Abstract
Quantum chemical methods now permit the prediction of many spectroscopic observables in proteins and related model systems, in addition to electrostatic properties, which are found to be in excellent accord with those determined from experiment. I discuss the developments over the past decade in these areas, including predictions of nuclear magnetic resonance chemical shifts, chemical shielding tensors, scalar couplings and hyperfine (contact) shifts, the isomer shifts and quadrupole splittings in Mössbauer spectroscopy, molecular energies and conformations, as well as a range of electrostatic properties, such as charge densities, the curvatures, Laplacians and Hessians of the charge density, electrostatic potentials, electric field gradients and electrostatic field effects. The availability of structure/spectroscopic correlations from quantum chemistry provides a basis for using numerous spectroscopic observables in determining aspects of protein structure, in determining electrostatic properties which are not readily accessible from experiment, as well as giving additional confidence in the use of these techniques to investigate questions about chemical bonding and chemical reactions.
Keywords: quantum mechanics, protein structure, protein electrostatics, nuclear magnetic resonance spectroscopy, Mössbauer spectroscopy, chemical shifts
1. Introduction
Quantum chemistry is a branch of chemistry involved with determining, computationally, the three-dimensional and electronic structures of molecules. In general terms, the basic problem is to solve the Schrödinger equation, Hψ=Eψ, where H is the Hamiltonian operator, ψ is the wave function and E is the energy. Many spectroscopic properties (such as nuclear magnetic resonance (NMR) chemical shifts and scalar couplings) are formal derivatives of the energy, whereas others (such as charge densities) are more directly related to ψ. Methods to evaluate E were first developed in the 1920s by Heisenberg, for the singlet and triplet states of the He atom (Heisenberg 1926), by Heitler & London (1927) for the H2 molecule and by Pauling (1928a) for the hydrogen molecule ion (H2+). The correct wave functions were found to correspond to sums of functions (resonance) since they yielded the lowest energies, and in the case of, for example, H2, this led to stable bond formation. The idea of resonance (hybridization) was then further developed by Pauling to explain the equivalence of the 2s and three 2p orbitals in methane (Pauling 1928b); then in 1932, Pauling described the double-bond character of amide groups, a key feature of protein structure, owing to resonance (Pauling 1932). Of course, at this time, the nature of protein ‘particles’ was quite obscure (Svedberg et al. 1939), but the observation of Bernal & Crowfoot (1934) of diffraction from a pepsin crystal helped to remove misconceptions about their colloidal nature, and giant molecules were then believed to exist. But what were their structures?
2. The early days
In the early 1950s, Pauling again used the theory of quantum mechanical (QM) resonance to develop structural models for the α-helix and parallel and anti-parallel pleated sheets in proteins, in which the planar nature of the peptide bond (and the electrostatic interactions between these structures) were essential ingredients (Pauling & Corey 1950, 1951), but still there were no high-resolution structures of proteins.
Shortly after these proposals were published, Bernal visited Madras and the interests of a young G. N. Ramachandran were shifted from crystal physics to protein structure determination (Ramakrishnan 2001). With G. Kartha, a proposal for the structure of collagen, a triple helix, was published (Ramachandran & Kartha 1954, 1955), the details of which were objected to on the basis that there were too many steric hindrances (Rich & Crick 1955). This led Sasisekharan and Ramachandran to investigate this topic in more detail. With knowledge of Pauling's postulate of the planar nature of the peptide bond, due to QM resonance, Ramachandran began the first computational chemical studies of protein structure and stability—albeit armed with only a desk calculator (Ramakrishnan 2001). The peptide models used contained just two amino acids, but given the trans nature of the central peptide bond, could be well described by just two torsion angles, ϕi and ψi−1. All atoms were described as rigid spheres with radii corresponding to their van der Waals radii and only hardcore repulsive interactions were considered. Nevertheless, the Ramachandran maps thus obtained clearly indicated regions of stability in ϕ, ψ space (Ramachandran et al. 1963) and successfully predicted that two non-helical residues in myoglobin must be glycines, which proved to be the case (Ramakrishnan 2001). With further improvements in the development of potential energy functions, such as the Buckingham and Lennard–Jones potentials, the accuracy of such Ramachandran maps improved, although these potentials were still of an empirical nature.
