Abstract
Brain mapping has evolved considerably over the last century. While most emphasis has been placed on coordinate-based spatial atlases, coordinate-independent parcellation-based mapping is an important technique for accessing the multitude of structural and functional data that have been reported from invasive experiments, and provides for flexible and efficient representations of information. Here, we provide an introduction to motivations, concepts, techniques and implications of coordinate-independent mapping of microstructurally or functionally defined brain structures. In particular, we explain the problems of constructing mapping paths and finding adequate heuristics for their evaluation. We then introduce the three auxiliary concepts of acronym-based mapping (AM), of a generalized hierarchy (GM ontology), and of a topographically oriented regional map (RM) with adequate granularity for mapping between individual brains with different cortical folding and between humans and non-human primates. Examples from the CoCoMac database of primate brain connectivity demonstrate how these concepts enhance coordinate-independent mapping based on published relational statements. Finally, we discuss the strengths and weaknesses of spatial coordinate-based versus coordinate-independent microstructural brain mapping and show perspectives for a wider application of parcellation-based approaches in the integration of multi-modal structural, functional and clinical data.
Keywords: brain atlas, cerebral cortex, localization, ontology, primate, spatial normalization
1. Introduction
Mapping the brain in its many different structural and functional aspects is at the basis of the quest to understand how the brain develops, works and adapts. Particularly complex and challenging are the architecture and functional organization of the cerebral cortex, which has been a subject of keen interest for more than 100 years, beginning with the work of Meynert (1868) and systematized by Brodmann (1903, 1905, 1909), the Vogts (Vogt 1903; Vogt & Vogt 1919), Campbell (1905) and Mauss (1908) among others. Over the last decade, brain mapping has seen remarkable improvements, such as observer-independent detection of microstructural borders in brain sections (e.g. Schleicher et al. 1999), reconstruction and spatial registration of brain volumes and surfaces (e.g. ICBM atlases; Mazziotta et al. 2001), deformation of brain volumes and surfaces for comparisons between individuals, groups and species (e.g. Ashburner & Friston 2000), as well as between different structural and functional features (e.g. Van Essen, Drury et al. 2001; Van Essen et al. 2001b; Van Essen 2004), to name just a few. The vast body of accumulated data, however, is not referenced to spatial coordinate systems but is shown in selected sections and schematic overviews, and described in terms of brain regions and maps defined by microstructural, macroscopical and functional features. The purpose of this article is to introduce the motivations, concepts, methods and perspectives of coordinate-independent microstructural mapping. We provide a number of examples from the mapping information collated in the CoCoMac database (Stephan et al. 2001; Kötter 2004) and provide details of three relevant techniques used to improve mapping based on nomenclatural, conceptual and topographical relationships. Finally, we point out perspectives of how coordinate-independent mapping can be applied to make the observations of a century of brain research referenced to brain maps accessible in a systematic and flexible fashion.
2. Coordinate-based versus coordinate-independent brain maps
Neuroinformatics approaches to brain mapping currently focus on the challenges and rewards of coordinate-based spatial representations, so-called digital brain atlases (e.g. Gorin et al. 2001; Toga & Thompson 2001; Van Horn et al. 2001). The challenges are many, starting from the vast amounts of data and the differences in spatial scales and resolutions to more specific problems arising from differences in coordinate systems, individual and reference brains, deformation methods, data modalities, and so on. It appears to be a major advantage of coordinate-based approaches that they can largely ignore the uncertainties and controversies surrounding the adequacy and correctness of different brain maps that have led to ongoing discussions ever since the publication of the first partitioning schemes of the cerebral cortex a century ago. But do coordinate-based approaches make this debate obsolete? Do we still need parcellation-based brain maps?
The essence of the answer can be found in the original motivation given by Brodmann for his cytoarchitectonic work: ‘Our final goal is therefore the creation of a comparative system of organs of the cerebral cortex, which is founded on its anatomical characteristics’ (Brodmann 1909; p. 1). From his view, the best approach to achieve this ‘system of organs’ was by ‘the regional dissection of the cerebral cortex into structural cortical fields or, what means the same, the division into homogeneous and among them distinctively built regions of the hemispherical surface. We call such distinct regions Areae anatomicae’ (Brodmann 1909; p. 8). Today, we might say that Brodmann was striving to discern the elementary units, which in their entirety, compose the cerebral cortex. These units could be recognized by morphological methods, but they would also represent computational units that interact to exert integrative cortical functions. This goal is as timely as ever. However, to find the elementary units we do not want to rely on morphological methods alone; we also want to take into consideration as many structural and functional observables as possible when delineating cortical areas.
It has proven difficult to establish a universally accepted parcellation of the cerebral cortex. Here, we put species differences aside, which raise additional profound questions concerning homology, similarity and development across temporal, spatial, behavioural and environmental scales, and focus on closely related species. Unfortunately, over the last century, the number of alternative parcellation schemes has increased rather than decreased, and a definitive universally recognized scheme is not on the horizon. The predominant reasons for this development are: (i) interindividual variability of brain shape (particularly in strongly gyrated human brains), (ii) interindividual differences in the size and location of brain structures, (iii) method-dependent detectability of microstructural differentiations, and (iv) observer-dependent assessments of complex differentiating features. Coordinate-based brain mapping effectively addresses the first two problems. By contrast, using microstructurally defined brain structures as the reference, we can abstract both from interindividual variability in shape and from differences in the location of differentiating features, such as areal boundaries. Despite remaining problems with the validity of different deformation procedures, it is the detection of interindividual variability and group differences in space where coordinate-based atlases celebrate their major successes (e.g. Ashburner & Friston 2000; Thompson et al. 2001; Sowell et al. 2003). It has to be realized, however, that datasets registered in spatial coordinates rarely provide direct evidence of areal boundaries (the most notable exception being functional mapping of receptive field properties; e.g. Sereno et al. 1995; Huk et al. 2002). The more common application is to detect variations in topographical landmarks, such as cortical folding or thickness (e.g. Thompson et al. 1996). Concerning the third problem (method dependence), coordinate-based mapping has a facilitating role. By eliminating the confound of interindividual variability in brain shape, we can hope to facilitate comparisons between results obtained with different delineation methods. The fourth problem (observer-dependence) may be alleviated by an enhancement of differentiations when viewed on a population basis but, of course, the appraisal of what are considered as relevant features remains with the observer.
Thus, we can conclude that coordinate-based mapping facilitates the purpose of parcellation-based mapping that Brodmann aimed for. Although both approaches are valuable in their own right, the ‘regional dissection of the cerebral cortex into homogeneous and among them distinctively built structural cortical fields’ has some remarkable advantages over coordinate-based approaches that are often overlooked.
First, because these cortical fields or areas are normally present in every individual brain of a species independent of its shape and size, they provide an efficient description of brain architecture. From this point of view, interindividual variation appears as noise obscuring the principal information and should be filtered out if possible. If we can dispense with coordinates and refer to areas instead, then we achieve a remarkable degree of data compression.
Second, by referring to areas, we can relate to the rich structural and functional context, which has been accumulated over a century of parcellation-based research and which allows us to interpret recordings as well as the effects of stimulations and lesions of areas to an extent that is not available for coordinate-based data.
The desire to relate to this coordinate-independent context explains the wide use of approximate mappings between coordinate systems and microstructurally defined areas as in the ‘Talairach atlas’ of the human brain (Talairach & Tournoux 1988) or Talairach Daemon (http://ric.uthscsa.edu/projects/talairachdaemon.html; Lancaster et al. 2000). Unfortunately, this atlas indicates a higher precision than is available because a microstructural parcellation was not obtained for the atlas brain but was manually transferred from the cortex parcellation depicted by Brodmann (1909) almost a century ago.
A rigorous alternative to such approximations is the creation of probabilistic coordinate-based atlases of microstructurally defined brain structures (e.g. Roland et al. 2001; Rademacher et al. 2002; Eickhoff et al. in press). The general idea is to take microstructurally defined areas from a population of brains and map them individually to a brain in standard space so that the probability that a coordinate coincides with a specific brain structure can be calculated, and vice versa. The spatial registration of microstructurally defined brain areas and fibre tracts is a valuable long-term goal with the current limitation that it requires post-mortem brains and very meticulous and time-consuming expert work (for in vivo imaging approaches see Johansen-Berg et al. 2004; Eickhoff et al. 2004). Therefore, it is still worthwhile to consider coordinate-independent parcellation-based data and to make optimal use of it.
3. Coordinate-independent brain mapping
In the previous section, we established the distinction between coordinate-based and coordinate-independent approaches to compare different brain maps. Clearly, the coordinate-independent mapping between multiple structural and/or functional features depends upon a carefully constructed ontology of areae anatomicae. In this section, we consider this ontology and some specific solutions we have developed in the context of our own work with brain maps. Briefly, we discuss some generic aspects of hierarchical relationships within any ontology. We then go on to discriminate between original, acronym, generic and regional ontologies, that each have special roles in integrating knowledge about function and structure in the brain. Where appropriate, we provide specific examples based on our experience with databases of such information.
Although microstructurally defined brain structures are described in the large majority of cases without reference to spatial coordinate systems, they still have defined topographical inter-relationships. These relationships are commonly known by alternative names (synonyms), related definitions or alternative classification schemes (brain maps; see Felleman & Van Essen 1991; Stephan et al. 2000). Moreover, there is information on neighbourhood relationships with common borders (topologies; e.g. Scannell et al. 1995, 1999) or relative spatial locations in an anatomical reference system (e.g. medial/lateral, ventral/dorsal, rostral/caudal; see Bota & Arbib 2004). In these parcellation-based reference systems, cortical areas are essentially regarded as 2D structures viewed from the pial surface. Therefore, coordinate-independent parcellation-based approaches relate particularly easily to surface-based representations (cerebral or cerebellar cortex), but they have also been applied to volume-based representations where they have to be compared with coordinate-based reference systems.
