Abstract
The sister chromatid cohesion essential for the bi-orientation of chromosomes on mitotic spindles depends on a multi-subunit complex called cohesin. This paper reviews the evidence that cohesin is directly responsible for holding sister DNAs together and considers how it might perform this function in the light of recent data on its structure.
Keywords: sister chromatid cohesion, cohesin, SMC and Kleisin proteins, chromosome segregation, mitosis, meiosis
1. Introduction
Cohesion between sister chromatids is essential for their bi-orientation on mitotic spindles as well for efficient post-replicative double-strand break repair. Cohesion between sisters persists from their generation during DNA replication until their bi-orientation during metaphase. The destruction of sister chromatid cohesion is necessary for the segregation of sister chromatids to opposite poles of the cell at the onset of anaphase. Genetic studies have shown which that sister chromatid cohesion requires the activity of four proteins, Smc1, Smc3, Scc1 and Scc3, which together form a multi-subunit complex called ‘cohesin’ (Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998; Toth et al. 1999). These four proteins form a complex stable enough to be purified from soluble extracts. They also invariably coexist along with the Pds5 protein at specific locations along chromosomal DNA (Hartman et al. 2000; Panizza et al. 2000). The ability of cohesins to associate with chromosomes and to confer sister chromatid cohesion depends on a largely separate complex composed of the Scc2 (Mis4, Nipped B) and Scc4 proteins (Michaelis et al. 1997; Furuya et al. 1998; Ciosk et al. 2000). A small fraction of Scc2/Scc4 complexes can be found associated with soluble cohesin complexes (Arumugan et al. 2003), but they are rarely, if ever, found at the same locations on chromosomes as cohesin (Ciosk et al. 2000; Lengronne et al. 2004). This article will briefly summarize the evidence that sister DNAs are physically held together by cohesin and not by other protein complexes or by the inter-twining of sister DNAs. It will then consider, in the light of what is now known about the structure of cohesin, potential mechanisms by which it might perform its function.
2. What is the evidence that cohesin holds sister DNAs together?
In addition to being required for sister chromatid cohesion, the proteins actually responsible for holding sister DNAs together must be present on chromosomes at the right place and time. Most evidence suggests that cohesin fulfils this criterion. In yeast, cohesin's Scc1 and Smc subunits are found tightly associated with specific sites along chromosomes. Interestingly, the sites along chromosome arms usually correspond to inter-genic regions at which transcription converges from both left and right flanking genes (Filipski & Mucha 2002; Glynn et al. 2004; Lengronne et al. 2004). Cohesin is, however, considerably more abundant within peri-centric regions spanning 50–60 kb surrounding kinetochores than it is along chromosome arms, which is due to an ability of kinetochores to stimulate cohesin's recruitment to their vicinity and not to properties of the peri-centric regions per se (Weber et al. 2004). The peri-centric regions within which cohesin is enriched due to kinetochore activity are considerably larger than kinetochores themselves. This raises the question of whether cohesin recruited by kinetochores moves large distances into their flanking regions, or whether kinetochores somehow influence (at some distance) the state of surrounding chromatin so that it is more probable to recruit cohesin. Cohesin's distribution along chromosomes in the fission yeast Schizosaccharomyces pombe is broadly similar to that in Saccharomyces cerevisiae, but its enrichment to peri-centric regions is known to depend on binding of the HP1-like Swi6 protein to trimethylated lysine 9 residues on histone H3 (Bernard et al. 2001; Nonaka et al. 2002).
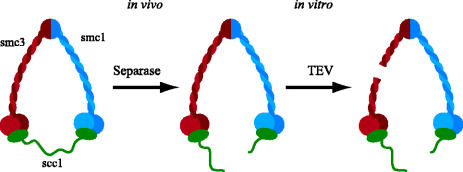
In S. cerevisiae, cohesin's Scc1 subunit disappears from both centromeric and chromosome arms regions at the onset of anaphase (Michaelis et al. 1997) due to its cleavage by a site-specific thiol protease called separase (Uhlmann et al. 1999, 2000). Crucially, mutation of Scc1's separase cleavage sites prevents Scc1's disappearance from chromosomes and blocks the loss of sister chromatid cohesion upon activation of separase. The latter is mediated by the anaphase-promoting complex or cyclosome (APC/C), a ubiquitin protein ligase that upon bi-orientation of all chromosomes on the mitotic spindle, promotes destruction of both cyclin B and an inhibitory separase chaperone called securin (Cohen-Fix et al. 1996; Funabiki et al. 1996). In addition, replacement of endogenous SCC1 genes by a version that expresses an Scc1 protein that can be cleaved by the foreign TEV protease makes it possible to remove Scc1 from chromosomes and trigger sister chromatid disjunction by induction of the TEV protease (Uhlmann et al. 2000). Cleavage of Scc1's orthologue in S. pombe Rad21 by separase is also essential for sister chromatid disjunction (Tomonaga et al. 2000), though whether this causes most cohesin to dissociate from chromosomes is not known.
The finding that most cohesin dissociates from chromosomes in animal cells during prophase and pro-metaphase initially cast some doubt as to whether cohesin could conceivably be responsible for sister chromatid cohesion during metaphase (Losada et al. 1998; Sumara et al. 2000), when sister chromatids remain associated not only at centromeres, but also along chromosome arms. Subsequent studies have detected small amounts of cohesin at centromeres during metaphase, either using antibodies against endogenous proteins in Drosophila melanogaster (Warren et al. 2000; Valdeolmillos et al. 2004) or by detecting epitope (myc or GFP) tagged proteins expressed at physiological levels from tetracycline inducible promoters in mammalian tissue culture cells (Waizenegger et al. 2000; Gimenez-Abian et al. 2004; Hauf et al. 2005). The disappearance of centromeric cohesin at the onset of anaphase in animal cells is accompanied by cleavage of its Scc1 subunit and indeed the latter is necessary for proper chromatid disjunction (Hauf et al. 2001). It has been much harder to detect cohesin along chromosome arms during metaphase, but circumstantial evidence suggests that it probably exists and is responsible for holding arms together (Gimenez-Abian et al. 2004).
What emerges from these studies is the notion that two distinct processes remove cohesin from chromosomes during mitosis in mammalian cells. The first process commences during prophase and involves phosphorylation of its Scc3-like subunits called SA1 and SA2 by the Polo-like kinase PLK1 (Hauf et al. 2005). This so-called ‘prophase pathway’ does not appear to involve cleavage of Scc1 by separase (Sumara et al. 2000, 2002). The second process only takes place at the onset of anaphase and involves cleavage of Scc1 by separase. The prophase pathway seems capable of removing all cohesin from chromosome arms when cells are arrested for prolonged periods in a mitotic-like state due to activation of the spindle assembly checkpoint by poisons that either stabilize (taxol) or de-stabilize (nocodazole) microtubules. The persistence of cohesin at centromeres under these circumstances leads to the formation of chromosomes whose sister chromatids are only connected at their centromeres. The recent finding that either inactivation of PLK1 (Gimenez-Abian et al. 2004) or expression of SA2 proteins that can no longer be phoshorylated by PLK1 (Hauf et al. 2005) prevents sister chromatid arm separation in the presence of nocodazole is a powerful argument that cohesin is indeed responsible for arm cohesion as well as that at centromeres.
