerrebrock et al. 1992). These could arise either from attachment of the kinetochore of single, separated, sister chromatids to both poles, a failure of kinetochore microtubule attachment, or a failure of poleward movement. Defects in kinetochore behaviour are consistent with the observed chromosome loss that occurs in the mutants in meiosis II.
The frequency of meiosis I missegregation events is between 10- and 30-fold below levels in meiosis II and is relatively constant between the 10 alleles, despite their range of effects in meiosis II. This suggests that MEI-S332 affects meiosis I in females via a distinct mechanism from its role in maintaining cohesion at sister-chromatid centromeres. At present, this function is unknown, but given the role of vertebrate Sgo1 in affecting spindle microtubule dynamics and the distinct requirements for spindle formation in female and male meiosis, an investigation of spindle properties in mei-S332 mutants is warranted.
6. Male and female specific MEI-S332 protein domains
An unexpected characteristic of the mei-S332 alleles that exhibit moderate levels of chromosome missegregation is that they differentially affect male or female meiosis (Kerrebrock et al. 1992). The sex predominance of these alleles is not a consequence of higher levels of MEI-S332 activity being required in one sex over the other, because two alleles predominantly affect females, whereas two alleles predominantly affect males. It is striking that the male-predominant alleles map to the N terminal domain of the protein and the female-predominant alleles map to the C terminal basic domain (Kerrebrock et al. 1995). The female-predominant mutations cause a failure of detectable centromere localization of the protein in both female and male meiosis, as do all point mutations in the C terminus (Tang et al. 1998). In mitosis, these mutant forms of the protein are able to localize during slower canonical mitotic cycles but not during the rapid divisions of early embryogenesis. The mechanism by which the sex-predominant alleles preferentially perturb meiosis in one sex rather than the other remain to be understood. These alleles do not show higher levels of non-disjunction in meiosis I than the other mei-S332 alleles. Therefore, it is not the case that these mutations affect the aspects of meiosis I unique to males or females (Kerrebrock et al. 1992).
It seems most likely that despite the mechanistic similarities in meiosis II in Drosophila females and males, some of the other proteins needed for sister-chromatid segregation are unique to each sex. The sex-predominant alleles of mei-S332 presumably differentially affect interactions with these sex-specific players. It is interesting that the microtubule-binding domain of the vertebrate Sgo1 protein is the conserved N terminal region where male-specific alleles map. The male meiotic spindle is similar to a mitotic spindle, but the female spindle is anastral and organized by the chromosomes. In addition, the components of the meiotic cohesin complex remain to be identified in Drosophila. A Rec8 orthologue has not been identified and it is possible that there are sex-specific components of the cohesin complex used in female and male meiosis. If MEI-S332 interacts directly with the cohesin complex, this would provide an appealing explanation for the sex-predominant alleles.
7. Regulation of MEI-S332 localization and delocalization
Using transient transfection of a functional MEI-S332–GFP fusion into mitotically dividing Drosophila S2 culture cells as an assay, we found that both the conserved N and C termini are required for centromere localization of MEI-S332 (Lee et al. 2004). The majority of the central domain of the protein could be deleted without affecting centromere binding. Previously, point mutations in the N terminus were found to have no effect on localization, despite their predicted disruption of a putative coiled-coil structure. Although these amino acid changes did not impinge on chromosomal association, deletion of the entire region demonstrated it to be essential.
Two distinct lines of evidence demonstrate that although MEI-S332 binds to the centromere, this binding is not sequence specific, and the protein has the capability to bind throughout the chromosome. First, overexpression of MEI-S332 following transfection into culture cells or upon high levels of induction in the developing fly revealed that the protein is not constrained intrinsically to bind solely to the centromere (LeBlanc et al. 1999; Lee et al. 2004). In cells with high levels of the protein, MEI-S332 was observed to be present along the length of the chromosomes. Similarly, when mei-S332 was mutated such that it could not bind or be phosphorylated by POLO kinase, the protein not only failed to dissociate from the centromeres in anaphase but frequently was present coating the chromosomes in subsequent interphases and metaphases (Clarke et al. 2005). Second, MEI-S332 localizes to the vicinity of the kinetochore on minichromosomes in which a centromere and kinetochore has formed de novo, that is, on DNA sequences derived from the euchromatin and subtelomeric heterochromatin of the X-chromosome that do not bind MEI-S332 on a normal X-chromosome (Lopez et al. 2000). Drosophila MEI-S332 also will bind to human centromeres following transfection (Lee et al. 2004). These results indicate that localization of MEI-S332 is dictated by chromatin structure rather than DNA sequence.