Coordinate-independent mapping procedures take advantage of the abundance of explicit statements on area topographical inter-relationships in scientific publications. These statements can be systematically extracted, collated and analysed although, at present, this process requires the manual work of human experts. Extensive listings have been provided repeatedly comparing different maps, area definitions and terminologies used (e.g. Felleman & Van Essen 1991; Young 1993; Scannell et al. 1995, 1999; Lewis & Van Essen 2000; Zilles 2004, tables 27.3–27.7). Formal classifications of parcellation-based brain maps and the area inter-relationships in combination with modern database technology and graph theoretical analyses can help to evaluate, compare and make better use of these statements (Stephan et al. 2000).
In the following subsections we will investigate the relevant concepts of brain maps and area inter-relations in more detail.
(a) Parcellation-based brain maps
A set of microstructurally or functionally defined entities as presented in a particular published work constitutes a ‘parcellation scheme’ or ‘brain map’. Compared with a spatial description of brain data in Euclidean 3D space, representing brain data based on parcellation schemes can be described as a non-Euclidean representation in the multidimensional space of microstructural-functional properties. For example, in a cytoarchitectonic study that defines areal boundaries on the basis of: (i) relative width of supra- and infragranular layers, (ii) staining intensity of layer IV, (iii) density of pyramidal cells in layer III, and (iv) density of pyramidal cells in layer V, these criteria define the four dimensions of the non-Euclidean feature space in which the delineated brain structures would be located.
Owing to the variety of applied methods and the high observer-dependency of many microstructural-functional criteria, a large number of parcellation schemes exist that differ in nomenclature, number, location, size, boundaries and representation of the delineated brain structures. This variability is particularly well known from the cerebral cortex, but can also be found with subcortical structures (parcellation of thalamus, amygdala, brain stem etc.). The combination of different methods and observer-dependent criteria can make comparing data described by two different parcellation schemes substantially difficult. Formally, such a comparison is equivalent to comparing two points from two non-Euclidean spaces whose dimensions are characterized by the microstructural-functional criteria of the two parcellation schemes. Depending on differences between these criteria, the two feature spaces have different numbers and/or different types of dimensions. The question of how difficult it is to compare data based on two parcellation schemes can thus be formally rephrased as, ‘Is there a determinable mapping between the two feature spaces?’ or, less formally, ‘Is there a way to relate (and maybe match) the two parcellation schemes so that they may be compared in a meaningful way?’.
For example, let us look at several parcellation schemes of premotor cortex. For the two studies of Brodmann (1909) and von Bonin & Bailey (1947), the experimental method (i.e. Nissl staining) and the criteria used for delineation (i.e. certain cytoarchitectonic properties determined by visual inspection) are very similar but of high observer-dependence. In spite of the resulting obvious differences between these two maps, one can still compare these parcellations relatively easily because the dimensions of the two microstructural spaces are of similar quality. For example, the general cytoarchitectonic features described by Brodmann for his area 6 (e.g. agranular cortex without Betz cells in layer V) are still valid for each of the premotor areas FB, FBA and FCBm recognized by von Bonin & Bailey, but the latter used additional criteria to distinguish these subdivisions within Brodmann's area 6. A third study by Vogt & Vogt (1919) also follows a classical architectonic approach using visually determined changes in myeloarchitecture to determine areal borders. How can this parcellation be compared with those of Brodmann (1909) or von Bonin & Bailey (1947) given that the dimensions of the respective microstructural feature spaces are of an entirely different quality? In other words, how can we determine whether a border determined by myeloarchitectonic criteria matches a border that has been determined by cytoarchitectonic criteria? The direct comparison is only possible if we can establish a clear match between particular myeloarchitectonic and particular cytoarchitectonic features. Without resorting to coordinate-dependent mapping, this comparison relies on studies that applied both criteria simultaneously (e.g. Barbas & Pandya 1987), so that we have a connecting link that allows us to make the comparison.
Beyond mere architectonic criteria, the dimensional mismatch between the feature spaces of parcellations becomes even larger if any of the structural studies above is compared with other studies of premotor cortex that apply electrophysiological criteria to distinguish brain structures on the basis of different response properties (e.g. Mitz & Wise 1987; Luppino et al. 1991). In such cases where the microstructural-functional feature spaces of two parcellations are not directly comparable, additional information is needed, which must arise from the parcellations themselves not from their underlying criteria. Commonly, this is topographic information, which results from a comparison between the two parcellation schemes with respect to relative size and position of brain structures and relative location to landmarks. This process is certainly highly observer-dependent and additionally problematic because of the interindividual differences in brain shape (see also the comments on the correspondence of myeloarchitectonic, cytoarchitectonic and stimulation results by Vogt & Vogt 1919). Nevertheless, approximate topographic information can always be obtained for two given parcellation schemes and may be the only criterion to perform any comparison at all between parcellation schemes with no direct comparability of their microstructural-functional feature spaces.
Both approaches—comparisons by microstructural-functional and topographic criteria—are frequently discussed in the literature. Unfortunately, very few authors explicitly point out the basis of their statements on relations between different brain maps. Often, a combination of both microstructural-functional and topographic comparisons seems to underlie such statements.
(b) Brain mapping based on comparative statements
Considering a pair of brain areas, they are either co-extensive (e.g. primary visual cortex V1, Brodmann's 1909 area 17, von Bonin & Bailey's 1947 area OC), form inclusions (the supplementary motor area is a part of the premotor cortex), overlap (e.g. Felleman & Van Essen's 1991 areas AITd and CITd with Seltzer & Pandya's 1978 areas TEa and TEm, or visual areas V6 and V6A with various definitions of area PO; Galletti et al. in press), or are disjoint (e.g. the supplementary somatosensory area in the mesial parietal cortex compared with ventro-laterally located secondary somatosensory cortex SII). We denote these relations between pairs of areas by Relation Codes (RCs): I (identity), S/L (subarea/larger area), O (overlap), D (disjoint; for a detailed description see Stephan et al. 2000).
Published mapping studies contain extensive information concerning the RCs between brain structures in different brain maps. Particularly when introducing a new parcellation scheme, authors make explicit statements on how the proposed new entities relate to relevant known ones. Despite the abundance of such statements, they concentrate on a few highly popular maps (in the macaque, e.g. Brodmann 1909; Walker 1940; von Bonin & Bailey 1947; Olszewski 1952), whereas many other relationships have not been explicitly stated. Based on available statements, however, further relations can be derived by concatenating known ones. For example, area 9 as defined by Brodmann (1909) comprises areas 9 and 46 of Walker (1940), and area 46 again has been divided into various subareas. Clearly, these subareas are also contained in area 9 as defined by Brodmann. Table 1 shows the list of results of all possible concatenations of relations.
Table 1.
Table of results of concatenation of relations. (I, identity; S, subarea; L, larger area; O, overlapping area; D, disjoint area. The first column denotes the state before mapping and the first row identifies the mapping step to be applied to that previous state. For the first mapping step start with I as the previous state. Other matrix entries list the set of resulting possibilities. The resulting set can then be transferred to the first column to apply the next mapping step.)
| I | S | L | O | D | |
|---|---|---|---|---|---|
| I | I | S | L | O | D |
| S | S | S | ISLOD | SOD | D |
| L | L | ISLO | L | LO | LOD |
| O | O | SO | LOD | ISLOD | LOD |
| D | D | SOD | D | SOD | ISLOD |
| SO | SO | SO | ISLOD | ISLOD | LOD |
| LO | LO | ISLO | LOD | ISLOD | LOD |
| SOD | SOD | SOD | ISLOD | ISLOD | ISLOD |
| LOD | LOD | ISLOD | LOD | ISLOD | ISLOD |
| ISLO | ISLO | ISLO | ISLOD | ISLOD | LOD |
| ISLOD | ISLOD | ISLOD | ISLOD | ISLOD | ISLOD |
Table 1 shows that mapping to a subarea (L) followed by a mapping to a larger area definition (S) has the set of four possible results: ISLO. By contrast, when mapping first to a coarser definition (S) and then to a subarea (L), the resulting set includes the possibility that the target region is disjoint from the initial one: ISLOD. Obviously, if this possibility is included then the mapping may be invalid. Therefore, it is not useful to continue this line of transformation. If we exclude all resulting sets that include the disjoint possibility (D) from the input for the next mapping step then we obtain a reduced mapping (table 2).
Table 2.
Reduced table of results of concatenation of relations after elimination of mapping results that do not guarantee at least overlap with the original brain area. (For explanations, see table 1.)
| I | S | L | O | |
|---|---|---|---|---|
| I | I | S | L | O |
| S | S | S | — | — |
| L | L | ISLO | L | LO |
| O | O | SO | — | — |
| SO | SO | SO | — | — |
| LO | LO | ISLO | — | — |
| ISLO | ISLO | ISLO | — | — |
The concatenation of area inter-relations delivers mapping paths. We have referred to the application of mapping paths to the problem of coordinate-independent brain mapping of structural and functional data as objective relational transformation (ORT; Stephan et al. 2000).
The network that consists of all areas as nodes and all relations (symmetric I, O relations and asymmetric S, L relations) as edges is called, in a theoretical context, a mixed graph (Wanke & Kötter 2004). The paths that include asymmetric relations of either only S or only L type (and any number of symmetric relations) are called oriented paths. Here, we will not go into the formal algorithmic details of finding oriented paths in mixed graphs. Although the problem of finding a simple oriented path in a mixed graph can be highly intractable for many special cases, a large number of paths can be found in an acceptable amount of time using heuristic algorithms. The paths in the mapping network that represent an interrelation between two areas, not including the disjoint possibility (D), form the basis for specifying the formally correct relationship between different areas and different brain maps.
The large set of paths obtained for original statements on area relationships frequently includes redundant and contradictory paths because of the uncertainty of specifying brain structures and their relations in different maps (see above). Therefore, we need heuristic methods that help us to select the ‘best’ paths. Two possibilities are presented here. The first method uses as its criterion the quality of the resulting set of states. If the resulting set contains only a single state, then we have a higher certainty about the relation of the resulting brain structure to the initial brain structure. Among the single state sets, the I-relationship is more precise than are the S and L relationships, and these are also more precise than the O relationship. Thus, we can construct a hierarchy of resulting state sets: I>S/L>O. The second heuristic method uses additional information that can be extracted with the literature statements that mention the relations. A useful criterion is the precision of the description of the relation using information provided in text and/or figures including the quality and correspondence of these. The logic of this method leading to precision of description codes (PDCs) is shown in figure 1. Since one can certainly argue about the relative merits of various heuristic methods and the order of the respective criteria, we consider it most important to make this decision process transparent, reproducible and adaptable to the specific situation (more details can be found in Stephan et al. 2001).