An important issue is what causes cohesin at centromeres to be refractory to the prophase pathway. Recent data suggests that proteins orthologous to the yeast Sgo1 (Kitajima et al. 2004; Rabitsch et al. 2004) and D. melanogaster MeiS332 proteins (Kerrebrock et al. 1995) are located in the peri-centric regions of mitotic chromosomes and are responsible for preventing dissociation of cohesin by the prophase pathway in HeLa cells (McGuinness et al. 2005). In conclusion, small amounts of cohesin can be detected at centromeres during metaphase in animal cells while little or none can be detected by the time cells have initiated anaphase. These findings are consistent with the notion that cohesin is responsible for holding sister chromatids together during their bi-orientation on mitotic spindles in animal as well as fungal cells. Lastly, if cohesin were responsible for sister chromatid cohesion during mitosis, then orthologues of its main subunits should be found encoded in all known eukaryotic genomes, which is indeed the case (Nasmyth & Schleiffer 2004).
Cohesin must also be found at the right places at the right time during meiosis. Owing to crossing over between maternal and paternal chromatids, sister chromatid cohesion along chromosome arms connects maternal and paternal centromeres together. The structures so created are called ‘chiasmata’ (Petronczki et al. 2003). This has the consequence that homologous maternal and paternal sister centromere pairs, and not sister centromeres themselves, are pulled in opposite directions by meiosis I spindle fibres. Sister chromatid cohesion along chromosome arms is therefore essential for meiosis I chromosome segregation, unlike mitosis where it is relatively unimportant (Rieder & Cole 1999). Loss of sister chromatid cohesion along arms is essential for the resolution of chiasmata and hence for the first meiotic division. Cohesion between sister centromeres is retained after meiosis I because it is necessary for bi-orientation of sister centromeres during meiosis II. Two key features should therefore distinguish meiotic from mitotic cohesin if it were indeed the substance that held sister chromatids together. First, it should be found in abundance on chromosome arms until metaphase I and disappear from this location at the onset of anaphase I. Second, it should persist at centromeres until the onset of anaphase II. This is clearly the case in both budding (Klein et al. 1999) and fission yeast (Watanabe & Nurse 1999), where cohesin containing Rec8, a meiosis-specific Scc1 variant, is present along the entire axis of chromosomes during meiosis I, disappears from chromosomes arms at the onset of anaphase, but persists at centromeres until the onset of anaphase II. Furthermore, cleavage of Rec8 by separase is essential for the removal of cohesin from chromosome arms at the onset of anaphase I and for the resolution of chiasmata in both S. cerevisiae (Buonomo et al. 2000) and S. pombe (Kitajima et al. 2003).
The situation in mammals is much more confusing. Both mitotic Scc1 and Rec8 are expressed during spermatogenesis. Rec8's distribution resembles that of its yeast orthologues, namely it is present along the axes of chromosomes from pre-meiotic DNA replication until the onset of anaphase I, whereupon it disappears from chromosome arms but persists around centromeres until the onset of anaphase II (Eijpe et al. 2003; Lee et al. 2003). However, inactivation of Rec8 in mice causes only a modest loss of sister chromatid cohesion (Bannister et al. 2004), as does inactivation of a meiosis-specific isoform of Smc1 called Smc1β (Revenkova et al. 2004). This suggests either that Scc1 also makes an important contribution to sister chromatid cohesion during meiosis, or that cohesin containing Scc1- or Rec8-like proteins is not in fact responsible for sister chromatid cohesion during mammalian meiosis. The former seems to be the more likely explanation, but it will be necessary to inactivate both Scc1 and Rec8 in meiotic cells to find out for sure. What is even more confusing are the claims by cytologists that neither Smc1 nor Smc3 proteins are present along chromosome arms at metaphase I (Revenkova et al. 2001; Eijpe et al. 2003). If true, this would imply either that Rec8 and/or Scc1 proteins mediate cohesion between sister chromatid arms in the absence of SMC proteins, or that cohesin is simply not required for meiotic sister chromatid cohesion. Neither possibility appears plausible in the light of what is known about the structure and function of cohesin in yeast. Our present picture of the distribution of Smc1 and Smc3 proteins during mammalian meiosis may therefore be incomplete. A fundamental problem with studies that merely use cytology, is that cohesin is notoriously difficult to detect on fixed chromatin, and any failure to detect one of its subunits can be attributed equally well to poor antibodies or to inappropriate fixation conditions as to any actual absence of the protein in question. Given our current understanding of the structure of Scc1-like proteins, known as α-kleisins, it seems improbable that they could act without binding to the ATPase heads of SMC proteins (Haering et al. 2004; #4773). The notion that α-klesisns might mediate sister chromatid cohesion without SMC partners does not seem probable.
The situation is equally confusing in Drosophila, where a meiosis-specific Rec8-like protein called C(2)M accumulates after pre-meiotic DNA replication and disappears long before the first meiotic division, a behaviour that is inconsistent with it having any role in holding homologues together during meiosis I (Heidmann et al. 2004). The simplest explanation for this observation is that the mitotic Scc1-like protein Rad21 is largely, if not solely, responsible for sister chromatid cohesion during meiosis as well as during mitosis in Drosophila. However, it is still unknown whether Rad21 is even present in meiotic cells in Drosophila. The only animal in which there is clear evidence that cohesin mediates sister chromatid cohesion during meiosis is C. elegans, where a meiosis-specific Rec8-like protein is both necessary for sister chromatid cohesion and is distributed on chromosomes in a pattern that resembles, at least superficially, that of Rec8 in yeast (Pasierbek et al. 2001). It is also known that separase is essential for the resolution of chiasmata in C. elegans (Siomos et al. 2001).
Another issue that is potentially relevant to this debate is whether cohesin's presence on chromosomes is necessarily correlated with sister chromatid cohesion. This is clearly not the case, but this does not mean that cohesin cannot mediate the connections between sister chromatids. Cohesin normally re-associates with chromosomes during telophase in animal cells (Losada et al. 1998; Sumara et al. 2000), that is, long before cells actually undergo DNA replication. Thus, in G1 cells, cohesin associates with chromatin in a manner that can have nothing to do with sister chromatid cohesion. Even in budding yeast, where cohesin's Scc1 subunit is only synthesized shortly before S phase, Scc1 expressed from the GAL promoter during G2 associates stably with chromosomes but does so without forming sister chromatid cohesion (Uhlmann & Nasmyth 1998; Haering et al. 2004). The implication is that cohesive structures, if indeed they are formed by cohesin, can only be formed during DNA replication. Clearly, cohesin can bind tightly to chromatin without forming cohesive structures. This phenomenon may help to explain the otherwise puzzling observation that during metaphase in budding yeast cells sister kinetochores are pulled apart by 200–500 nm even though cohesin remains tightly associated with them (Goshima & Yanagida 2000; He et al. 2000; Tanaka et al. 2000). It has been suggested that this observation casts doubt on the notion that cohesin holds sister chromatids together. It is more likely, though by no means proven, that the cohesin detected at yeast kinetochores during metaphase is not actually participating in holding sister chromatids together, just like the cohesin that binds to chromosomes in G1 in animal cells or the cohesin that only associates with chromosomes after DNA replication in yeast.
In summary, cohesin's location during mitosis and meiosis is consistent with it conferring the connections between sister chromatids needed for chromosome segregation, but its distribution and roles during meiosis in animal cells definitely requires clarification. One cannot at present exclude the possibility, albeit unlikely, that cohesin has only a minor role in mediating sister chromatid cohesion during mammalian meiosis. It also remains to be shown that cohesin is present at precisely the point where sister DNAs are connected.