In these early days, it was not possible to carry out electronic QM structure calculations on molecules as large as peptides or even amino acids. The speed of an electronic structure calculation strongly depends on the number of atoms in the system, or more precisely, the number of functions, basis functions, which are used to describe the atomic orbitals that make up the molecular orbitals in the molecule. For Hartree–Fock self-consistent field (SCF) calculations, the length of a calculation varies as ca N3.5, where N is the number of basis functions, while for higher-level calculations the length of a calculation can be as high as ca N5–N6. However, as computer speeds have increased, such calculations are now becoming tractable. In addition, recent developments in an alternative approach to electronic structure calculations, density functional theory (DFT), are somewhat faster, scaling as ca N3 (Kohn et al. 1996). Armed with these tools, it is now a relatively straightforward matter to evaluate Ramachandran maps for energies using first principles methods, and as discussed below, many other properties of interest can now also be predicted. The results of energy calculations are particularly important since they can then be used to parameterize semi-empirical (molecular mechanics (MM)) approaches to structure prediction and refinement, and such derived ‘force fields’ are now commonly used in X-ray crystallographic and NMR structural investigations. Moreover, such MM methods can be combined with QM methods to investigate very large systems, such as enzymes (Gogonea et al. 2001; Gao & Truhlar 2002). But just how large a system can be investigated by using purely quantum chemical methods? And what can we learn? Is this approach important? Cannot we now just measure everything of interest?
3. The energy
Evaluating the total eigenenergy of a molecule is the first critically important step to evaluating a wide range of properties, many of which (such as the charge density ρ(r), the charge density at the nucleus ρ(0) and the electrostatic potential Φ(r)) cannot be readily determined experimentally. Likewise, many spectroscopic properties, such as NMR chemical shifts, NMR hyperfine shifts, Mössbauer isomer shifts and quadrupole splittings, quadrupole couplings in NMR and nuclear quadrupole resonance (NQR), can be readily measured, but without quantum chemistry they cannot be interpreted in much detail. In addition, of course, there are no direct experimental measurements of molecular orbitals or electronic structure; rather, they have to be deduced from an electronic wave function whose quality has to be assessed by its ability to successfully predict a series of observables. By way of example, I consider first the energy of a protein molecule and how this is related to observable structural properties.
In its simplest embodiment, the energy of a molecule having a given structure can be computed by using variational or SCF techniques in which coefficients describing the composition of various atomic orbitals are varied—the linear combination of atomic orbitals method, such that the lowest eigenenergy for the system is obtained (Hehre et al. 1986). From such an electronic structure calculation, it is then possible to deduce properties of interest, such as those outlined above. In most cases, it is not necessary to carry out a calculation on an entire protein, since the property of interest is essentially a local one, such as an NMR chemical shift. In other cases, however, this is not the case. The most important and most obvious example of this exception is that of the structure of the protein itself.
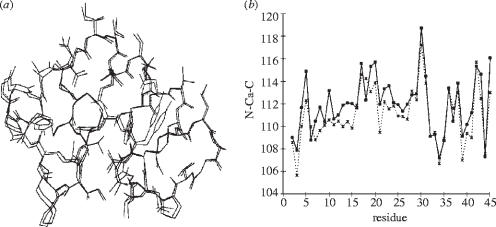
In recent work, it has been shown to be possible to carry out electronic structure calculations on entire proteins by using ab initio techniques. The results of both single-point (single-geometry) energy calculations on the proteins endothelin, charybdotoxin and p53, and of ab initio Hartree–Fock geometry optimization calculations on crambin, a small hydrophobic protein of 46 residues, have recently been reported (Challacombe & Schwegler 1997; Van Alsenoy et al. 1998). The crambin study used a multiplicative integral approximation method, which when combined with the direct SCF approach results in linear scaling with system size in building the Fock matrix, enabling the investigation of very large systems in which many body interactions are taken into account in a direct fashion (Van Alsenoy et al. 1998). This approach is expected to be more accurate than using force fields based on the parameterization of two-body interactions, and should lead to direct evaluation of many electrostatic and energy derivative properties, which can be evaluated from the computed wave functions. In the case of crambin, 79 cycles of geometry optimization were carried out, and the resulting structure was found to be remarkably close to that obtained crystallographically, as shown in figure 1a. The results of these theoretical calculations revealed basically the same deviations from planarity of the peptide groups as those seen crystallographically, even in the absence of correlation (instantaneous electron–electron) effects, and the N–C(α)–C′ three atom backbone bound angles were also found to be highly correlated with the crystallographic values—corresponding to only a 1.5° r.m.s. deviation, as shown in figure 1b. Based on these results, it seems likely that quantum chemical geometry optimization will play a significant role in structure refinement in NMR and in crystallography in the not too distant future.