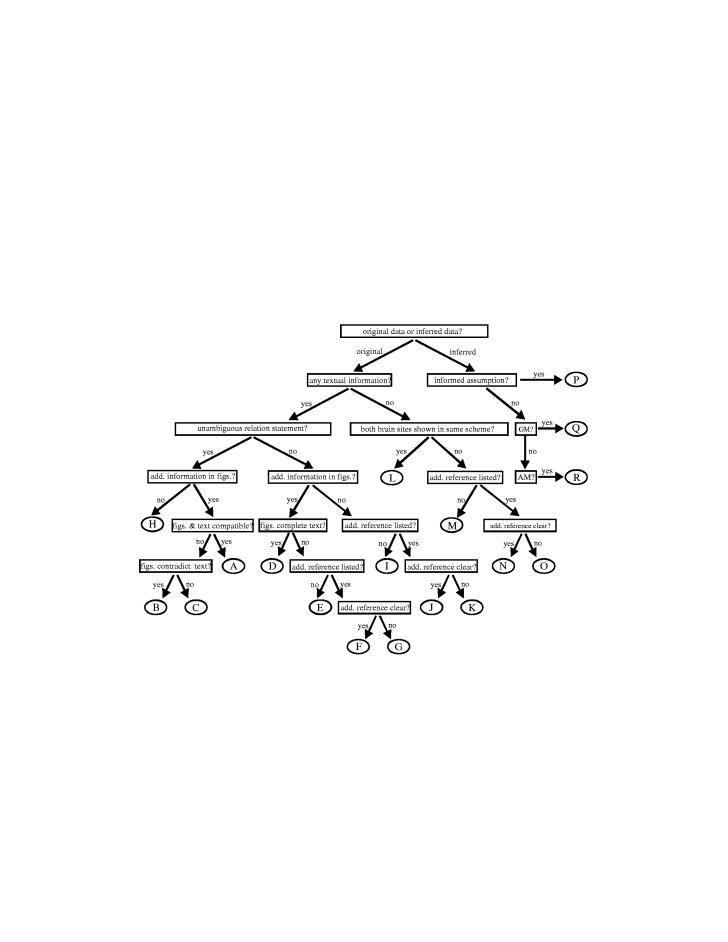
Figure 1.
Decision tree for determination of PDCs for relations between brain areas. Starting at the top, a series of binary decisions is made until a discrete case (i.e. a leaf of the tree, represented by an encircled capital letter) is reached. Note that although the individual cases are labelled by consecutive uppercase letters, the alphabetical order does not imply a predefined hierarchy among PDCs. The hierarchy currently used in CoCoMac is A>C>H>L>D>F>J>N≫B>G>E>K>I>O>M>P>Q>R and can be adjusted according to personal preferences. AM, Acronym map; GM, General map. Further details about the decision criteria are specified in the CoCoMac data entry manual, which is available for download from http://www.cocomac.org.
Following the main theme of this paper, we now describe three further procedures that enhance the power of coordinate-independent mapping where published statements on the relationship between areas are missing. The next section addresses those who are interested in the details of coordinate-independent mapping and is dispensable for the general reader. We suggest that those with a more general interest jump to §5 and refer to table 4 for an overview.
Table 4.
Properties of acronym map, general map and regional map.
| acronym map (AM) | general map (GM) | regional map (RM) |
|---|---|---|
| AM contains algorithmically created BrainSites (see figure 2). No changes are possible for the data collator | the BrainSites in GM are predefined (see table 3). Extensions and changes are possible if consensus is reached | the RM BrainSites are broadly topographically defined (see figure 3). Changes depend on further insights |
| AM includes one BrainSite for each acronym that is used by at least two original BrainSites | GM offers standardized BrainSites for certain regions of the brain only | RM provides a species-independent topographic/functional parcellation of the entire cerebral cortex |
| the BrainSites of AM have no pre-defined hierarchical relations specified by the data collators | the BrainSites of GM are defined according to a predefined hierarchy | RM constitutes a full cortical map of contiguous BrainSites |
| in AM, BrainSites can refer to equivalent brain structures (e.g. AM-Caud, AM-Cd, AM-C#3, AM-CA#1 all refer to the caudate nucleus) | in GM, there is no semantic overlap between its elements except for hierarchical relations (e.g. there is only one BrainSite in GM referring to the caudate nucleus as an entity) | in RM, overlaps and gaps are excluded. Semantic overlap through external relations can occur at the borders |
| neither intrinsic nor external relations of AM need to be specified by the data collator. Instead, they are generated algorithmically | both internal and external relations of GM must be specified by the data collator | only external relations of RM occur and must be specified by the data collator |
4. Special mapping procedures
The concepts of brain maps and mapping paths, as described above, can be applied to a vast majority of mapping problems and they are particularly suitable for brain regions where a variety of different histological and/or physiological definitions of areas and nuclei coexist. Good examples of such regions with parcellation-defined brain structures are the cerebral cortex, the thalamus and the amygdala. In other parts of the brain, differences between parcellations are less frequent, either because of obvious demarcations of structures by macroscopic features (e.g. demarcation by white matter) or because concepts of fine-grained parcellations have not yet been considered important. In both cases, many authors refer to brain structures without explicit definitions or reference to a specific brain map (e.g. they refer to ‘the caudate nucleus’, ‘the pons’, ‘the granular layer’ of ‘the cerebellum’ etc.). One may thus speak of generalized brain structures. A similar lack of specificity is sometimes encountered in the cerebral cortex, where definitions are either unanimous (e.g. for primary visual cortex) or where broad topographical regions are being referred to (e.g. dorsolateral prefrontal cortex). These situations are not adequately covered by the procedures described above because explicit parcellation schemes and corresponding statements expressing RCs are missing. To address these particular problems, we will discuss three additional coordinate-independent mapping procedures, which are (i) acronym-based mapping, (ii) generalized ontology-based mapping and (iii) mapping with recourse to topographic regions. In the following, we use the acronym OM (original map) to refer either to the entire set of brain maps defined by original articles or to one specific such original brain map. In contrast, AM denotes the acronym map, GM the general map, and RM the regional map. These concepts have been implemented enhancing the ORT procedure (Stephan et al. 2000), which we routinely apply to connectivity data from invasive tracing studies (Stephan et al. 2001). Whenever we refer to terms of our specific implementation, then the term is shown in italics.
(a) Acronym map
One common problem with the procedure of relation-based mapping is posed by brain structures (BrainSites) that are likely to have the same acronym or the same meaning (i.e. delineation) and where the literature provides no explicit statements on their identity. For example, articles describing connectivity of the visual cortex often mention an area ‘V1’ without referring to any definition of this area (i.e. these are cases of adoption of an unspecified pre-existing parcellation and nomenclature). The tacit assumption made in these articles is that all definitions of ‘V1’ refer to the same topographic structure and are therefore identical. This assumption is likely to be correct given the wide agreement on the definition of primary visual cortex and the ease of its delineation. Based on the considerations in §3a could state the assumed identity relationships explicitly for each pair of structures called V1. This procedure has the practical disadvantage that one formally has to keep track of large numbers of relations that could be automatically generated by a simple rule: whenever there are two OM BrainSites with identical acronyms and no explicit information on their relation, generate an I-relation for these BrainSites and define a special PDC (P; see figure 1) to express the fact that we make an assumption. Manually dealing with these relations is not only tedious but also error-prone.
The concept of AM tackles this problem. Defined briefly, AM is an artificially introduced brain map that can be created algorithmically from all BrainSites whose acronym is used by at least two OM BrainSites. For each of these acronyms, there is one element of AM that represents the acronym as a general brain site. At first sight, this procedure may appear to be a burden on conceptual and computational efficiency as it adds an additional artificial brain map with a large set of additional BrainSites to the existing profusion. If used properly, however, AM considerably simplifies the computational representation of BrainSites and inter map relations in a neuroinformatics system and contributes to reducing the number of BrainSites that effectively have to be processed.
(i) Definition and use of AM
The user does not have to define the elements of AM or create relations to its elements. Instead, AM is generated algorithmically each time the ORT process is performed. The following principles apply (terms in italics refer to the specific implementation, e.g. tables, fields etc. in the CoCoMac database; Stephan et al. 2001; Kötter 2004).
For each acronym A in the list of BrainMaps_BrainSiteAcronyms that is used by at least two BrainSites, an element AM-A in BrainMaps_BrainSites is created. If the same acronym refers to obviously different brain structures, then indices are appended to distinguish them and these indices are also included in the ID of AM-BrainSites. For example, for the four acronyms P (P#1–P#4 representing putamen, nucleus parafascicularis thalami, periamygdaloid cortex and area prostriata, respectively), the corresponding elements of AM in BrainSites_BrainSites would be AM-P#1, …, AM-P#4.
For each element AM-A in the AcronymMap, I-relations between AM-A and all corresponding BrainSites OMi-A (OMi, original brain map i; A, acronym) of BrainMaps_BrainSites are created. These relations are assigned a new PDC (PDC_Relation=R: this PDC denotes a relation automatically created on grounds of identical acronyms; see figure 1). For example, for the BrainSites BDU91-V1 (visual area 1 according to Boussaoud et al. 1991) and FV91-V1 (visual area 1 according to Felleman & Van Essen 1991), the relations BDU91-V1→AM-V1 and FV91-V1→AM-V1 are created with RC=I (see figure 2). Note that no BrainSite is created in AM for an acronym (including index) that is used by only a single BrainSite.
Placing the AM-specific PDC_Relation (R) at the bottom of the PDC hierarchy ensures that all specific relations will override any relations that are created automatically via AM.
Figure 2.