Another important question when considering the case for cohesin is whether proteins other than cohesin might form the actual bridge between sister DNAs. For example, Furuya et al. (1998) have suggested that proteins orthologous to Scc2 and Mis4 and not cohesin itself might form the structures that hold sister chromatids together. The authors proposed ‘a simple model in which Mis4 associates with chromosomes throughout the cell cycle and forms the connection between sister chromatids directly or indirectly through another linking protein. Disruption of the link might occur by splitting of the oligomeric Mis4 complex into halves or removal of the linking protein from chromosomes in anaphase while Mis4 remains associated with chromosomes. Post-translational modification such as protein de-phosphorylation may be important in disconnecting the linkage.’ It was for this reason that it was suggested to call this class of proteins adherins to distinguish them from cohesins. There are several problems with this proposal. The first is that unlike cohesin, which is essential not only for building cohesion during DNA replication but also for maintaining it until the onset of anaphase, neither Scc2 nor Scc4 appear to be necessary to maintain cohesion between sister chromatids after DNA replication has been completed (Ciosk et al. 2000). This conclusion needs, however, to be qualified because only temperature sensitive alleles have been used to inactivate Scc2 and Scc4 in post-replicative cells, and it is conceivable that the mutations do not fully inactive the Scc2/Scc4 complex upon shifting post-replicative cells to the restrictive temperature. A second problem is that the Scc2/Scc4 complex is essential for loading cohesin onto chromosomes both in yeast (Ciosk et al. 2000) and in Xenopus extracts (Gillespie & Hirano 2004; Takahashi et al. 2004). The most parsimonious explanation of this finding is that the sister chromatid cohesion defects of scc2 and mis4 mutants are due to their failure to load cohesin onto chromosomes. Lastly, there is so far no strong evidence that the distribution of Scc2/Scc4 complexes on chromosomes tracks with cohesion between sister chromatids during mitosis and meiosis, let alone any clue as to how the bridges it is purported to build might be broken at the onset of anaphase.
Another candidate that has repeatedly been proposed as a substitute for cohesin is the class of proteins orthologous to MeiS332 in D. melanogaster and Sgo1 in yeast. To explain their finding that little or no cohesin was bound to mitotic chromosomes in animal cells, Losada et al. (2000, p. 406) suggested as an alternative to the notion that cohesin was merely undetectable on mitotic chromosomes, that ‘the cohesin complex participates in holding the sister chromatids together from S phase to G2 but it does not play a major role in cohesion from prophase to metaphase. A different protein component, such as MeiS332 in D. melanogaster could function as a mitosis-specific chromatid glue.’ Likewise, according to Salic et al. (2004, p. 575) ‘it is not clear that cohesin alone is responsible for centromeric cohesion in vertebrate metaphase; it is possible that part of this function is passed to another protein (such as Sgo) or even DNA topology (such as catenation or Holliday junctions) during prophase, when much of the cohesin is removed from chromosomes’. It is true that Sgo1/MeiS332 proteins are usually found within peri-centric regions during mitosis and disappear from this location simultaneous with the loss of sister centromere cohesion, whether during mitosis or meiosis II (Moore et al. 1998; Salic et al. 2004). It is also true that they are required to maintain cohesion between sister centromeres during mitosis in tissue culture cells (Salic et al. 2004; McGuinness et al. 2005) and between meiosis I and meiosis II in D. melanogaster (Kerrebrock et al. 1992) and in yeast (Kitajima et al. 2004; Rabitsch et al. 2004). Despite these impeccable credentials, there are several reasons why Sgo1/MeiS332-like proteins cannot form the bridges that actually hold sister chromatids together. First and foremost, these proteins are not essential for mitosis in either yeast (Katis et al. 2004; Marston et al. 2004) or in D. melanogaster (LeBlanc et al. 1999). Second, they do not appear to associate with centromeres until the onset of mitosis in animal cells (Moore et al. 1998; Salic et al. 2004; McGuinness et al. 2005). Other proteins would have to hold sister chromatids together from S phase until prophase and Sgo1/MeiS332 would have to build connections anew at prophase, which all seems rather unlikely. Last but not least, recent evidence implies that Sgo1 actually functions to control cohesin's association with chromosomes. In Hela cells, inactivation of Sgo1 by RNA interference causes the loss of cohesin from centromeres before cells have had time to trigger anaphase. However, this precocious loss of centromeric cohesin is suppressed by expression of a non-phosphorylatable Scc3-SA2 (Hauf et al. 2005; McGuinness et al. 2005). The implication is that Sgo1's presence at centromeres might be required to prevent the prophase pathway from removing cohesin from this chromosomal location. If Sgo1 regulates cohesin in this fashion, then it is improbable that it also builds bridges between sisters that are independent of cohesin. Even if such bridges are built, they cannot be essential for chromosome segregation.
Another suggestion that repeatedly crops up is the notion that sister chromatids are not in fact held together by proteinaceous structures, but instead by the inter-twining of sister DNAs that is a legacy of DNA replication (Murray & Szostak 1985; Morrison et al. 2003; Salic et al. 2004). According to this way of thinking, cohesin might merely help to hold sister chromatids together by preventing de-catenation of sister DNAs by topo-isomerase II. There appear to be two incontrovertible reasons why this hypothesis cannot be valid, at least in the budding yeast S. cerevisiae. First, gel electrophoresis has shown that circular mini-chromosomes known to be held together by virtue of cohesin function in cells blocked in a mitotic-like state by nocodazole are clearly not inter-catenated (Koshland & Hartwell 1987). Second, cells arrested in metaphase due to depletion of the APC/C's activator protein Cdc20 are capable of separating most sister chromatid DNA when Cdc20 is induced from the GAL promoter in mutants supposedly lacking any topo-isomerase activity (Sullivan et al. 2004). There are even theoretical grounds for doubting whether cohesion mediated by inter-catenation of sister DNAs would be a good idea even in principle. Failure to de-catenate any single inter-twining of sister DNAs will lead to chromatid breakage during anaphase, a scenario that could only be rendered more probable by the existence of mechanisms that would inhibit de-catenation until anaphase onset (Nasmyth & Schleiffer 2004).
In summary, there is little or no hard evidence inconsistent with the notion that cohesin physically holds sister DNAs together. There are currently no realistic alternatives to this hypothesis. Nevertheless, the issue of whether cohesin really does perform this task can only be satisfactorily settled once we know how cohesin actually holds sister chromatids together. How then, might cohesin perform this extraordinary function?
3. How might cohesin hold sister chromatids together?
Cohesin exists as a stable soluble complex as well as one tightly associated with chromosomes. Our understanding of the structure of soluble cohesin complexes or subcomplexes is, needless to say, much more complete than that of structures formed by cohesin on chromosomes, which are difficult, if not impossible, to purify without running the risk of changing their nature. It is for this reason that the evolution of ideas as to how cohesin might hold sister chromatids together largely, but not exclusively, reflects advances in our knowledge as to the structure of soluble cohesin complexes.
The first major advance was the realization that cohesin's Smc1 and Smc3 subunits most probably form V-shaped heterodimers whose arms are composed of 50 nm long coiled coils, at the ends of which are globular ATP-binding heads. This stemmed from the analysis of electron micrographs of SMC proteins from bacteria, which form homodimers composed of anti-parallel coiled coils (Melby et al. 1998). The discovery that the coiled coils of bacterial SMC or MukB proteins were anti-parallel was surprising because all previously characterized coiled coils with dimensions similar to those of SMC proteins were parallel and not anti-parallel. It was assumed without any direct evidence that the coiled coils of SMC dimers were inter-molecular and that as a consequence their globular ATP-binding heads were composed of an N-terminal domain containing a Walker A motif from one SMC protein juxtaposed with a C-terminal domain containing a Walker B motif from a separate SMC polypeptide. According to this model, two intact polypeptide chains ran in anti-parallel directions from one head through the central hinge to the other head (Melby et al. 1998).