Figure 1.
Comparison between experimental and ab initio computational results on crambin. (a) Superposition of the crystallographic (pdblcnr) and HF/4-21G optimized heavy-atom framework of crambin. (b) Comparison of crystallographic (solid line; pdblcnr) and HF4-21G optimized N–C(α)–C′ angles (dotted line) in crambin. Reprinted in part with permission from Van Alsenoy et al. (1998). Copyright 1998 American Chemical Society.
4. Electrostatic properties
Energy is not a measurable quantity, although energy differences and derivative properties are. Likewise, electrostatic properties, such as charges (or more specifically, charge densities), electrostatic potentials, electrostatic fields and electric field gradients (EFGs) are not direct observables, although many such properties can be deduced from experimental (NMR, diffraction) results, at least for small molecule systems. If these electrostatic properties can be successfully predicted for systems such as amino acids and peptides, it seems reasonable to suppose that similar calculations on proteins will also yield accurate results.
The electric field, E, is a vector property and is defined as
where ϕ is the electrostatic potential, and from Gauss's Law, the divergence of the vector E is given by the scalar ρ, the charge density, as
where ϵ0 is the permittivity of free space, and it follows that
where ∇2ϕ is the Laplacian of the potential. The EFG, V, is defined as
and the gradient of the vector is a tensor of rank two. To evaluate the electric potential, ϕ, more commonly called the molecular electrostatic potential, an SCF calculation is first carried out to determine the wave function ψ, with the electron probability density or the charge density then being evaluated from |ψ|2. The molecular electrostatic potential at a point can then be obtained by integrating over the charge density. Once the potential is known, electric fields (−∇ϕ) and field gradient tensors (∇E) can be computed. With this brief introduction, we next ask the questions: are these methods applicable to amino acids, peptides and proteins? Are the methods accurate? And what can we learn about protein structure?
At present, there have been few reports of ab initio calculations of ρ(r) or Φ(r) values in complete proteins. But for smaller systems (amino acids, peptides and some protein prosthetic groups), numerous reports of such calculations have been published. But how does one know that these calculated results are accurate? The answer of course is that in each case the computed properties need to be rigorously compared with the results of experiments in which these properties are either directly measured, or where the properties of interest are related in a known way to the experimental property.
Arguably, the simplest property to extract from a computed wave function is the charge density, ρ(r), since this is proportional to |ψ|2, the probability of finding an electron at a given position. This approach can be used to compute three-dimensional plots of charge density isosurfaces, which can then be compared with ρ(r) surfaces extracted from X-ray diffraction data, since the scattering of an X-ray beam is a function of the electron distribution. Likewise, the electrostatic potential can be evaluated from the quantum chemical results by integrating over the charge density
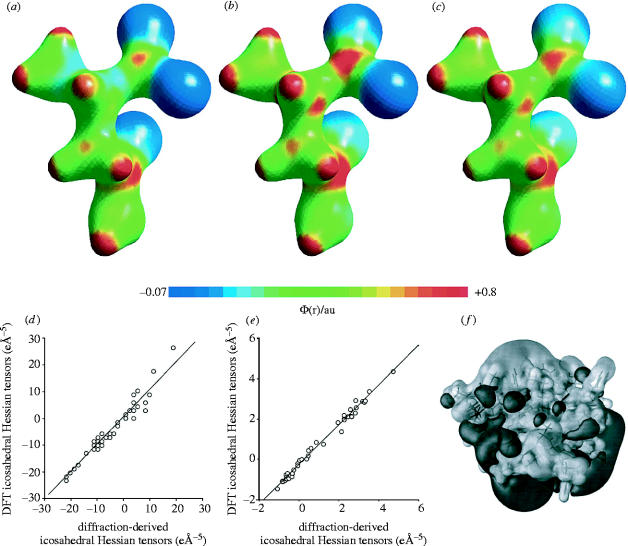