The trans-acting factors necessary for MEI-S332 localization largely remain to be identified. Ablation of the centromere-specific form of histone H3 CID (the Drosophila CenpA homologue) either by RNAi or by antibody injection eliminated MEI-S332 centromere localization (Blower & Karpen 2001). This could reflect a requirement for CenpA itself for MEI-S332 binding by an unclear mechanism given the distinct, non-overlapping sites of localization of the two proteins within the centromere region. An alternative possibility exists, however, because ablation of CenpA also causes loss of localization of the BubR1 spindle assembly checkpoint protein (Blower & Karpen 2001). In fission yeast, localization of Sgo1 to the centromere requires Bub1 (Kitajima et al. 2004). The dependency of MEI-S332 on the Bub1 or BubR1 checkpoint pathway remains to be elucidated.
Dissociation of MEI-S332 from the chromosomes does not occur in separase or polo mutants (Lee et al. 2004; Clarke et al. 2005). The retention of MEI-S332 in separase mutants indicates that anaphase promoting complex/cyclosome (APC/C) activity is not sufficient to delocalize MEI-S332 because the APC is active in separase mutants (Philp & Glover 1997). This may differ from other organisms in which Sgo1 family members have been shown to be APC/C substrates, but these studies have not investigated whether APC/C affects centromere localization or degrades pools of the Sgo1 protein at anaphase (Kitajima et al. 2004; Salic et al. 2004). It is possible that degradation of the proteins by the APC/C follows centromere dissociation and is not causal for delocalization. Similarly, although levels of S. pombe Sgo1 decline normally in separase mutants, centromere localization of the protein has not been examined (Kitajima et al. 2004). The requirement for separase function for MEI-S332 delocalization in Drosophila is not fully understood, but it is unlikely that MEI-S332 is a direct substrate for separase. Deletion of the two best candidate separase cleavage sites in MEI-S332 does not affect its association or dissociation from the centromere (Lee et al. 2004). It also is not known whether separase is needed to delocalize MEI-S332 in meiosis as well as mitosis. If so, then MEI-S332 itself would require some protection against separase action at the metaphase I/anaphase I transition. In contrast to separase, POLO appears to promote dissociation of MEI-S332 by direct binding and phosphorylation (Clarke et al. 2005).
MEI-S332 also persists on the centromeres into anaphase in deco and separation anxiety mutants (Williams et al. 2003). These genes encode acetyltransferases, and deco is the Drosophila orthologue of the yeast Eco1 gene necessary to establish cohesion by activating the cohesin complex. In Drosophila, functional san and deco genes are needed for accumulation of the Rad21 cohesin subunit at the centromeres but not for localization of MEI-S332. In these mutants, the spindle assembly checkpoint remains active and sister chromatids separate in a separase-independent manner. At present, it is not clear whether san and deco have a direct effect on MEI-S332 dissociation, whether inactivation of the spindle assembly checkpoint is a prerequisite for MEI-S332 delocalization, or whether the failure of MEI-S332 dissociation is because separase is not activated in these mutants.
8. Inactivation of MEI-S332
In polo mutants, MEI-S332 is retained on the centromeres in anaphase of mitosis and anaphase II of meiosis (Clarke et al. 2005). Despite this, sister chromatids can separate from each other, although non-disjunction occurs. These observations reveal another layer of MEI-S332 regulation: there exists a mechanism to inactivate the protein that is independent of delocalization of the protein from the centromere. This conclusion is supported further by the observation that in two other mutants, sister chromatids can separate and still retain MEI-S332 localization. In ord mutants, the sister chromatids precociously separate early in meiosis I, but MEI-S332 is retained on the centromeres (Bickel et al. 1998). In mitosis, in the absence of the condensin subunit dCAP-G, the sister-chromatid centromeres lose their cohesion while retaining localized MEI-S332 (Dej et al. 2004). Similarly, other family members can be inactivated while retaining centromere association. If the S. pombe Sgo1 protein is ectopically expressed through meiosis II, then it binds to the centromeres but does not block dissociation of the sister chromatids at the metaphase II/anaphase II transition (Rabitsch et al. 2004). The control of MEI-S332 activity will be an important area of investigation.
9. Conclusions
Drosophila MEI-S332 is the founding member of a recently identified protein family whose members are needed for the maintenance of cohesion at sister-chromatid centromeres in mitosis and meiosis. Future work will uncover whether these proteins protect the cohesin complex from cleavage by the separase protease and, if so, whether this is via a direct physical interaction. Precise positioning of the proteins relative to the kinetochore and centromeric pools of cohesin at the level of the electron microscope is likely to be informative both in understanding the relationship between MEI-S332 (or the Sgo proteins) and cohesin, and also to the effect of some family members on kinetochore activity and structure. The domains of MEI-S332 that are required for centromere localization have been mapped, but further studies are needed to define the regions that function to confer cohesion. It will be interesting to test for additional roles of MEI-S332 and other family members, such as in controlling spindle microtubule dynamics. Unravelling the regulatory mechanisms by which the protein is inactivated will be an important future goal.