Example of how six BrainSites of primary visual cortex are represented and processed, (a) without and (b) with the AM procedure. Bold black arrows denote I-relations that have to be specified explicitly. Except for RC=I for mapping between B09-17 and FV91-V1, which is clearly stated in the literature (PDC: H), these relations are assumed and thus PDC=P. Dotted grey arrows represent I-relations with PDC=R that are algorithmically inserted for BrainSites with identical acronyms but unknown relations. Dashed black arrows represent I-relations that are created automatically by ORT during the optimization of the transformation path. Black BrainSites enter the optimization of the transformation graph and all subsequent stages of ORT. Grey BrainSites do not enter ORT. (a) Without AM, the data collator is required to create at least five relations to ensure that transformation paths are eventually created between all six BrainSites, all of which have to enter the graph optimization routine. (b) Using AM, the data collator only needs to create a single relation (the one that is documented in the literature), still all possible transformation paths are created. Another advantage compared with (a) is that only four BrainSites need to enter ORT. The BrainMaps referred to are: B09 (Brodmann 1909), B88 (Barbas 1988), BDU91 (Boussaoud et al. 1991), FV91 (Felleman & Van Essen 1991), PHT00 (Paxinos et al. 2000) and UD86 (Ungerleider & Desimone 1986).
Thus, AM summarizes general notions of how brain structures are related to each other simply by nomenclature and not by underlying specific parcellations. All BrainSites with identical acronyms (including qualifiers differentiating obviously different BrainSites) are automatically linked by I-relations to the same element of AM unless there is a specific relation statement in the literature for this pair of areas that overrides it. The most important effect of automatic linking via the AM is that it prevents failure of mapping between identically named BrainSites, for which no relationship had been explicitly stated in the literature.
(ii) Limitations of AM
The use of AM is open to the problem that names are often loosely defined and thus the same name and acronym can be used by two different investigators to refer to differently defined and topographically different BrainSites. Obvious examples are parieto-occipital area, PO, and middle temporal area, MT, for which several spatially and conceptually different definitions exist. This is a major concern in the AM approach in that without a high-level supervisor of brain structure assignments, names alone are likely to produce errors. In our experience, however, AM does not introduce more inaccuracy in the assignments than is present in the statements collated from the published literature. The reason becomes clear when one considers AM in the context of known inter-map relations for two BrainSites A and B with identical names/acronyms: (i) if an explicit statement exists that the two BrainSites differ, then our PDC-based method guarantees that AM is not used; (ii) if, contrary to reality, only identity statements exist, then AM is no worse than existing statements, and it would still not be used; and (iii) only if no explicit statement is present, then AM comes to bear. This typically occurs when authors consider the identity relationship to be self-evident.
Examples for the three cases are now presented: area MT as described by Van Essen et al. (1981) is smaller (RC=S; PDC=H) than what is also called MT by Ungerleider & Mishkin (1979). The most likely reason is that the heavily myelinated zone described by Van Essen et al. does not extend to the dorsal border of MT as determined by the reversal of the progression of receptive field centres or by the projections from striate cortex (Gattass & Gross 1981, p. 636). Later, Lewis & Van Essen (2000, table 2) commented that an area MT that is identical (RC=I, PDC=H) to the one from Van Essen et al. (1981) overlaps (RC=O; PDC=H) with areas MT of Cusick et al. (1995) and Hof & Morrison (1995). Here, we have an impressive case of method-related differences between at least three areas that are called MT. However, the explicit comparative statements according to our heuristics override AM (RC=I, PDC=R) so that no error is made.
The second case can be illustrated also with reference to area MT because the literature contains several identity statements (RC=I, PDC=H) for pairs of MT definitions: Desimone & Ungerleider (1986)—MT→Ungerleider & Desimone (1986)—MT→Van Essen et al. (1981)—MT→Tanaka et al. (1986)—MT→Ungerleider & Mishkin (1979)—MT. These statements can be concatenated to a mapping path followed by Ungerleider & Mishkin (1979)—MT being larger (L) then Seltzer & Pandya (1978)—OAa–I, which is identical (I) to Barnes & Pandya (1992)—OAa. According to table 2, the overall result for mapping from Desimone & Ungerleider (1986)—MT to Barnes & Pandya (1992)—OAa is RC=L. Barnes & Pandya (1992, p. 224), however, make the explicit statement that their area OAa is larger (RC=L) than Desimone & Ungerleider's (1986)—MT, which is the inverse relation. Because the two L-relations are convincingly demonstrated, it follows that one or more of the I-relations must be inaccurate, most likely involving slightly different definitions of the extent of area MT. Although this degree of contradiction is an exceptional case, it illustrates several important points. Notably, even explicit comparative statements of identity do not guarantee a precise topographical match. Such inaccuracy is inherent in qualitative coordinate-independent mapping and is not a danger specific to using AM. In addition, even trained experts have difficulties picking up slight changes in the topographical extent of cortical area definitions so that expert supervision of mapping relationships can still fail. Therefore, a heuristic that penalizes the length of concatenated mapping statements may be equally helpful.
Finally, mapping statements are rare for areas called SI, 3a, 3b, 1, 2, 4 or MI, and these areas of the primary somatosensory and motor cortices can be reasonably assumed to have very similar extents in different maps (this cannot be said for areas SII and SMA, and explicit statements to the contrary exist).
In conclusion, AM is a simple but powerful technique that enhances coordinate-independent mapping procedures that are based on comparative statements. In addition to our current use of PDC heuristics, which exploit the precision of the mapping description, it may be feasible to formalize a hierarchy of further criteria for area assignment. The divergence and number of mapping statements appears to be a useful indicator of the degree of controversy of a mapping. It may also be possible to use method-related confidence levels (e.g. cyto-, myelo-, chemoarchitectonics), which would have to be adapted as methodological progress is made.
Acronym mapping uses I-relationships only. It does not infer hierarchical relationships. For example, it does not recognize MDmc as a subarea (S) of MD (i.e. that the mediodorsal nucleus of the thalamus has a magnocellular part). Although the naming of substructures using prefixes or suffixes is a common procedure, the potential gain by exploiting such regularities seems to be offset by the complications of having to deal with different relationships. What this reasoning could be good for, however, is the generation of a list of relations that is sorted by plausibility and suggested to the user for manual confirmation or rejection.
Finally, since the capabilities of AM are limited to equating identity of BrainSites with identically called structures, this procedure cannot recognize whether several BrainSites are identical in spite of having different acronyms (and names). For example, the CoCoMac database contains four different acronyms extracted from the literature for the caudate nucleus: Caud, Cd, C, CA (ignoring indices). When AM is generated, these four acronyms are represented as four different BrainSites of AM. Identity between them is only recognized if at least a certain subset of all these relations has been entered manually by the data collator. This problem is addressed by the GM, which is described in §4b.
(b) General map
While AM efficiently deals with missing relations between similar brain structures that carry the same acronym, the coexistence of different acronyms or variations of spelling for obviously identical structures (at least at the coarse level of resolution that they are normally referred to) requires a different approach. For example, the primary motor cortex, the caudate nucleus or the superior colliculus usually have identical definitions between authors, but various acronyms are being used for their designation. In the CoCoMac database, we find five different acronyms for the primary motor area (4, F1, FA, M1, MI), four terms for the caudate (Caud, Cd, C, CA), and three different types of shorthand for the superior colliculus (SC, SuC, sup.col.). The assumed I-relations between all the BrainSites that use these acronyms have to be entered manually into the database. This process is not only tedious but also embodies the risk that there may be disconnected clusters of identical, yet differently named, BrainSites if relations between these groups are missing entirely. As a solution to this problem, we present in this section the concept of the GM.
(i) Definition of GM
The GM is a predefined set of BrainSites that represent general definitions for brain structures under unified names. The main purpose of GM is to offer BrainSites for those regions of the brain where brain structures are commonly referred to: (i) without explicit definitions, and (ii) by different names in the literature although they are identical (or at least very similar). Thus, the GM can be regarded as an idealized brain map of standardized brain structures. BrainSites in GM should have widely recognized names and acronyms since they form a standardized set of nomenclature. The names could in fact be recruited from the terminologia anatomica (the successor of the nomina anatomica including English in addition to Latin terms; Whitmore 1998) where they contain a comprehensive set of acceptable terms with adequate granularity. Two parts of the GM definition from CoCoMac are shown in table 3.
Table 3.
Excerpts from the general map as used in the CoCoMac database. (General map names and acronyms for the lateral geniculate nucleus and the caudate nucleus. The underscores mark the level of hierarchy in relation to the entire nucleus. The hash indicates nomenclature by Norden & Kaas (1978). The major division of the caudate nucleus is based on anatomical landmarks as suggested by Yeterian & Pandya (1991).)
| Name | GM-acronym |
|---|---|
| lateral geniculate nucleus | GM-GL |
| dorsal part | GM-GLd |
| magnocellular | GM-GLd_mc |
| laminar zone | GM-GLd_mc_l |
| external layer | GM-GLd_mc_l_ext (# I) |
| internal layer | GM-GLd_mc_l_int (# II) |
| interlaminar zone | GM-GLd_mc_il |
| parvocellular | GM-GLd_pc |
| … | GM-GLd_pc_… |
| superficial (s)-layer | GM-GLd_s (# 0 or k1) |
| ventral part | GM-GLv |
| nucleus caudatus | GM-Cd |
| head | GM-Cd_h |
| medial | GM-Cd_h_m |
| ventral | GM-Cd_h_m_v |
| central | GM-Cd_h_m_c |
| dorsal | GM-Cd_h_m_d |
| intermediate | GM-Cd_h_i |
| … | GM-Cd_h_i_… |
| lateral | GM-Cd_h_l |
| … | GM-Cd_h_l_… |
| body | GM-Cd_b |
| … | GM-Cd_b_… |
| genu | GM-Cd_g |
| … | GM-Cd_g_… |
| tail | GM-Cd_t |
| … | GM-Cd_t_… |
In addition to the predefined terminology, GM has a predefined hierarchy of ‘internal’ relations between the BrainSites contained in this map. Recall that a hierarchy may occur also in an OM; it is not specific to GM or limited to relations between brain maps of different granularity. For example, the map defined by Barbas (1988) contains prefrontal area 12 (B88-12) as well as its orbital (B88-12o) and lateral (B88-12‐l) subdivisions. Apart from subdivisions (i.e. BrainSites related through internal S/L-relations), there is no semantic overlap among the BrainSites in GM (i.e. the BrainSites represent disjoint concepts), which effectively abandons the use of O-relations. For example, there is only one BrainSite in GM referring to the superior colliculus (GM–SC), but GM–SC has L-relations to each of the seven layers of superior colliculus (GM–SC_I, …, GM–SC_VII). While all internal relations of GM are predefined, external GM-relations (i.e. relations between GM-BrainSites and OM-BrainSites) must be specified explicitly (see §4b(ii)).