where ρ2 is the charge density, dV2 is the volume element at point 2, and r12 is the distance between points 1 and 2, resulting in electrostatic potential surfaces Φ(r), which can again be compared with results extracted from diffraction data as shown, for example, in figure 2a–c, for l-asparagine·H2O. However, these graphical comparisons are not satisfactory for making rigorous comparisons between computed and experimentally derived properties. One solution to this problem is to use the atoms-in-molecules (AIM) theory method developed by Bader (1990). This method uses the idea that associated with every chemical bond is a point, rb, at which the first derivative of the charge density, ρ(r), is zero. At this so-called bond critical point (BCP) there are three non-zero principal curvatures in ρ(rb), two negative and one positive. The curvature in the charge density at a BCP is described by a real symmetric tensor known as the Hessian-of-ρ(r), H, which has nine elements of the form ∂2ρ/∂rirj. When diagonalized, H is expressed by three elements, λ1–3, which correspond to curvatures in ρ(rb) along three principal axes. The Laplacian of the charge or electron density at a BCP, ∇2ρ(rb), is simply the sum of these three principal curvatures. In recent work, it has been found that there is excellent agreement between λi and ∇2ρ(r) values determined via quantum chemistry and those derived from X-ray diffraction data, for a series of amino acids and peptides (Dittrich et al. 2000; Arnold et al. 2000a; Li et al. 2002). However, the principal curvatures in ρ(r) represent only the magnitudes of the Hessian tensors, H, not their orientations. Tensors are commonly expressed in a principal axis system (PAS) as three principal values and the corresponding direction cosines with respect to a molecular reference frame. Thus, to compare the orientations of such tensors, one must compare the angles between each PAS and the molecular reference frame, and experimental errors in individual Cartesian tensor elements must be indirectly translated into errors in degrees or radians. A comparison of tensors in an icosahedral representation, however, is much more convenient, since both the magnitudes and the orientations of the tensors are quantitatively evaluated at the same time by comparing the six icosahedral tensor elements, χ1–6. Also, the six icosahedral elements are equally weighted in any coordinate frame, making this representation ideal for fitting a least-squares line through theoretical-versus-experimental tensor data, an approach developed previously by Alderman and colleagues, for investigating chemical shielding tensors (Alderman et al. 1993). Using this approach, there has been shown to be very good agreement between theory and experiment for both the magnitudes and the orientations of these Hessian tensors. For example, again investigating l-asparagine·H2O, a slope of 1.09 and R2=0.96 for a comparison between the Hessian-of-ρ(r) tensors in the icosahedral representation at all heavy atom BCPs has been demonstrated (Arnold et al. 2000a; figure 2d). Even better accord is found at hydrogen bond BCPs (Arnold et al. 2000a; figure 2e). These results show that ρ(r), λi, ∇2ρ(r) and Hessian-of-ρ(r) properties can now all be accurately predicted using quantum chemical techniques. This helps validate their use on proteins, such as p53 and charybdotoxin (Challacombe & Schwegler 1997), and an interesting example of this approach is shown in figure 2f for Φ(r) for charybdotoxin in which the positive Φ(r) surface thought to be responsible for binding to the K+-channel can clearly be seen (on top, light grey).
Figure 2.
Electrostatics results for asparagine and charybdotoxin. (a–c) Molecular electrostatic potentials for asparagine, Φ(r), mapped from −0.07 to +0.8 e/4πe0a0, onto a 0.11 eÅ−3 isosurface of the electron density. (1 Å=1×10−10 m.) (a) X-ray diffraction. (b) HF/6-311++G(3df,3pd) calculation. (c) B3LYP/6-311++G(3df,3pd) calculation. (d) Experimental (X-ray) versus theoretical icosahedral Hessian-of-ρ(rb) tensors for asparagine at BCPs. (e) As (d) but at H-BCPs. (f) Electrostatic potential surface for charybdotoxin. The light surface corresponds to a value of 0.0035 a.u., the dark surface −0.0035 a.u. Reprinted from Challacombe & Schwegler (1997) with permission. Reprinted in part with permission from Arnold et al. (2000a). Copyright 2000 American Chemical Society.