At around this time, cohesin had just been shown to contain equimolar amounts of two different SMC proteins, namely Smc1 and Smc3 (Losada et al. 1998; Toth et al. 1999). This suggested that cohesin might contain a Smc1/Smc3 heterodimer with a geometry similar to that of bacterial SMC proteins. If so, then each of the two heads of the predicted V-shaped Smc1/Smc3 heterodimers would be composed of Smc1 and Smc3 sequences while their arms would be composed of coiled coils formed between alpha helical sections of Smc1 and Smc3 (figure 1a). It seemed unlikely that DNA would be bound directly by coiled coils and because there was only a single hinge region that would be incapable of bivalent interactions, it was suggested that cohesin might hold sister chromatids together by the association of each of its two ATP-binding heads with sister DNAs (Toth et al. 1999). According to this scheme, the connection between sister chromatids would be conferred by the continuous polypeptide chains running from a head associated with one DNA molecule to its companion associated with its sister DNA (see figure 1a). There have been several variants of this type of model. The concept was adapted to the subsequent finding that the coiled coils of SMC proteins are in fact intra- and not inter-molecular (Haering et al. 2002). In this case, it was envisioned that one DNA was bound by the head of a Smc1 molecule, while its sister was bound by the head of a Smc3 molecule associated with Smc1 exclusively via their hinge dimerization domains (figure 1b). The proposal that cohesin's Scc1 subunit connects the Smc1 and Smc3 heads can also be accommodated by this type of model. It was suggested that Scc1 would provide a second (reinforcing) link holding the two heads and thereby sister DNAs together (figure 1c; Campbell & Cohen-Fix 2002).
Figure 1.
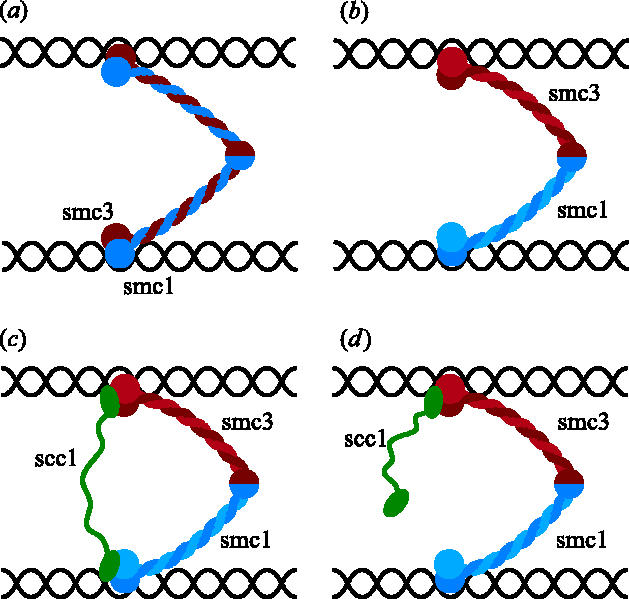
Models for sister chromatid cohesion according to which SMC ATPase heads bind DNA.
There are two fundamental problems with this model. First, it provides no explanation for how cohesin's two Smc heads would bind stably to DNA. Indeed, there is little or no evidence that Smc heads can perform this function. Second, the model provides no clear explanation for how Scc1 cleavage would cause cohesin to dissociate from chromosomes and destroy sister chromatid cohesion. A variation of this type of model envisions that the link between sister DNAs is provided not by an individual Smc1/Smc3 heterodimer, but by a pair of them held together by Scc1 molecules that connect the Smc1 head of one heterodimer to the Smc3 head of its partner heterodimer (figure 2; Anderson et al. 2002; Hirano 2002). This version of the model suffers from the same fundamental problems as earlier versions. It is nevertheless distinguished from them by proposing that the cohesin complexes engaged in holding sister chromatids together function as dimers.
Figure 2.
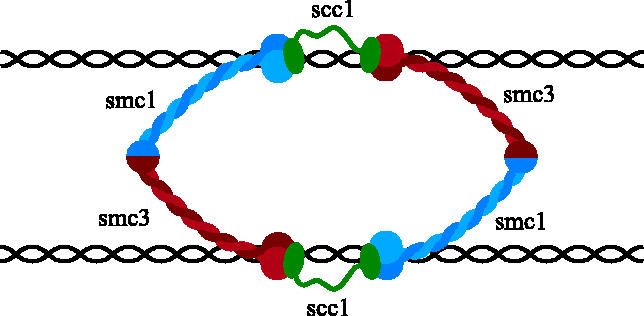
Scc1 inter-connects two Smc1/Smc3 heterodimers that bridge sister DNAs.
The lack of any convincing evidence that Smc1 or Smc3 heads bind DNA has not discouraged other models in which SMC heads are responsible for binding DNA. Before it became clear that Scc1 binds to Smc1 and Smc3 via their heads, it was suggested that the two heads of a single Smc1/Smc3 heterodimer bound the same DNA molecule and that Scc1 connected its hinge region to that of a separate Smc1/Smc3 heterodimer bound to its sister DNA (figure 3a; Uhlmann et al. 1999). This model had the merit of explaining why cohesion would be destroyed by Scc1 cleavage but still relied on hypothetical and potentially non-existent interactions between Smc heads and DNA. Moreover, the model was proved conclusively wrong by the finding that Scc1 binds Smc1/Smc3 heterodimers via their heads and not via their hinge domains (Haering et al. 2002, 2004). This type of model has recently been resurrected as the so-called ‘snap model’ (Milutinovich & Koshland 2003), according to which, cohesion between sister DNAs is mediated by two Smc1/Smc3 heterodimers bound to sister DNAs that associate with each other due to interactions between their coiled coils (figure 3b). A shortcoming of this model is that there is so far no evidence for a tight interaction between the coiled coils of different cohesin complexes. Indeed, there is little or no evidence that cohesin forms any kind of stable dimeric structure whether on or off chromosomes (Haering et al. 2002; Weitzer et al. 2003).
Figure 3.
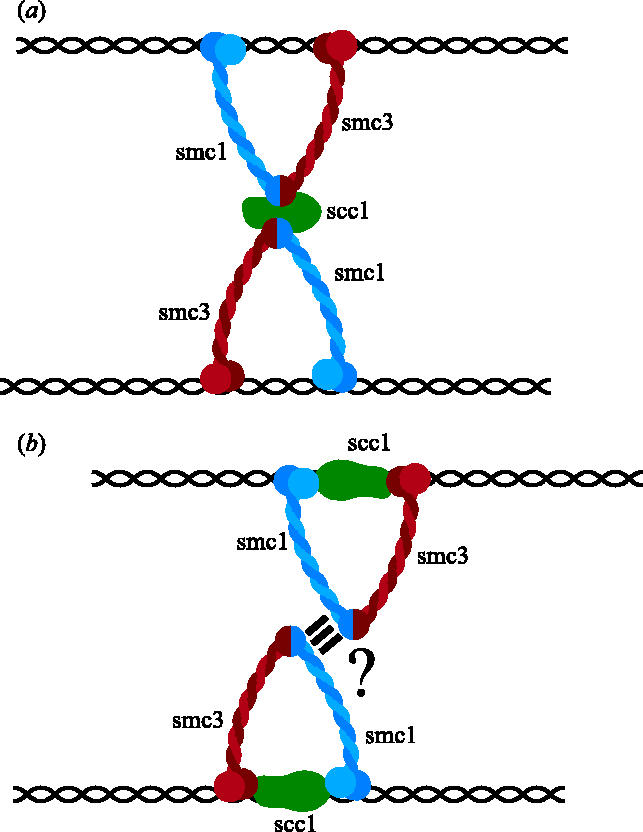
Might interactions between SMC hinges or between SMC coiled coils mediate the connection between sister DNAs?