(ii) Linking brain structures and GM
For reasons of transparency, we allow only I-relations to link OM-BrainSites with corresponding GM-BrainSites. This means that relations between OM and GM are only allowed for BrainSites at the same conceptual level (e.g. directly linking a nucleus in GM to a subnucleus in OM is excluded). Seen from another perspective, this also means that within the brain regions for which it has been defined, GM must offer a BrainSite for each conceptual level of data description that is commonly used in the original publications. Overall, the exclusive use of I-relations ensures that OM-BrainSites that have different acronyms but identical meaning, and that are linked to the same GM-BrainSite, will be considered identical.
(iii) GM, vocabularies, ontologies
The construction of a hierarchy between the BrainSites contained in GM extends the standardized vocabulary of neuroanatomical nomenclature into (an incomplete) neuroanatomical ontology. An ontology has been defined as ‘the specification of relationships between words and the operationally defined structural concepts they represent’ (Bowden & Dubach 2003). Ontologies play a major role not only in defining standardized vocabularies but, more importantly, in providing a framework for the classification of concepts and associated terms so that data from a variety of sources can be related irrespective of their specific origin, context and terminology. In particular, published digital ontologies, such as UMLS (http://www.nlm.nih.gov/research/umls/), provide a convenient starting point for automated data mining in vast amounts of complex neuroscience data. Efficient linking between separate data sources can be achieved if the data are classified according to such a generally accepted ontology. It has been recognized, however, that the present variability of data contents, contexts and concepts, as well as different user preferences, cannot be ignored or easily diminished. Therefore, methods for dealing with synonyms (different terms having the same meaning), homonyms (the same term having unrelated meanings), or polysemes (the same term having different but related meanings) are required. The linking of OM-BrainSites and their relationships with the ontology of GM-BrainSites described here exemplifies a strategy that provides an alternative method with significant advantages over a centrally prescribed vocabulary including approved synonyms and so on. In our scheme, the user is free to use whatever nomenclature and conceptual framework that appear most suitable in the specific context (in CoCoMac: the OMs and OM inter-map relationships). Through establishing relationships with the ontology (here: GM), these concepts can be linked to all other concepts that also relate to the same ontology. Since relations are inserted automatically for identical BrainSite_Acronyms through AM, this effort is no greater than the identification of synonyms for central storage. A potential loss of precision of the mapping (which is excluded in our case through the requirement of I-relationships) is balanced by a maximal increase in flexibility, which has the capability of accommodating new concepts.
Another feature that distinguishes our GM concept from most vocabularies and ontologies is the focus on brain regions and structures with unspecified parcellation schemes. At present, we do not intend to provide a complete classification for the entire brain, but we follow the needs of data collation. Although it appears useful to add GM-BrainSites for the most comprehensive subdivisions, so that paths exist between all pairs of GM-BrainSites, there is no need to insert GM-BrainSites for those brain structures that are covered by OM-BrainSites and that are clearly related through original literature statements about the relations between them.
In summary, GM allows for correct mapping between brain structures with different acronyms but identical semantics. The correct use of GM requires an additional effort by the data collator since one has to decide for each new OM-BrainSite whether it needs to be linked to a corresponding GM-BrainSite or not. This extra effort, however, can be used to construct further mapping paths, which enhance the power of coordinate-independent mapping. In addition, further population of GM with additional BrainSites creates important opportunities to carry out hierarchical searches within the database and to relate to the contents of other databases.
(c) Regional map
As explained above, the microstructural feature spaces used for parcellation cannot always be mapped to one another directly so that topographical landmarks are used (such as the position relative to sulci), which provide indirect links. Note that the use of topographical landmarks for comparison between microstructural features is conceptually distinct from the creation of topographical parcellation schemes (e.g. Crespo-Facorro et al. 2000). Although the two approaches refer to the same class of macroscopical features, microstructural parcellation is conceptually independent of topographical landmarks whereas, by definition, a topographical parcellation relies on them. Practically, one can argue about the precision of using topographical features to assess the similarity of parcellations with different microstructural feature spaces. The precision will depend on the variability of both brain shape and the location of microstructurally defined areas relative to the topographical landmarks. Where both types of variability are minimal, the comparison can be made with a precision that approaches the precision in establishing microstructural borders. At least in human brains, however, both types of variability are large enough to leave room for a spatial imprecision of up to 1 cm (e.g. Rademacher et al. 1993; Amunts et al. 2000; Geyer et al. 2000; Morosan et al. 2001; Gaser & Schlaug 2003).
Despite this spatial imprecision, it is not unreasonable that many researchers compare the location of structural and functional features (e.g. activations in functional imaging) between different studies, and even between different species with reference to topographical features, for several reasons. First, until recently, almost all imaging studies involve the pooling of data across different individuals. Although spatial deformation procedures reduce the shape differences between individual brains, they do not necessarily put corresponding brain regions in the same spatial position. Second, even during the imaging of a single subject, movements occur that degrade data quality and are not perfectly corrected by current registration techniques. Third, the spatial resolution of the blood oxygen level dependent signal evaluated in fMRI studies is limited by the geometry of the blood vessels and resolves microstructural borders only under exceptional circumstances (e.g. Duong et al. 2001). Finally, in the comparison between structural and functional data, the variability in the functional imaging data noted above combines with the variability of anatomical locations in the probabilistic atlases. Given that most functional imaging studies apply a smoothing kernel of 4–8 mm, it follows that under current conditions, a spatial inaccuracy of the functional imaging procedure matches quite well with the accuracy of locating microstructural borders with reference to topographical features of the cerebral cortex. Therefore, for the majority of imaging applications, a topographical representation of coordinate-independent microstructural parcellations is sufficiently accurate so long as the range of uncertainty near the borders of cortical areas is respected.
Based on these methodological considerations, we provide an aid to structure–function comparisons in primate cortical areas, which we refer to as the regional map (RM). The purpose of the RM parcellation is to make a wealth of microstructural-functional data, which have been described in a coordinate-independent manner, amenable to a rational comparison with coordinate-based structural and functional data.
(i) Definition of RM
The RM is a coarse parcellation scheme of the cerebral cortex with respect to a combination of microstructural, functional and topographic features. It consists of a full map of distinct BrainSites that can be used in the same way as other OMs, for example, the ones delineated by Brodmann (1909) or Petrides & Pandya (1994). Because of its broad topographic definition, RM can be drawn on an individual or a schematic brain (see figure 3). In a spatial framework, approximate coordinates could be given for its constituent BrainSites as done for the Talairach atlas (Lancaster et al. 2000). The BrainSites of RM are supposed to be non-overlapping, but a small amount of overlap is bound to occur due to the relations of RM with a variety of standard brain maps that differ themselves in the precise definition and location of cortical areas.
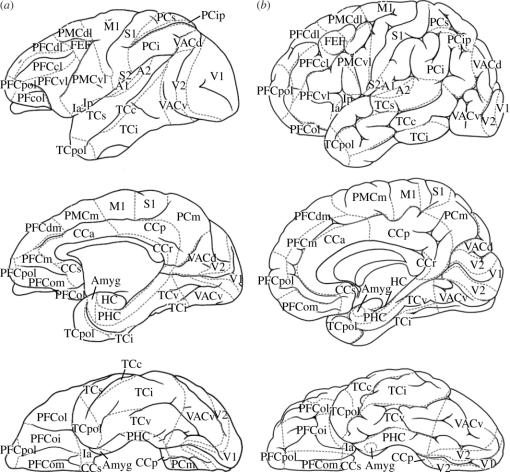
Figure 3.
Regional map of primate cerebral cortex superimposed on the cortical surface both of a macaque (a) and a human brain (b) seen from the lateral (top), mesial (middle) and ventral aspect (bottom). Areal borders are indicated by grey hatched lines. Hidden regions of the primary (A1) and secondary auditory cortex (A2), anterior (Ia) and posterior insula (Ip), as well as of the amygdala (Amyg), are shown as projections in approximate locations. Further abbreviations: CCa, anterior cingulate cortex; CCp, posterior cingulate cortex; CCr, retrosplenial cingulate cortex; CCs, subgenual cingulate cortex; FEF, frontal eye field; HC, hippocampus; M1, primary motor cortex; PFCcl, centrolateral prefrontal cortex; PFCdl, dorsolateral prefrontal cortex; PFCdm, dorsomedial prefrontal cortex; PFCm, medial prefrontal cortex; PFCoi, intermediate orbital prefrontal cortex; PFCol, orbitolateral prefrontal cortex; PFCom, orbitomedial prefrontal cortex; PFCvl, ventrolateral prefrontal cortex; PFCpol, polar prefrontal cortex; PHC, parahippocampal cortex; PMCdl, dorsolateral premotor cortex; PMCm, medial (supplementary) premotor cortex; PMCvl, ventrolateral premotor cortex; S1, primary somatosensory cortex; S2, secondary somatosensory cortex; PCi, inferior parietal cortex; PCip, cortex of the intraparietal sulcus; PCm, medial parietal cortex; PCs, superior parietal cortex; TCc, central temporal cortex; TCi, inferior temporal cortex; TCs, superior temporal cortex; TCpol, polar temporal cortex; TCv, ventral temporal cortex; V1, primary visual cortex; V2, secondary visual cortex; VAC, anterior visual cortex. The detailed mapping relations of RM to OM-BrainSites can be downloaded from http://www.cocomac.org/regionalmap.pdf.