Acknowledgments
We thank George Bell for assistance with analysis of the Drosophila mei-S332 genes and Jacqueline Lopez for the micrographs in figure 1. This work was supported by grant MCB 0132237 from the National Science Foundation to T.O.-W.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Chromosome segregation’.
Present address: Kansai Advanced Research Center, 588-2, Iwaoka-cho, Iwaoka, Nishi-ku, Kobe, Hyogo 651-2492, Japan.
References
- Balicky E.M, Endres M.W, Lai C, Bickel S.E. Meiotic cohesion requires accumulation of ORD on chromosomes before condensation. Mol. Biol. Cell. 2002;21:3890–3900. doi: 10.1091/mbc.E02-06-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S.E, Wyman D.W, Orr-Weaver T.L. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–1331. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S.E, Moore D.P, Lai C, Orr-Weaver T.L. Genetic interactions between mei-S332 and ord in the control of sister-chromatid cohesion. Genetics. 1998;150:1467–1476. doi: 10.1093/genetics/150.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D, Karpen G.H. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo S.B.C, Clyne R.K, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Clarke A.S, Tang T.T.-L, Ooi D.L.-Y, Orr-Weaver T.L. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev. Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Clayberg C. Cytogenetic studies of precocious meiotic centromere division in Lycopersicon esculentum Mill. Genetics. 1959;44:1335–1346. doi: 10.1093/genetics/44.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. Genetic analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. Mol. Gen. Genet. 1971;113:251–272. doi: 10.1007/BF00339546. [DOI] [PubMed] [Google Scholar]
- Dej K.J, Ahn C, Orr-Weaver T.L. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168:895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A.E, Cantley L.C, Yaffe M.B. Proteomic screen finds pSer/pThr binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Goldstein L.S.B. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner C.F. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- Katis V.L, Galova M, Rabitsch K.P, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W, Miyazaki W.Y, Birnby D, Orr-Weaver T.L. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W, Moore D.P, Wu J.S, Orr-Weaver T.L. MEI-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S, Kawashima S.A, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- LeBlanc H.N, Tang T.T.-L, Wu J.S, Orr-Weaver T.L. The mitotic centromere protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
- Lee B.H, Amon A, Prinz S. Spo13 regulates cohesin cleavage. Genes Dev. 2002;16:1672–1681. doi: 10.1101/gad.989302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y, Orr-Weaver T.L. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 2001;17:753–777. doi: 10.1146/annurev.cellbio.17.1.753. [DOI] [PubMed] [Google Scholar]
- Lee J.Y, Dej K.J, Lopez J.M, Orr-Weaver T.L. Control of centromere localization of the MEI-S332 cohesion protection protein. Curr. Biol. 2004;14:1277–1283. doi: 10.1016/j.cub.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Lopez J.M, Karpen G.H, Orr-Weaver T.L. Sister-chromatid cohesion via MEI-S332 and kinetochore assembly are separable functions of the Drosophila centromere. Curr. Biol. 2000;10:997–1000. doi: 10.1016/s0960-9822(00)00650-3. [DOI] [PubMed] [Google Scholar]
- Marston A.L, Tham W.H, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Moore D.P, Page A.W, Tang T.T.-L, Kerrebrock A.W, Orr-Weaver T.L. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J. Cell Biol. 1998;140:1003–1012. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Philp A.V, Glover D.M. Mutations affecting chromatid separation in Drosophila: the fizzy metaphase arrest persists in pimples fizzy and fizzy three rows double mutants. Exp. Cell Res. 1997;230:103–110. doi: 10.1006/excr.1996.3396. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P, Petronczki M, Javerzat J.P, Genier S, Chwalla B, Schleiffer A, Tanaka T.U, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P, Gregan J, Schleiffer A, Javerzat J.P, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters J.C, Mitchison T.J. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Shonn M.A, McCarroll R, Murray A.W. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 2002;16:1659–1671. doi: 10.1101/gad.975802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.T.-L, Bickel S.E, Young L.M, Orr-Weaver T.L. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch K.P, Galova M, Schleiffer A, Buonomo S.B.C, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 2003;13:R104–R114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Warren W.D, et al. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Williams B.C, Garrett-Engele C.M, Li Z, Williams E.V, Rosenman E.D, Goldberg M.L. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Xu T, Harrison S.D. Mosaic analysis using FLP recombinase. In: Goldstein L.S.B, Fyrberg E.A, editors. Drosophila Melanogaster: practical uses in cell and molecular biology. Academic Press; San Diego: 1994. pp. 655–681. [DOI] [PubMed] [Google Scholar]