Given the key part played by cleavage of Scc1 by separase in triggering sister chromatid disjunction at the onset of anaphase, the position of this subunit within cohesin is of crucial importance. An important step in establishing this was the discovery that the coiled coils of SMC proteins must be intra- and not inter-molecular (Haering et al. 2002; Hirano & Hirano 2002). The coiled coils of both Smc1 and Smc3 are formed by the folding back of each polypeptide on itself, forming rod shaped proteins with a globular ABC-like ATPase head at one end and a heterodimerization domain at the other, through which Smc1 and Smc3 interact to form stable V-shaped heterodimers (figures 1b and 4a). This development was important for understanding how Scc1 interacts with the Smc1/Smc3 heterodimer because it turns out to interact with the SMC ATPase head domains, whose identity would have been very different (i.e. composed of sequences from two different polypeptides) had the coiled coils proven to be inter-molecular.
Figure 4.
Cohesin forms a ring structure that might entrap DNA.
Co-expression of different cohesin subunits in insect cells showed that of the two non-SMC subunits Scc1 and Scc3, only the former forms direct stable contacts with the Smc1/Smc3 heterodimer. Thus Scc3 is recruited to the complex by its binding to Scc1, which in turn binds the Smc1/Smc3 heterodimer (Haering et al. 2002; figure 4a). A special feature of Scc1's association with Smc1/Smc3 heterodimers is its bivalent nature. Scc1's conserved N-terminal domain, which is predicted to form a helix turn helix domain, binds to Smc3's ATPase heads when both proteins are co-expressed in insect cells while its conserved C-terminal domain, which forms a winged helix turn helix domain, binds to a pair of β-strands at the bottom surface of Smc1's ATPase head (Haering et al. 2002, 2004). Scc1 is therefore envisioned to connect directly the Smc1 and Smc3 ATPase heads through a mechanism that is (potentially) independent of their association via hinge domains (figure 4a). This arrangement implies that cohesin's Smc1, Smc3 and Scc1 subunits form a gigantic ring or triangle, with Scc1 forming one side of the triangle and the coiled coils of Smc1 and Smc3 forming the other two sides (figure 4a). One of this triangle's corners (that known as the hinge) is formed by the association of Smc1 and Smc3's dimerization domains, while the other two are formed by the association of N- and C-terminal domains of Scc1 with Smc3 and Smc1 ATPase heads, respectively.
Because formation of a stable ring structure has potentially important implications as to the mechanism by which cohesin might associate with DNA, it is worth reviewing in detail the evidence for the three types of subunit interaction that create cohesin's ring. Association between Smc1 and Smc3 dimerization domains has been reconstructed in vitro and its dissociation constant determined to be in the low nanomolar range. Both domains are clearly homologous with the equivalent domains of bacterial SMC proteins, one of which has been crystallized and its homodimeric structure determined at atomic resolution (Haering et al. 2002). The monomer is a novel fold composed of two domains, related by a pseudo-twofold symmetry operation, linked by a long ordered loop (figure 5). The doughnut-shaped dimer is formed by combining three β-strands from domain I of one monomer with five β-strands from domain II of the other to form two eight-stranded β-sheets. As expected, the coiled coil forming α-helices emerging from the dimer are anti-parallel and crucially they originate from the same polypeptide chain, which proves without doubt that the coiled coils of SMC proteins are intra-molecular. Mutagenesis of glycine residues within Smc1 and Smc3 at sites predicted to lie at the interface between their hinge dimerization domains destroys their interaction in vitro. Because of the dimeric hinge's doughnut-shaped structure, there exist two independent interaction surfaces, and interestingly, mutation of both is required to abolish their interaction, at least in vitro (Chiu et al. 2004). The effect of mutations that specifically disrupt the two interaction surfaces have not yet been investigated in vivo. Important questions for the future are whether both interaction surfaces within the hinge dimerization domain are required for cohesin to function in vivo, how high an affinity is required for in vivo function, and whether the hinge has roles other than merely to join tightly together Smc1 and Smc3 proteins. It has been suggested, for example, that the angles subtended by Smc1 and Smc3 arms within cohesin are much wider than those made by Smc2 and Smc4 proteins within the related condensin complex and that the hinge might have some role in determining this angle (Anderson et al. 2002).
Figure 5.
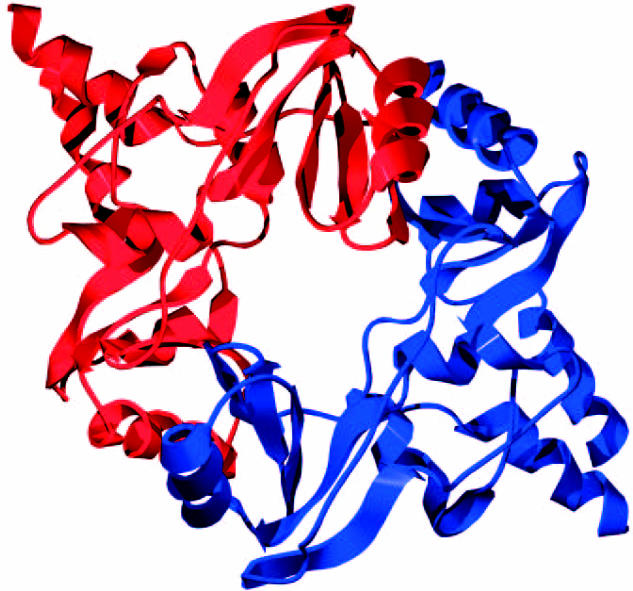
Crystal structure of a dimeric hinge domain from Thermatoga maritima (Haering et al. 2002).
The association of Smc1's ATPase head with that of Scc1's C-terminal domain has also been reconstructed in vitro and a crystal structure of a Smc1/Scc1-C complex has recently been determined to 2.9 Å resolution (Haering et al. 2004). Scc1's C-terminal domain forms a winged helix domain that interacts with a pair of β-strands at the bottom surface of the Smc1 head; that is, at the surface opposite to where its coiled coils enter the head (figure 6). Mutation of interaction residues, either within Smc1's β-strands or within Scc1's winged helix, abolish not only interaction of Scc1's C terminus with Smc1, but also all association of Scc1 with Smc1/Smc3 heterodimers, at least in yeast cells. The implication is that association of Scc1's C-terminal domain with Smc1 heads is essential for the subsequent formation of stable contacts between Scc1's N-terminal domain and the Smc3 head, assuming that is, that the latter really exists (see below).
Figure 6.
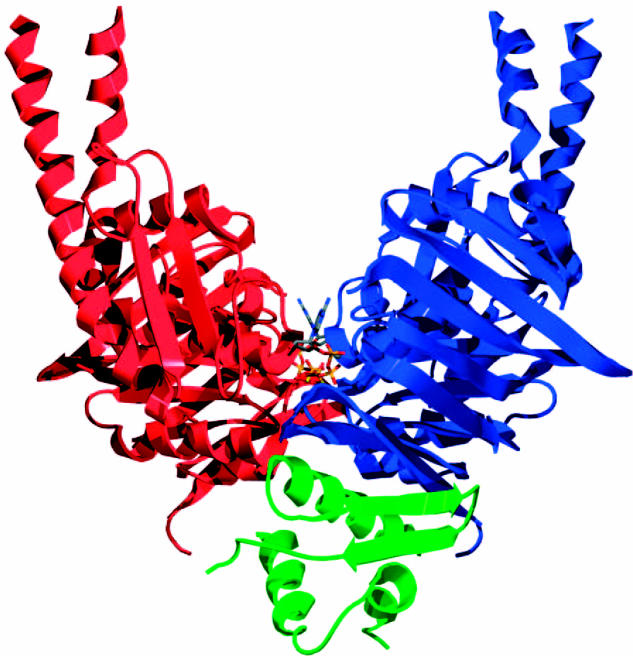
Crystal structure of a dimeric Smc1 ATPase head complexed with Scc1's C-terminal winged helix (Haering et al. 2004).