A particular feature of RM is its generalized terminology, which allows its application to a wide range of primates including man and macaque monkey. We deliberately stay away from controversial issues, such as the presence of an equivalent to Broca's area in the monkey or the choice among competing pre-existing terminologies. More precisely, RM uses general functional (e.g. V1, M1, FEF), structural (e.g. amyg, HC) or topographic (e.g. PFCdl, PCip, TCpol) area names that are widely recognized and convenient to use.
Since most coordinate-dependent data have been gathered in human subjects, whereas the majority of relevant microstructural data come from invasive experiments in non-human primates, the desire for comparison suggests that we map the RM to the cerebral cortex of both the macaque monkey and the human brain using the same terminology. For methodological reasons, the borders in the RM are closely related to accepted microstructural boundaries in the macaque that have been established with a high level of confidence and in increasing detail over a century of brain mapping. In the human, we rely largely on topographical features and recent non-invasive functional data. The divisions of RM are shown in figure 3, and the region names are derived as far as possible from a consistent set of species-independent topographical or basic functional terminology. In this way, we avoid some of the nomenclatorial confusion, for example, concerning the relative location of Brodmann's areas 5 and 7 in the macaque and the human, respectively. In the macaque, the intraparietal sulcus separates the superior parietal area 5 from the inferior parietal area 7. In the human brain, both areas are dorsal of the intraparietal sulcus, whereas the inferior parietal lobule contains areas 39 and 40, which Brodmann did not discern in the macaque (Brodmann 1909; see Zilles 2004, p. 1020). While our RM does not distinguish further rostral and caudal parietal regions, it does recognize the intraparietal sulcus as a separate, although complex, territory. Equally interesting is the resemblance of areas of the middle temporal gyrus in the human with what is the ventral bank of the superior temporal sulcus in macaques. On another matter, all major cytoarchitectonic parcellations of the macaque orbito-frontal cortex suggest at least three divisions, which are roughly separated by the medial and lateral orbital sulci. We have preferred this tripartite scheme to the purely topographic bipartite division often encountered in human functional imaging studies.
(ii) Linking RM
The specification of the RM (http://www.cocomac.org/regionmalmap.pdf) lists approximate human and macaque Brodmann areas, as well as relationships with other brain maps, which allows an effective conversion of original research data to the RM parcellation scheme. These relations have to be inserted manually as other maps are being collated but, in practice, a large set of maps are effectively linked through mapping paths that comprise areas from the standard maps. As opposed to these external relations, internal relations between the BrainSites of RM do not occur. These are not necessary because of the optimized design of the RM parcellation and because alternative area definitions would usually be linked through external relations. We found, however, that it may be useful on some occasions to define ‘combination areas’ from the existing ones (e.g. dorsal premotor cortex from dorsomedial and dorsolateral premotor cortices to represent observations that span the dorsal convexity).
(iii) Significance of RM
The RM bridges the gap between coordinate-based atlases and coordinate-independent parcellation schemes and addresses the need to provide an appropriate framework for mapping data between brains of related species with relatively similar topography and comparable parcellation schemes. In contrast to the surface-based mapping tool CARET (Van Essen et al. 2001a), which lends coordinates to the macaque brain maps of Brodmann (1909) and von Bonin & Bailey (1947), for example, whose coordinates can only roughly be specified based on surface views and a few sections, the RM is a new map that is related to previous parcellation schemes with the degree of precision that coordinate-independent mapping provides. The challenges and limitations of mapping between macaque and human brains using surface-based atlases have been discussed in more detail elsewhere (e.g. Van Essen et al. 2001b), and a unified coordinate-independent parcellation scheme based on comparative architectonic analyses of the frontal cortex has been proposed previously (Petrides & Pandya 1994). With the approach of the RM, we are less ambitious concerning spatial resolution and adherence to existing nomenclature. We believe, however, that this approach will be of greater use in mediating between macaque and human studies, whose combination is required to obtain a better understanding of brain function in health and disease.
5. Comparing special mapping procedures
The concepts of the three procedures for mapping brain structures, AM for mapping by acronym, GM for mapping with reference to a hierarchically organized standard vocabulary, and RM for topographical mapping based on microstructural and functional similarities, are perhaps best understood when directly contrasted. Table 4 illustrates their main features and differences.
6. Conclusions and perspectives
In this paper, we have explored the use of coordinate-independent mapping procedures, which allow us to make better use of the wealth of neuroscience data that have been related to microstructural-functional parcellations but never to spatial coordinate systems, or that must be compared only with reference to macroscopical landmarks for other reasons. The most important aspect of coordinate-independent mapping is the exploitation of published statements on area relationships for the construction of mapping pathways that enable us to relate between data that are registered to different parcellation schemes. Where such statements are not available, the auxiliary methods of AM, GM and RM provide additional strategies as well as enhancements in the efficiency of data representation and mapping, in the inter-operability between data resources and vocabularies, and in the relation between species and between spatial and coordinate-independent mapping techniques. The latter leads us once more to the comparison between these two unequal mapping techniques.
Given the amount of data only available with reference to microstructural parcellation schemes, it is surprising that coordinate‐independent brain mapping is almost entirely overlooked in recent books and reviews, which otherwise cover almost everything that could be mapped to spatial coordinates. There is an obvious reason for this ignorance; a century of brain mapping has still not resolved the plan of cortical organization, and too many neuroanatomists spend time on disentangling the different definitions and nomenclatures created, adapted and misunderstood. In addition, microstructural delineations operate on an approximate scale with regard to macroscopically visible brain structures, whereas spatial mapping bears the promise to be quantitative and exact. In combination with the non-invasiveness and repeatability of most brain imaging studies, it appears self-evident that the latter is the way to go. Are spatial mapping approaches really superior to coordinate-independent mapping procedures?
We have already mentioned the spatial uncertainty incurred with functional imaging techniques for technical reasons. We will now extend this uncertainty to the anatomical aspect. As an example, figure 4 shows the spatial extent of primary auditory cortex (A1) from 10 brains resulting in a spatial probability map of its location on a standard brain. These images were registered in the Montreal Neurological Institute (MNI) standard space and superimposed on the single subject MNI standard brain. The resulting picture gives the impression that A1 does have a low, but non-zero, probability of being partially located in the parietal operculum and the insular cortex. However, microstructurally, A1 is exclusively located on the dorsal aspect of the superior temporal gyrus and has never been reported to encroach onto the parietal operculum. In fact, this is also what is shown in the spatial cytoarchitectonic maps of all of the individual brains.
Figure 4.
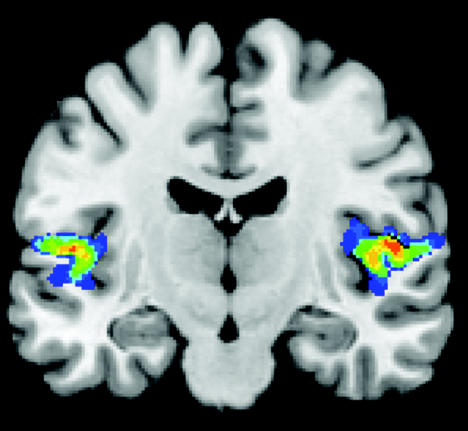
Spatial probability map of cytoarchitectonically identified primary auditory cortex (A1) superimposed on a frontal in vivo MRI section of the single subject Montreal Neurological Institute reference brain. The left hemisphere is on the left. Colours indicate the number of brains where A1 was present in that voxel (dark blue: 1 to red: 10; 10 individual brains were studied). Note that in this representation of a single brain, it appears as if A1 had a non-zero probability of being located in parietal operculum and insula, which was not the case in any of the individual brains before deformation. The figure was kindly provided by Patricia Morosan and Karl Zilles, IME, FZ Jülich.
What has gone wrong? Clearly, a single subject brain has its individual shape and cannot represent the variability among individual brains even if it has the typical textbook pattern of cortical folding. For comparing between several different brains, spatial normalization is required, in this case to the template brain shown in the section. The result of this normalization procedure depends on the method used. Volume-matching methods have no concept of sulcal anatomy and, therefore, can displace brain structures to nearby, but topographically incorrect, locations. This could have been avoided by using a normalization method that explicitly matched the lateral fissure (see Brett et al. 2002 for a detailed discussion). As an alternative, the probability map could have been displayed on an average structural image of the individual brains. This, however, would look blurred so that it does not allow us to discern the precise anatomical relations. It turns out that not only the degree of interindividual variability shown depends strongly on the method used for spatial normalization, but also that the probability space of 10 individual brains is an abstract concept, which cannot be visualized in an anatomically correct way.
Another example of the conceptual difficulties with probabilistic maps resulting from spatial mapping is illustrated in Mazziotta et al. (2001, fig. 9). When microstructurally defined areas 44 or 45 from 10 individuals were superimposed on a single brain template, the highest probability that the respective area is found at any voxel was 7 out of 10 brains. Therefore, using probabilistic atlases, we may not have even a single voxel that can be unambiguously attributed to a specific microstructural area. Consequently, we also have to accept spatial imprecision and misattribution of function to structure with coordinate-based mapping unless we go to the single subject level and eliminate deformations, which effectively means to limit quantitative comparisons to those data modalities that can be obtained in the same individual (for a review see Brett et al. 2002).
Altogether, a detailed inspection of spatial and coordinate-independent mapping techniques confirms our earlier conclusion that the two approaches have their respective strengths and weaknesses and each must be applied in the correct way to produce valid results. More important, however, is that the two approaches answer different questions. Spatial mapping is concerned with variability of structural and functional brain attributes in space. It is also a prerequisite for quantitative comparisons of data obtained from different brains, although it may fail to achieve a full correction. Coordinate-independent mapping relies on the unambiguous identification of brain structures and allows for their efficient and meaningful representation. It is indispensable for meta-studies where data were not, or could not, be spatially compared, such as most clinical data in humans and in vivo tracing data in primates.