5. Mössbauer spectroscopy and electrostatics
How else might the results of quantum chemical calculations of charge densities be verified? One possibility is to investigate spectroscopic observables that are directly related to the charge density. The so-called isomer shift in Mössbauer spectroscopy is one such example. Mössbauer spectroscopy is a technique widely used to investigate the structures of metalloproteins, metalloporphyrins and other model systems, and is a potentially powerful tool with which to deduce both geometric and electronic structures. 57Fe Mössbauer spectra have frequently been studied in proteins, and are typically dominated by two interactions: the quadrupole splitting, ΔEQ, which arises from the non-spherical nuclear charge distribution in the I*=3/2 excited state in the presence of an EFG at the 57Fe nucleus; and an isomer (or chemical) shift, δFe, which arises from differences in the charge density at the nucleus between the absorber (A, the molecule or system of interest) and a reference compound (usually α-Fe at 300 K). This interaction is given by
| (5.1) |
where Z represents the atomic number of the nucleus of interest (iron) and R, R* are average nuclear radii of the ground and excited states of 57Fe. Since is a constant, the isomer shift (from Fe) can be written as
| (5.2) |
where α is the so-called calibration constant and ρtot(0) is the computed charge density at the nucleus. In recent work, quantum chemical methods have been used to compute ρ(0) charge density values for a wide variety of haem proteins and model systems, and it has been found that the R2 values between experimental and computed δFe values are typically greater than 0.98, and r.m.s. errors between experimental and theoretical isomer shifts are only ca 3–4% (Zhang et al. 2002a). These results are interesting since they mean that even charge densities at the nucleus can now be quite well described by using quantum chemistry. Moreover, the Mössbauer isomer shift can begin to be used in structure determination, since it is found to be highly sensitive to local geometry.
The other Mössbauer observable is the quadrupole splitting, ΔEQ, which is related to the components of the EFG tensor (Vxx, Vyy, Vzz) at the nucleus, another electrostatic property, as follows:
where e is the electron charge, Q is the quadrupole moment of the nuclear excited state, and the principal components of the EFG tensor are labelled according to the convention: with the asymmetry parameter, η, being given by
That is, the Mössbauer quadrupole splitting is another direct spectroscopic probe of electrostatics, since it is directly related to the gradient of the electric field vector, E. Once again, the availability of a good wave function enables an immediate evaluation of all nine components of the EFG tensor, together with their orientations in a molecular axis system (Zhang et al. 2002b). The tensor is necessarily symmetric and traceless (Laplace equation) so only three elements (and their orientations) are needed to fully describe the tensor. As might now be anticipated, there is found to be excellent accord between theory and experiment for ΔEQ in a wide range of haem proteins and model systems when using an appropriate level of theory and large basis sets (typically, ca 500–1000 basis functions, for a haem model). For example, a theory versus experiment correlation having an R2 of ca 0.98, a slope of ca 1.12 and an error of ca 0.26 mm s−1, when considering 23 different systems covering a 5.63 mm s−1 range in ΔEQ is obtained, corresponding to a 4–5% overall error (Zhang et al. 2002b). Thus, 57Fe Mössbauer quadrupole splittings in proteins and related systems can now all be predicted quite accurately and provide another important test of quantum chemical methods in predicting ‘electrostatic’ properties.
6. EFGs and NMR spectroscopy
It should also be noted that the EFG parameter is not restricted to Mössbauer spectroscopy. All nuclei in non-spherically symmetric environments will have an associated EFG, and if the nucleus of interest has a non-zero quadrupole moment, then there will be an interaction, of strength (eq)(eQ)/h, where eq=Vzz and eQ is the quadrupole moment of the nucleus. In magnetic resonance spectroscopy, if e2qQ/h is small compared with the nuclear Zeeman effect, there are typically perturbations of the NMR line shape, from which EFG tensor information can be determined, while for large e2qQ/h values, transitions between quadrupolar levels can often be observed directly, by means of pure NQR. These nuclear quadrupole interactions are generally difficult to measure in proteins, although they have been determined for 2H (LiWang & Bax 1997) and 17O (Park et al. 1991), by NMR. But in amino acid or peptide crystals, both tensor magnitudes and orientations have been measured experimentally and provide another important test of the accuracy of quantum chemical calculations. As an example, the 14N EFG tensors in l-asparagine have been determined by NMR on a single crystal sample (Naito & McDowell 1984). Such single crystal studies yield both the magnitudes and the orientations of the 14N EFG for both the amide and ammonium nitrogens, and both properties have been calculated by using DFT (Arnold et al. 2000a). Since the EFG is a tensor of rank two, the three principal EFG tensor components for each site, together with their orientations (direction cosines), can be combined using the icosahedral representation to yield six icosahedral tensor elements for each site: a total of 12 observables. When the experimental results were compared with the results of large basis DFT calculations, they were found to be highly correlated (R2=0.94) with a slope of 1.14 (Arnold et al. 2000a). Thus, both Mössbauer and NMR EFG observables can now be accurately predicted by using quantum chemistry, which suggests that in the future these properties may be used in protein structure determination, since again they are very sensitive to local geometry, as described below in more detail.