In contrast to the Smc1/Smc3 hinge interactions and the Smc1 head/Scc1-C interactions, which have been confirmed by crystal structures, the nature of the contact between Scc1's N-terminal domain and Smc3 heads is less certain. This interaction was first detected after co-expression of yeast Smc3 and Scc1 subunits in insect cells. Scc1's N-terminal domain associated with Smc1/Smc3 heterodimers lacking Smc1's head domain but not with Smc1/Smc3 heterodimers lacking Smc3's head domain (Haering et al. 2002). In the same series of experiments, Scc1's C-terminal domain had the opposite preference, namely for Smc1/Smc3 heterodimers containing Smc1's head. Co-expression studies also revealed that Scc1's N- and C-terminal domains also associated with isolated Smc3 and Smc1 ATPase domains, respectively. Importantly, Rec8's N- and C-terminal domains behave in an identical fashion (Gruber et al. 2003), suggesting that all Scc1-like proteins might possess an ability to bind Smc3 heads and thereby enable Smc1/Smc3 heterodimers to form stable tripartite ring structures. However, in contrast to the interaction between Scc1's C-terminal winged helix domain with Smc1 heads, the interaction between Scc1's N-terminal domain with isolated Smc3 heads has so far not been reproduced in vitro. Furthermore, when expressed alone in yeast, Scc1 N-terminal fragments fail to associate with endogenous Smc1/Smc3 heterodimers (Arumugan et al. 2003). These observations along with the finding that the interaction of Scc1 with Smc1/Smc3 heterodimers in yeast depends on the interaction between its C-terminal domain and Smc1 heads imply that the interaction between Smc3 heads and Scc1's N-terminal domain is not a simple one. It is nevertheless tempting to speculate that Scc1's N-terminal domain binds to the Smc3 head in a manner that is similar to that of its C-terminal domain to the Smc1 head.
The issue of whether cohesin's Smc1 and Smc3 heads really are stably inter-connected by Scc1 is complicated by the fact that the two heads are ABC-like ATPases that are known to associate with each other in the presence of ATP even in the absence of other factors. The phosphates of ATP bound to Walker A and B motifs on one subunit are contacted by a so-called signature motif on another. Hydrolysis of ATP is only possible after it has been sandwiched between two subunits in this manner. This dimeric structure was initially discovered with Rad50 (Hopfner et al. 2000), which is a close relative of SMC proteins and also possesses a 50 nm long coiled coil inserted into the middle of an ABC-like ATPase head. Similar dimeric structures have since been documented in several other ABC-like ATPases (Higgins & Linton 2004), including a Smc1 ATPase head complexed with the C terminus of Scc1, which forms homodimers in the presence of ATP (Haering et al. 2004; figure 6) and a bacterial SMC ATPase head (Lammens et al. 2004).
Given that Smc1 is attached to Smc3, it is probable that ATP usually induces interaction of the Smc1 heads not with another Smc1 head from a separate Smc1/Smc3 heterodimer but rather with the Smc3 head belonging to the same heterodimer. If, as seems probable, Smc1 and Smc3 heads interact directly in a manner mediated by ATP, then what would be the function of any additional inter-connection of the two heads mediated by Scc1? It is important to point out that the interaction between ABC ATPase heads may not be a particularly stable one. First, it would be de-stabilized by ATP hydrolysis, which is thought to drive ABC-like ATPase heads apart (figure 7), and second, it is not even clear how strong the interaction is even in the absence of ATP hydrolysis. Future experiments are clearly needed to address the affinity of Smc1 and Smc3 heads carrying mutations within their Walker B motifs that permit ATP binding but cannot support its hydrolysis. It is therefore quite possible that the proposed binding of Scc1's N-terminal domain to Smc3 heads provides a far more stable and long-lasting inter-connection between Smc1 and Smc3 ATPase heads than that provided by ATP binding alone.
Figure 7.
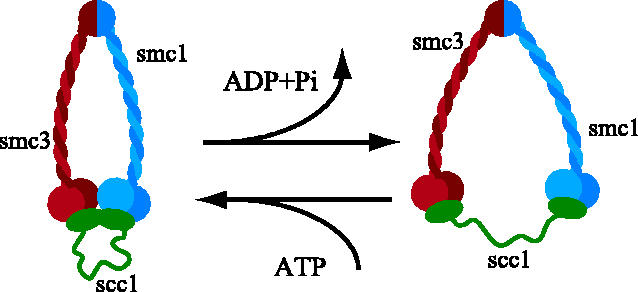
ATP binding might bring Smc1 and Smc3 heads together while its hydrolysis would drive them apart.
Because Smc1 and Smc3 heads have the potential to interact directly, at least in the presence of ATP, it was clearly important to measure the association Smc1 and Smc3 heads within cohesin and thereby address whether this interaction depends on the integrity of Scc1. Electron micrographs of soluble cohesin complexes from Xenopus egg extracts or from human tissue culture cells, suggest that the two arms of Smc1/Smc3 heterodimers are often bowed apart with a greater gap between the middle of the arms than at their extremities (heads), which are usually close to what appear to be non-SMC proteins, presumably Scc3 and Scc1 (Anderson et al. 2002). Such pictures neither delineate the position of Scc1 nor gauge the strength of direct Smc1 and Smc3 head interactions. The micrographs do, however, A show that cohesin's two SMC heads are frequently not closely juxtaposed, which is consistent with the notion that head–head interactions may not be very stable and leaves open the possibility that the two heads are nevertheless inter-connected by Scc1.
If Scc1's N-terminal domain does indeed bind stably to Smc3 heads, then cleavage of Scc1, either by separase or TEV-cleavable Scc1 variants by the TEV protease, should leave Scc1's C terminal domain associated with cohesin's Smc1's head and Scc1's N-terminal domain associated with its Smc3 head. Thus, the two cleavage fragments should remain associated with each other by virtue of being associated with the same Smc1/Smc3 heterodimer. This is indeed what is found either when Scc1 from soluble cohesin complexes is cleaved by TEV protease or when cleavage of Scc1 by separase causes cohesin to dissociate from chromosomes at the onset of anaphase (Gruber et al. 2003). If, in addition, cohesin's Smc1 and Smc3 heads did not remain tightly associated after Scc1 cleavage, then the association of Scc1 cleavage fragments should depend on the integrity of the Smc1/Smc3 heterodimer's coiled coils. To test this, TEV cleavage sites were inserted into short loops that break up both strands of Smc3's coiled coil about a third of the way between Smc3's head and its dimerization domain. Crucially, TEV-induced severance of Smc3's arm in this manner caused the release of Scc1's N-terminal domain together with the Smc3 ATPase head from the remaining parts of cohesin (figure 8), namely, the upper two-thirds of the Smc3 arm, Smc1 and Scc1's C-terminal cleavage fragment (Gruber et al. 2003). There are two important conclusions from this experiment. The first is that Scc1's N terminus does indeed bind stably to Smc3 ATPase heads, both within soluble cohesin complexes and within those that had been stably associated with chromosomes before their cleavage by separase. The second conclusion is that direct interaction of Smc1 and Smc3 heads is insufficient to hold them stably together at least after Scc1 cleavage.
Figure 8.
Evidence that Scc1's N-terminal domain binds to cohesin's Smc3 head. Separase activity in vivo cleaves Scc1 and releases cohesin from chromosomes, while severing Smc3's coiled coils in vitro releases the Smc3 head and Scc1's N-terminal cleavage fragment from the rest of cohesin.