What are the prospects of coordinate-independent brain mapping in the future? First, the power of coordinate-independent mapping could be significantly enhanced if large amounts of high quality data were submitted to databases. A system for automated extraction of neuroanatomical compound terms and acronyms from text (e.g. following optical character recognition) is currently under construction (Srinivas et al. 2003). This does not address the data that are provided in figures showing labelling on brain sections or surface views, but even such figures could be evaluated automatically if the brain regions were clearly identified and their borders marked.
Second, the heuristic methods for appraising the quality of mapping and relation statements can be improved. The present RC and PDC heuristics were the first formal indices developed for this specific purpose (see Stephan et al. 2000, 2001). Contents-based criteria could be added based on an appraisal of the reliability and objectivity of different delineation methods. Already, the number and diversity of relation statements for the same pair of areas is useful for identifying controversial brain regions and potential errors. Such methods can be combined with the technique of inferring mapping pathways, which will lead to a higher sensitivity.
Third, the multitude of parcellation schemes can be turned into an advantage if information concerning regions of differential overlap is to be retrieved: instead of retrieving data for a single scheme with abrupt borders, one could superimpose all available schemes in their rough topographical position. Selecting a certain region or coordinate could then trigger simultaneous queries in all areas that contain this location irrespective of the details of the parcellations used. Thereby, we obtain a higher spatial resolution with smoother transitions than is provided by any single parcellation scheme with the added benefit of cross-validation between the data obtained for each scheme.
Finally, if we use the parcellation schemes to retrieve associated microstructural–functional data on the brain regions in question, then we can devise high-throughput data analysis procedures that will identify interesting structure–function relationships in the associated datasets (Koslow & Hirsch 2004). These could potentially also lead to multidimensional evaluations of parcellation schemes indicating which borders represent truly important divisions.
What would Brodmann, Campbell and other pioneers of cortical mapping do if they lived today? Most probably, they would be excited by the present techniques of quantitative, multi-modal, and in vivo neuroanatomy. Perhaps they would also be amazed by the persistent uncertainties in search of a unique system of cortical structures. Would they dump the concept of cortical areas and look instead at columns and layers as meaningful units, or turn their interest to voxel-based analyses that are not contaminated by these conceptual problems? Extrapolating their comparative approach, they might develop better techniques to find clues in phylogenetic and ontogenetic studies of brain parcellation and try to understand how structure enables function and how function shapes structure.
Acknowledgments
This work was supported by the DFG Graduate School 320. We thank the students of the Computational | Systems | Neuroscience group for their contributions to the approaches presented here. In particular, the initial foundations of the concepts of AM, GM and RM were laid by Klaas E. Stephan as part of his Ph.D. work with R.K.
Glossary
- AM
acronym map
- GM
general map
- OM
original map
- ORT
objective relational transformation
- MT
middle temporal area
- PDC
precision of description codes
- PO
parieto-occipital area
- RC
relation codes
- RM
regional map
Footnotes
One contribution of 12 to a Theme Issue ‘Cerebral cartography 1905–2005’.
References
- Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann's areas 17 and 18 brought into stereotaxic space—where and how variable? NeuroImage. 2000;11:66–84. doi: 10.1006/nimg.1999.0516. doi:10.1006/nimg.1999.0516 [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K.J. Voxel-based morphometry—the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Barbas H. Anatomic organization of basoventral and mediodorsal visual recipient prefrontal regions in the rhesus monkey. J. Comp. Neurol. 1988;276:313–342. doi: 10.1002/cne.902760302. [DOI] [PubMed] [Google Scholar]
- Barbas H, Pandya D.N. Architecture and frontal cortical connections of the premotor cortex (area 6) in the Rhesus monkey. J. Comp. Neurol. 1987;256:211–228. doi: 10.1002/cne.902560203. [DOI] [PubMed] [Google Scholar]
- Barnes C.L, Pandya D.N. Efferent cortical connections of multimodal cortex of the superior temporal sulcus in the rhesus monkey. J. Comp. Neurol. 1992;318:222–244. doi: 10.1002/cne.903180207. [DOI] [PubMed] [Google Scholar]
- Bota M, Arbib M.A. Integrating databases and expert systems for the analysis of brain structures: connections, similarities, and homologies. Neuroinformatics. 2004;2:19–58. doi: 10.1385/NI:2:1:019. [DOI] [PubMed] [Google Scholar]
- Boussaoud D, Desimone R, Ungerleider L.G. Visual topography of area TEO in the macaque. J. Comp. Neurol. 1991;306:554–575. doi: 10.1002/cne.903060403. [DOI] [PubMed] [Google Scholar]
- Bowden D.M, Dubach M. Neuronames 2002. Neuroinformatics. 2003;1:43–60. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Brett M, Johnsrude I.S, Owen A.S. The problem of functional localization in the human brain. Nat. Rev. Neurosci. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Beiträge zur histologischen Lokalisation der Großhirnrinde. I. Mitteilung: Die Regio Rolandica. J. Psychol. Neurol. 1903;2:79–107. [Google Scholar]
- Brodmann K. Beiträge zur histologischen Lokalisation der Großhirnrinde. III. Mitteilung: Die Rindenfelder der niederen Affen. J. Psychol. Neurol. 1905;4:177–226. [Google Scholar]
- Brodmann K. Barth; Leipzig: 1909. Vergleichende Lokalisationslehre der Großhirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. [Google Scholar]
- Campbell W.W. Cambridge University Press; Cambridge: 1905. Histological studies on the localisation of cerebral function. [Google Scholar]
- Crespo-Facorro B, Kim J, Andreasen N.C, Spinks R, O'Leary D.S, Bockholt H.J, Harris G, Magnotta V.A. Cerebral cortex: a topographic segmentation method using magnetic resonance imaging. Psychiatry Res. 2000;100:97–126. doi: 10.1016/s0925-4927(00)00072-x. [DOI] [PubMed] [Google Scholar]
- Cusick C.G, Seltzer B, Cola M, Griggs E. Chemoarchitectonics and corticocortical terminations within the superior temporal sulcus of the rhesus monkey: evidence for subdivisions of superior temporal polysensory cortex. J. Comp. Neurol. 1995;360:513–535. doi: 10.1002/cne.903600312. [DOI] [PubMed] [Google Scholar]
- Desimone R, Ungerleider L.G. Multiple visual areas in the caudal superior temporal sulcus of the macaque. J. Comp. Neurol. 1986;248:164–189. doi: 10.1002/cne.902480203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T.Q, Kim D.S, Ugurbil K, Kim S.G. Localized cerebral blood flow response at submillimeter columnar resolution. Proc. Natl Acad. Sci. USA. 2001;98:10 904–10 909. doi: 10.1073/pnas.191101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S, Walters N.B, Schleicher A, Kril J, Egan G.F, Zilles K, Watson J.D, Amunts K. High-resolution MRI reflects myeloarchitecture and cytoarchitecture of human cerebral cortex. Hum. Brain Mapp. 2004;24:206–215. doi: 10.1002/hbm.20082. (ePublication ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B, Stephan K.E, Mohlberg H, Grefkes C, Fink G.R, Amunts K, Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. In press doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Felleman D.J, Van Essen D.C. Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Galletti C, Gamberini M, Kutz D.F, Baldinotti I, Fattori P. The relationship between V6 and PO in macaque extrastriate cortex. Eur. J. Neurosci. In press doi: 10.1111/j.1460-9568.2005.03911.x. [DOI] [PubMed] [Google Scholar]
- Gaser C, Schlaug G. Brain structures differ between musicians and non-musicians. J. Neurosci. 2003;23:9240–9245. doi: 10.1523/JNEUROSCI.23-27-09240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattass R, Gross C.G. Visual topography of striate projection zone (MT) in posterior superior temporal sulcus of the macaque. J. Neurophysiol. 1981;46:621–638. doi: 10.1152/jn.1981.46.3.621. [DOI] [PubMed] [Google Scholar]
- Geyer S, Schormann T, Mohlberg H, Zilles K. Areas 3a, 3b, and 1 of human primary somatosensory cortex. Part 2. Spatial normalization to standard anatomical space. NeuroImage. 2000;11:684–696. doi: 10.1006/nimg.2000.0548. doi:10.1006/nimg.2000.0548 [DOI] [PubMed] [Google Scholar]
- Gorin F, Hogarth M, Gertz M. The challenges and rewards of integrating diverse neuroscience information. Neuroscientist. 2001;7:18–27. doi: 10.1177/107385840100700106. [DOI] [PubMed] [Google Scholar]
- Hof P.R, Morrison J.H. Neurofilament protein defines regional patterns of cortical organization in the macaque monkey visual system: a quantitative immunohistochemical analysis. J. Comp. Neurol. 1995;352:161–186. doi: 10.1002/cne.903520202. [DOI] [PubMed] [Google Scholar]
- Huk A.C, Dougherty R.F, Heeger D.J. Retinotopy and functional subdivision of human areas MT and MST. J. Neurosci. 2002;22:7195–7205. doi: 10.1523/JNEUROSCI.22-16-07195.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens T.E, Sillery E, Ciccarelli O, Thompson A.J, Smith S.M, Matthews P.M. Functional-anatomical validation and individual variation of diffusion tractography-based segmentation of the human thalamus. Cereb. Cortex. 2004;15:31–39. doi: 10.1093/cercor/bhh105. doi:10.1093/cercor/bhh105 [DOI] [PubMed] [Google Scholar]