7. Spin densities and hyperfine shifts
In what we have discussed so far, we have not explicitly considered the case of paramagnetic species—molecules containing unpaired electron spins. For paramagnetic systems, quantum chemical methods typically describe these molecules as having what are essentially two subsets of interacting electrons and nuclei, each set having electrons with opposite spins (Hehre et al. 1986). The molecular orbitals obtained have either spin up (α) or spin down (β) electrons and consequently two different sets of charge densities, ρ(α) and ρ(β), are obtained. Their sum gives the overall charge density ρ(r) discussed above, but their difference gives a new property, the so-called spin density
The spin density is an important property since, in NMR, the Fermi contact or hyperfine shift δFC is directly proportional to ραβ:
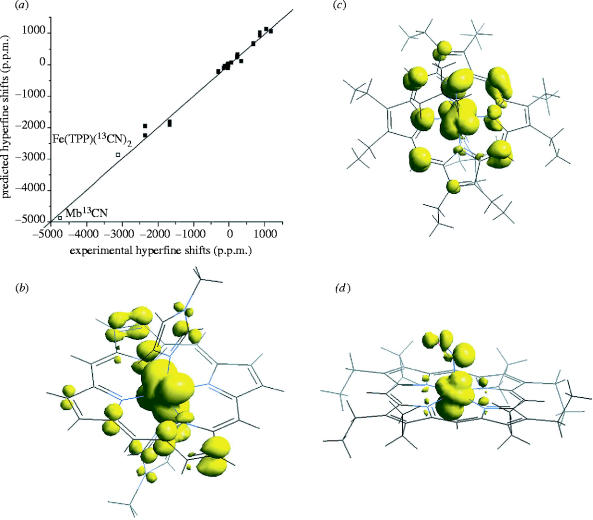
where μ0 and μβ are the nuclear and electron moments, ge is the Landé factor and S is the spin quantum number. The Fermi contact interaction is also of importance in mediating scalar or J-couplings in NMR, as well as hyperfine couplings in electron paramagnetic resonance (EPR). The Fermi contact shift in NMR can be very large—up to ca 6000 p.p.m. for 13C, and represents another major challenge for theory to reproduce. In early work, 2H and 15N shifts of amino acid residues bound to a FeS4 cluster in the protein rubredoxin, covering a ca 800 p.p.m. range of ‘chemical’ shifts, were quite accurately reproduced by using DFT methods (Wilkens et al. 1998). More recently, the entire ca 6000 p.p.m. range of hyperfine (contact) shifts seen in a variety of haem proteins and model systems, having S=1/2, 2 and 5/2, has been successfully reproduced (Mao et al. 2002). The R2 values were again ca 0.99 (N=39 compounds; p<0.0001) with a standard deviation of ca 100 p.p.m. over the 6000 p.p.m. total shift range, as shown in figure 3a. These results give considerable confidence in the use of quantum chemical methods to predict spectroscopic observables, even in paramagnetic proteins, and make it possible to begin to use NMR contact shifts in protein structure determination. In addition, they give added confidence in the use of the MO results to describe details of structure and bonding. For example, the spin density in a ferric myoglobin model is found to have the distribution shown in figure 3b, in sharp contrast to that found in the more unusual Fe(III) species, shown in figure 3c (Mao et al. 2002), while in an NO–haem system the unpaired electron is localized primarily in a dz2 orbital (figure 3d). These representations also help visualize the associated contact shifts, which depend on ραβ.
Figure 3.
Spin density and hyperfine shifts in heme model systems and proteins. (a) Graph showing experimental NMR hyperfine shifts over the entire ca 6000 p.p.m. range seen in proteins and model systems plotted as a function of the hyperfine shifts computed by using the DFT/B3LYP method. Open squares represent the highly shifted resonances in a 13CN-labelled metalloporphyrin and [13CN]− sperm whale cyanometmyoglobin. The line has a slope of 0.99 and an R2 value of 0.990. (b–d) Total spin density isosurfaces for: (b) [Fe(TMP)(1-MeIm)2]+; (c) [Fe(OEP)(t-BuNC)2]+; and (d) Fe(OEP)(NO). Reprinted in part with permission from Mao et al. (2002) (a) and Zhang et al. (2002b) (b–d). Copyright 2002 American Chemical Society.