In conclusion, there is strong evidence that the N terminus of Scc1 does indeed bind stably to cohesin's Smc3 head and that as a consequence Scc1 inter-connects Smc1 and Smc3 ATPase heads. Nevertheless, a crystal structure will be required both to define the nature of the Smc3–Scc1 interaction as well as to probe its physiological significance through mutagenesis of residues involved in the contact. Moreover, it will be necessary to reconstruct the interaction in vitro.
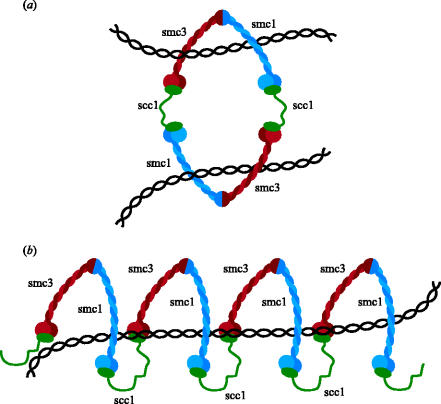
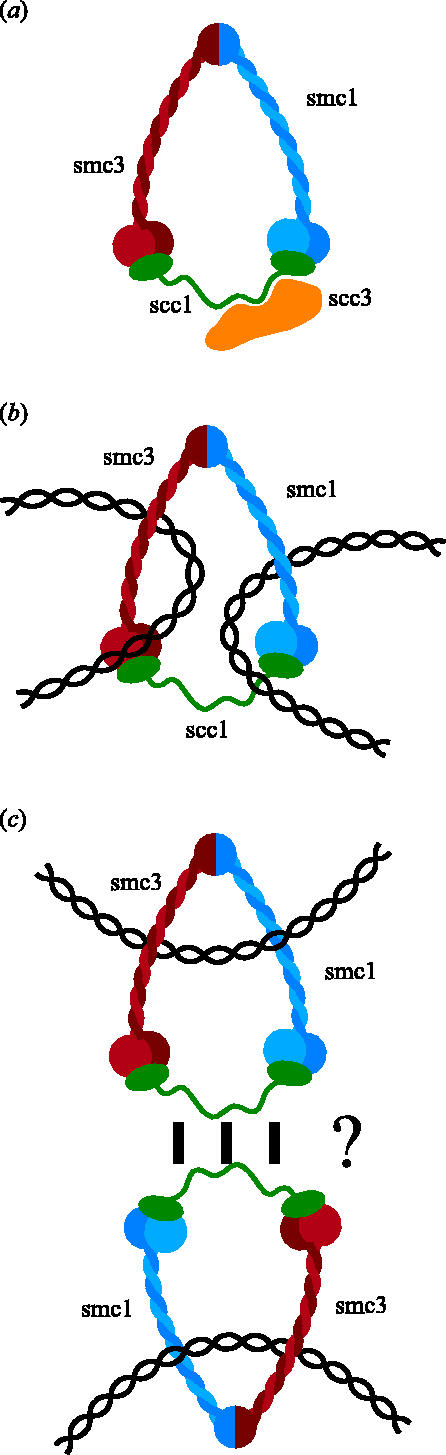
The discovery that cohesin's Smc1, Smc3 and Scc1 subunits together create a gigantic ring whose integrity is destroyed by Scc1 cleavage at the onset of anaphase has led to a radically different type of model for how cohesin might associate with chromatin and hold sister chromatids together (Haering et al. 2002). Cohesin's ring structure raises the possibility that DNA's double helices are trapped inside cohesin's ring and that sister DNAs might be held together by virtue of being entrapped within the same cohesin ring (figure 4b). This model has the merit of explaining cohesin's tight association with chromatin without invoking specific DNA binding domains, as well as providing an explanation for sister chromatid cohesion without invoking hitherto undetected subunit interactions. It also explains how cleavage of Scc1 by separase (Uhlmann et al. 1999) or Smc3's coiled coil by the TEV protease (Gruber et al. 2003) would cause both cohesin's dissociation from chromosomes and the dissolution of sister chromatid cohesion. There are several variants of the ring model. Thus, it is conceivable that sister DNAs are trapped by separate cohesin rings that interact with each other (figure 4c). The problem with this version is that we have no idea whether, not to mention how, cohesin rings stably interact. Another variant of the ring hypothesis envisions that Scc1 inter-connects Smc1 and Smc3 heads from a different not the same Smc1/Smc3 heterodimer. Scc1 could thereby generate either dimeric rings (figure 9a) or multimeric filaments that could entrap sister DNAs (figure 9b). A crucial prediction of this proposal is that individual cohesin subunits should be stably associated with subunits of the same type; that is, Smc1 with Smc1 and Scc1 with Scc1, and so on. Such associations have been sought for but not hitherto detected in cohesin complexes released from chromosomes in vitro owing to nuclease digestion (Haering et al. 2002). It is clear that a crucial issue for future research is to determine whether sister DNAs are held together by individual cohesin complexes or by two more complexes that interact with each other.
Figure 9.
Models in which Scc1 links Smc1 and Smc3 ATPase heads from different Smc1/Smc3 heterodimers, either forming a dimeric ring (a) or a filament (b).
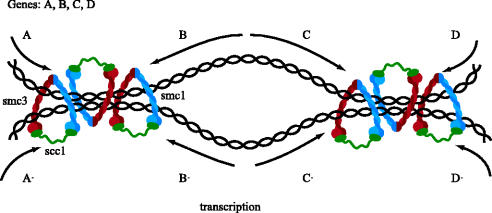
The notion that cohesin associates stably with chromosomes by trapping DNA inside rings formed by interactions between Smc1, Smc3 and Scc1 makes a number of key predictions. One of the most important is that cohesin should be capable of sliding along chromatin. Cohesin rings would be predicted to have a diameter of around 30 nm which is large enough to accommodate two 10 nm chromatin fibres. To test this, we have recently developed methods to purify small circular mini-chromosomes from yeast that remain stably associated with cohesin. This should enable us to address whether cleavage of DNA as well as that of Scc1 or Smc3 is sufficient to release cohesin from DNA. Meanwhile, however, the striking accumulation of cohesin within intergenic regions at which transcription converges from both sides, indicates that cohesin rings might slide along chromatin in vivo (Filipski & Mucha 2002; Glynn et al. 2004; Lengronne et al. 2004). A simple explanation of this pattern is that cohesin rings are pushed along chromatin by RNA polymerase as it transcribes DNA. If indeed this is what is happening, and if sister DNAs were trapped within individual cohesin rings, then one could also explain how cells ensure that sister DNAs are kept in register when cohesed. This is a potentially important issue as sister chromatid cohesion has an important role in double strand break repair (Sjoegren & Nasmyth 2001) and it is important that cohesin should hold broken ends close to homologous sequences on the unbroken chromatid that will be used to repair the break via homologous recombination. If individual cohesin rings encircling sister DNAs were continually pushed to sites of convergent transcription, then this would ensure that identical sequences on each sister would be held together by the cohesin complexes that cluster to these sites (figure 10).
Figure 10.
Transcription might drive cohesin to regions of convergent transcription and thereby hold sister chromatids in register.