- Koslow S.H, Hirsch M.D. Celebrating a decade of neuroscience databases. Looking to the future of high-throughput data analysis, data integration, and discovery neuroscience. Neuroinformatics. 2004;2:267–269. doi: 10.1385/NI:2:3:267. [DOI] [PubMed] [Google Scholar]
- Kötter R. Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics. 2004;2:127–144. doi: 10.1385/NI:2:2:127. [DOI] [PubMed] [Google Scholar]
- Lancaster J.L, et al. Automated Talairach Atlas labels for functional brain mapping. Hum. Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.W, Van Essen D.C. Mapping of architectonic subdivisions in the macaque monkey, with emphasis on parieto-occipital cortex. J. Comp. Neurol. 2000;428:79–111. doi: 10.1002/1096-9861(20001204)428:1<79::aid-cne7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Luppino G, Matelli M, Camarda R.M, Gallese V, Rizzolatti G. Multiple representations of body movements in mesial area 6 and the adjacent cingulate cortex: an intracortical microstimulation study in the macaque monkey. J. Comp. Neurol. 1991;311:463–482. doi: 10.1002/cne.903110403. [DOI] [PubMed] [Google Scholar]
- Mauss T. Die faserarchitektonische Gliederung der Großhirnrinde bei niederen Affen. J. Psychol. Neurol. 1908;13:263–325. [Google Scholar]
- Mazziotta J, et al. A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM). Phil. Trans. R. Soc. B. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. doi:10.1098/rstb.2001.0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynert T. Engelmann; Leipzig: 1868. Der Bau der Großhirnrinde und seine örtlichen Verschiedenheiten, nebst einem pathologisch-anatomischen Corollarium. [Google Scholar]
- Mitz A.R, Wise S.P. The somatotopic organization of the supplementary motor area: intracortical microstimulation mapping. J. Neurosci. 1987;7:1010–1021. doi: 10.1523/JNEUROSCI.07-04-01010.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosan P, Rademacher J, Schleicher A, Amunts K, Schormann T, Zilles K. Human primary auditory cortex: cytoarchitectonic subdivisions and mapping into a spatial reference system. NeuroImage. 2001;13:684–701. doi: 10.1006/nimg.2000.0715. doi:10.1006/nimg.2000.0715 [DOI] [PubMed] [Google Scholar]
- Norden J.J, Kaas J.H. The identification of relay neurons in the dorsal lateral geniculate nucleus of monkeys using horseradish peroxidase. J. Comp. Neurol. 1978;182:707–725. doi: 10.1002/cne.901820409. [DOI] [PubMed] [Google Scholar]
- Olszewski J. Karger; Basel: 1952. The thalamus of Macaca mulatta. [Google Scholar]
- Paxinos G, Huang X.-F, Toga A.W. Academic; San Diego, CA: 2000. The rhesus monkey brain in stereotaxic coordinates. [Google Scholar]
- Petrides M, Pandya D.N. Comparative architectonic analysis of the human and macaque frontal cortex. In: Boller F, Grafman J, editors. Handbook of neuropsychology, vol. 9, ch. 2. 1994. pp. 17–58. [Google Scholar]
- Rademacher J, Caviness V.S, Jr, Steinmetz H, Galaburda A.M. Topographical variation of the human primary cortices: implications for neuroimaging, brain mapping, and neurobiology. Cereb. Cortex. 1993;3:313–329. doi: 10.1093/cercor/3.4.313. [DOI] [PubMed] [Google Scholar]
- Rademacher J, Bürgel U, Zilles K. Stereotaxic localization, intersubject variability, and interhemispheric differences of the human auditory thalamocortical system. NeuroImage. 2002;17:142–160. doi: 10.1006/nimg.2002.1178. [DOI] [PubMed] [Google Scholar]
- Roland P, et al. A database generator for human brain imaging. Trends Neurosci. 2001;24:562–564. doi: 10.1016/s0166-2236(00)01924-x. [DOI] [PubMed] [Google Scholar]
- Scannell J.W, Blakemore C, Young M.P. Analysis of connectivity in the cat cerebral cortex. J. Neurosci. 1995;15:1463–1483. doi: 10.1523/JNEUROSCI.15-02-01463.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell J.W, Burns G.A.P.C, Hilgetag C.C, O'Neil M.A, Young M.P. The connectional organization of the cortico-thalamic system of the cat. Cereb. Cortex. 1999;9:277–299. doi: 10.1093/cercor/9.3.277. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Amunts K, Geyer S, Morosan P, Zilles K. Observer-independent method for microstructural parcellation of cerebral cortex: a quantitative approach to cytoarchitectonics. NeuroImage. 1999;9:165–177. doi: 10.1006/nimg.1998.0385. [DOI] [PubMed] [Google Scholar]
- Seltzer B, Pandya D.N. Afferent cortical connections and architectonics of the superior temporal sulcus and surrounding cortex in the rhesus monkey. Brain Res. 1978;149:1–24. doi: 10.1016/0006-8993(78)90584-x. [DOI] [PubMed] [Google Scholar]
- Sereno M.I, Dale A.M, Reppas J.B, Kwong K.K, Belliveau J.W, Brady T.J, Rosen B.R, Tootell R.B. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- Sowell E.R, Peterson B.S, Thompson P.M, Welcome S.E, Henkenius A.L, Toga A.W. Mapping cortical change across the human life span. Nat. Neurosci. 2003;6:309–315. doi: 10.1038/nn1008. doi:10.1038/nn1008 [DOI] [PubMed] [Google Scholar]
- Srinivas P.R, Gusfield D, Mason O, Gertz M, Hogarth M, Stone J, Jones E.G, Gorin F.A. Neuroanatomical term generation and comparison between two terminologies. Neuroinformatics. 2003;1:177–192. doi: 10.1007/s12021-003-0004-z. [DOI] [PubMed] [Google Scholar]
- Stephan K.E, Zilles K, Kötter R. Coordinate-independent mapping of structural and functional cortical data by objective relational transformation (ORT) Phil. Trans. R. Soc. B. 2000;355:37–54. doi: 10.1098/rstb.2000.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K.E, Kamper L, Bozkurt A, Burns G.A.P.C, Young M.P, Kötter R. CoCoMac: advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Phil. Trans. R. Soc. B. 2001;356:1159–1186. doi: 10.1098/rstb.2001.0908. doi:10.1098/rstb.2001.0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Thieme; Stuttgart: 1988. Co-planar stereotaxic atlas of the human brain. 3-D proportional system: an approach to cerebral imaging. [Google Scholar]
- Tanaka K, Hikosaka K, Saito H, Yukie M, Fukada Y, Iwai E. Analysis of local and wide-field movements in the superior temporal visual areas of the macaque monkey. J. Neurosci. 1986;6:134–144. doi: 10.1523/JNEUROSCI.06-01-00134.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M, Schwartz C, Lin R.T, Khan A.A, Toga A.W. Three-dimensional statistical analysis of sulcal variability in the human brain. J. Neurosci. 1996;16:4261–4274. doi: 10.1523/JNEUROSCI.16-13-04261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M, et al. Genetic influences on brain structure. Nat. Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. doi:10.1038/nn758 [DOI] [PubMed] [Google Scholar]
- Toga A.W, Thompson P.M. Maps of the brain. New Anat. 2001;265:37–53. doi: 10.1002/ar.1057. [DOI] [PubMed] [Google Scholar]
- Ungerleider L.G, Desimone R. Projections to the superior temporal sulcus from the central and peripheral field representations of V1 and V2. J. Comp. Neurol. 1986;248:147–163. doi: 10.1002/cne.902480202. [DOI] [PubMed] [Google Scholar]
- Ungerleider L.G, Mishkin M. The striate projection zone in the superior temporal sulcus of Macaca mulatta: location and topographic organization. J. Comp. Neurol. 1979;188:347–366. doi: 10.1002/cne.901880302. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C. Surface-based approaches to spatial localization and registration in primate cerebral cortex. NeuroImage. 2004;23:S97–S107. doi: 10.1016/j.neuroimage.2004.07.024. doi:10.1016/j.neuroimage.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Van Essen D.C, Maunsell J.H, Bixby J.L. The middle temporal visual area in the macaque: myeloarchitecture, connections, functional properties and topographic organization. J. Comp. Neurol. 1981;199:293–326. doi: 10.1002/cne.901990302. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C, Drury H.A, Dickson J, Harwell J, Hanlon D, Anderson C.H. An integrated software suite for surface-based analyses of cerebral cortex. J. Am. Med. Inform. Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C, Lewis J.W, Drury H.A, Hadjikhani N, Tootell R.B.H, Bakircioglu M, Miller M.I. Mapping visual cortex in monkeys and humans using surface-based atlases. Vision Res. 2001;41:1359–1378. doi: 10.1016/s0042-6989(01)00045-1. [DOI] [PubMed] [Google Scholar]
- Van Horn J.D, Grethe J.S, Kostelec P, Woodward J.B, Aslam J.A, Rus D, Rockmore D, Gazzaniga M.S. The functional magnetic resonance imaging data center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Phil. Trans. R. Soc. B . 2001;356:1323–1339. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt O. Zur anatomischen Gliederung des cortex cerebri. J. Psychol. Neurol. 1903;2:160–180. [Google Scholar]
- Vogt C, Vogt O. Ergebnisse unserer hirnforschung. 1.-4. Mitteilung. J. Psychol. Neurol. 1919;25:279–461. [Google Scholar]
- von Bonin G, Bailey P. University of Illinois Press; Urbana: 1947. The neocortex of Macaca mulatta. [Google Scholar]
- Walker E.A. A cytoarchitectural study of the prefrontal area of macaque monkey. J. Comp. Neurol. 1940;73:59–86. [Google Scholar]
- Wanke E, Kötter R. Oriented paths in mixed graphs. In: Fleischer R, Trippen G, editors. Proceedings of the 15th International Symposium on Algorithms and Computation (ISAAC). Lecture notes in computer science. Springer; Berlin: 2004. 2004. pp. 17–58. [Google Scholar]
- Whitmore I. Thieme; Stuttgart: 1998. Terminologia anatomica—international anatomical terminology, federative committee of anatomical terminology (FCAT) [Google Scholar]
- Yeterian E.H, Pandya D.N. Prefrontostriatal connections in relation to cortical architectonic organization in rhesus monkeys. J. Comp. Neurol. 1991;312:43–67. doi: 10.1002/cne.903120105. [DOI] [PubMed] [Google Scholar]
- Young M.P. The organization of neural systems in the primate cerebral cortex. Proc. R. Soc. B. 1993;252:13–18. doi: 10.1098/rspb.1993.0040. [DOI] [PubMed] [Google Scholar]
- Zilles K. Architecture of the human cerebral cortex. Regional and laminar organization. In: Paxinos G, Mai J.K, editors. The human nervous system. 2nd edn. Elsevier; San Diego, CA: 2004. pp. 997–1055. [Google Scholar]