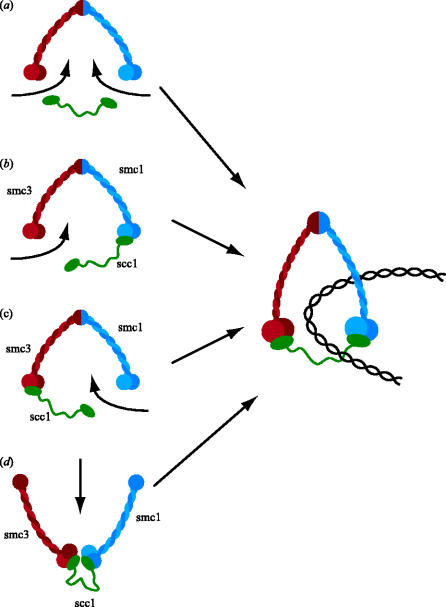
Any model for sister chromatid cohesion must not only postulate clearly the structure responsible for holding sister chromatids together, but also specify how such a structure is actually created. The simplest version of the ring model, with sister DNA molecules entrapped within a single cohesin ring, specifies fairly precisely what structure is created. Even when/if we know that such structures are made by cells, it will still be necessary to understand how the structures are created. There are several types of mechanism by which DNA could in principle be entrapped by cohesin rings. Either DNA is transiently cleaved by a topo-isomerase during entry, or one or more of the connections between Smc1, Smc3 and Scc1 must be at least transiently broken. One possibility (the bicycle lock model) is that DNA penetrates the gap between the arms of Smc1/Smc3 heterodimers before association of Scc1 with their heads (figure 11a). A second possibility (the karabiner model), is that either the N- or the C-terminal domain of Scc1 transiently dissociates from a Smc head (figure 11b,c). A third possibility is that the hinge of Smc1/Smc3 heterodimers transiently opens owing to dissociation of their hinge dimerization domains (figure 11d).
Figure 11.
Potential mechanisms by which DNA might be trapped by cohesin rings.
In summary, there is good evidence that the heads of cohesin's Smc1/Smc3 heterodimers are connected by Scc1 whose N- and C-terminal domains bind to the ATPase heads of Smc3 and Smc1, respectively, thereby creating a gigantic ring structure. The nature of the interaction between Scc1's N-terminal domain and Smc3 has yet to be defined. It is clearly not a simple one as it depends on the prior connection of Scc1's C-terminal domain with Smc1's ATPase head. Cleavage of Scc1 in the region connecting its N- and C-terminal domain opens the cohesin ring and triggers its dissociation from chromosomes and the dissolution of sister chromatid cohesion. These findings raise the possibility that the interaction between cohesin and chromatin fibres is a topological and not a physical one; that is, sister DNAs could be held together by being entrapped inside a single cohesin ring. Important issues for the future are whether cohesin acts as a monomer, whether DNAs are really trapped inside its ring, and if so, how they enter the ring.
Even if all the models outlined here prove incorrect, any credible future model will have to explain a number of key facts. It must explain the dependence of cohesin's association with chromosomes on the Scc2/Scc4 complex (Ciosk et al. 2000) and on hydrolysis of ATP bound to Smc1 and Smc3 heads (Arumugan et al. 2003), the persistence of cohesive structures for long periods of time after DNA replication (Haering et al. 2004) and how cleavage of Scc1 destroys sister chromatid cohesion (Uhlmann et al. 1999, 2000). In addition, it must clearly specify by what structure cohesin holds sister chromatids together and ultimately, explain how this structure comes into being.
Acknowledgements
Many thanks to Christian Haering and Hannes Tkadletz from the IMP for help with graphics. K.N's work is funded by the IMP, which is part of Boehringer Ingelheim International as well as the Austrian Science Fund (FWF) and the Austrian Industrial Research Promotion Fund (FFF).
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Chromosome segregation’.
References
- Anderson D.E, et al. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell. Biol. 2002;28:28. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugan P, et al. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bannister L.A, et al. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Buonomo S.B, et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Campbell J.L, Cohen-Fix O. Chromosome cohesion: ring around the sisters? Trends Biochem. Sci. 2002;27:492–495. doi: 10.1016/s0968-0004(02)02194-1. [DOI] [PubMed] [Google Scholar]
- Chiu A, et al. DNA interaction and dimerization of eukaryotic SMC hinge domains. J. Biol. Chem. 2004;279:26 233–26 242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- Ciosk R, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, et al. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Eijpe M, et al. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell. Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipski J, Mucha M. Structure, function and DNA composition of Saccharomyces cerevisiae chromatin loops. Gene. 2002;300:63–68. doi: 10.1016/s0378-1119(02)00848-x. [DOI] [PubMed] [Google Scholar]
- Funabiki H, et al. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Furuya K, et al. Faithful anaphase is ensured by Mis4, a sister chromatid cohesin molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie P.J, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian J.F, et al. Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Glynn E.F, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Gruber S, et al. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Guacci V, et al. A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H, et al. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haering C.H, et al. Structure and stability of cohesin's Smc1–kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hartman T, et al. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, et al. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Hauf S, et al. In press. Dissociation of cohesin from chromosome arms and loss of arm cohesion during prophase depends on phosphorylation of SA2. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, et al. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Heidmann D, et al. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- Higgins C.F, Linton K.J. The ATP switch model for ABC transporters. Nature Struct. Mol. Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K.P, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Katis V.L, et al. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W, et al. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W, et al. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S, et al. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S, et al. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Klein F, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Koshland D, Hartwell L. The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science. 1987;238:1713–1716. doi: 10.1126/science.3317838. [DOI] [PubMed] [Google Scholar]
- Lammens A, et al. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- LeBlanc H.N, et al. The mitotic centromeric protein MEI-S332 and its role in sister-chromatid cohesion. Chromosoma. 1999;108:401–411. doi: 10.1007/s004120050392. [DOI] [PubMed] [Google Scholar]
- Lee J, et al. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J. Cell Sci. 2003;116:2781–2790. doi: 10.1242/jcs.00495. [DOI] [PubMed] [Google Scholar]
- Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, et al. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/SCC3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A.L, et al. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McGuinness B.E, et al. In press. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T.E, et al. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell. Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, et al. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Milutinovich M, Koshland D.E. Molecular biology. SMC complexes—wrapped up in controversy. Science. 2003;300:1101–1102. doi: 10.1126/science.1084478. [DOI] [PubMed] [Google Scholar]
- Moore D.P, et al. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J. Cell Biol. 1998;140:1003–1012. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, et al. Sister chromatid cohesion and genome stability in vertebrate cells. Biochem. Soc. Trans. 2003;31:263–265. doi: 10.1042/bst0310263. [DOI] [PubMed] [Google Scholar]
- Murray A.W, Szostak J.W. Chromosome segregation in mitosis and meiosis. A. Rev. Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Schleiffer A. From a single double helix to paired double helices and back. Phil. Trans. R. Soc. B. 2004;359:99–108. doi: 10.1098/rstb.2003.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell. Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Panizza S, et al. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, et al. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, et al. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P, et al. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Revenkova E, et al. Novel meiosis-specific isoform of mammalian SMC1. Mol. Cell. Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nature Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Rieder C.L, Cole R. Chromatid cohesion during mitosis: lessons from meiosis. J. Cell Sci. 1999;112:2607–2613. doi: 10.1242/jcs.112.16.2607. [DOI] [PubMed] [Google Scholar]
- Salic A, et al. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Siomos M.F, et al. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 2001;11:1825–1835. doi: 10.1016/s0960-9822(01)00588-7. [DOI] [PubMed] [Google Scholar]
- Sjoegren C, Nasmyth K. Sister chromatid cohesion is required for post-replicative double strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Sullivan M, et al. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sumara I, et al. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell. Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Takahashi T.S, et al. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nature Cell. Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tanaka T, et al. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, et al. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, et al. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1p. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, et al. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Valdeolmillos A, et al. Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol. Cell. 2004;96:457–462. doi: 10.1016/j.biolcel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Waizenegger I, et al. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Warren W.D, et al. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Weber S.A, et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, et al. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]