8. Derivative properties
So far, discussion has focused on what might conveniently be thought of as electrostatic properties: ρ(r), λi, ∇2ρ(r), the Hessian-of-ρ(r), Φ(r), ∇E, ΔEQ, δFe and ραβ, since they are all related to |ψ|2 or the charge density. There are, however, a second set of properties that are, formally, energy derivatives:
where μ is a nuclear magnetic moment, B is an external magnetic field and F is an external electric field component. These, and other higher-order terms (hyper-polarizabilities), can all be evaluated by using quantum chemical methods and in most cases commercial software packages are available to enable the necessary calculations. Again, the question arises as to how accurate the results are. Also, can we apply these methods to proteins—which at present, for practical purposes, means: are the interactions of short enough range that small molecular fragments can be used to calculate the observables that are measured experimentally?
9. Scalar or J-couplings in NMR spectroscopy
The scalar or J-coupling property is perhaps the most well known since it has been investigated since the 1960s, where J-coupling calculations were pioneered by Karplus (1996). With the development of two-dimensional NMR techniques in the 1980s, a large body of experimental data on J-couplings in proteins became available, and this prompted the use of quantum chemical methods to predict J-couplings in proteins. These calculations involve the evaluation of diamagnetic and paramagnetic spin–orbit contributions to J, in addition to paramagnetic Fermi contact and spin–dipole contributions (Helgaker et al. 2000). The use of DFT methods with hybrid exchange correlation functionals has met with considerable success, and it has been possible to accurately reproduce all six 3J couplings, which involve the ψ backbone torsion angle. Moreover, there was an improvement in the quality of the predictions when the effects of molecular motion were taken into account, by using ‘snapshots’ from a molecular dynamics trajectory, on the protein ubiquitin (Case et al. 2000).
In addition to these ‘classical’ J couplings, associated with protein backbone torsion angles, there has also recently been considerable interest in using quantum chemical methods to investigate hydrogen-bond scalar couplings (Cornilescu et al. 1999), focusing on answering the question: what is the nature of such hydrogen bond scalar couplings? Do they represent covalent bonding? One way to investigate these questions is to use AIM theory and to investigate the topology of ρ(r) at the hydrogen BCP. The topology of ρ(r) at a BCP is described by the real, symmetric, second-rank Hessian-of-ρ(r) tensor, and the trace of this tensor is related to the bond interaction energy by a local expression of the virial theorem (Bader 1990):
where G(r) is the kinetic energy density and V(r) is the potential energy density (Bader 1990). Since G(r) must always be positive and V(r) must always be negative in stable, bound, stationary states, the sign of ∇2ρ(r) at a BCP is determined by which energy density is in excess over the virial average of 2 : 1 kinetic-to-potential energy. A negative Laplacian reveals excess potential energy at the BCP, meaning that electronic charge is concentrated into a bond. This is the case in all shared-electron (covalent) interactions. A positive BCP Laplacian reflects an excess of kinetic energy in a bond, and a relative depletion of electronic charge along a bond path. This is the case in all closed-shell (electrostatic) interactions. For every backbone hydrogen bond examined, ∇2ρ(r) was found to be positive and characteristic of a closed-shell or electrostatic interaction (Arnold & Oldfield 2000). Although each term increased exponentially as |3hJNC′| increased, the kinetic term was a slightly stronger exponential, and outpaced the increase in the potential term. That is, the net amount of excess kinetic energy density was greatest for the largest couplings. This could not be the case if such couplings were dependent upon, or indicative of, covalent character in the bonds.
Although the kinetic energy term provides the dominant virial contribution, resulting in a net closed-shell, electrostatic interaction, it is also necessary to consider whether these interactions represent a closed-shell limit by evaluating the total energy density, H(r), at the BCP (Cremer & Kraka 1984)
Bonds with any degree of covalent character (any amount of potential energy stabilization resulting from the accumulation of charge in the internuclear region) must have a BCP H(r) that is less than zero. Quantum chemical results for the PGB1 protein backbone hydrogen bonds provided no evidence of partial covalent character, H(r)>0 in all cases (Arnold & Oldfield 2000). Backbone hydrogen bonds thus appear to represent purely closed-shell, electrostatic interactions.
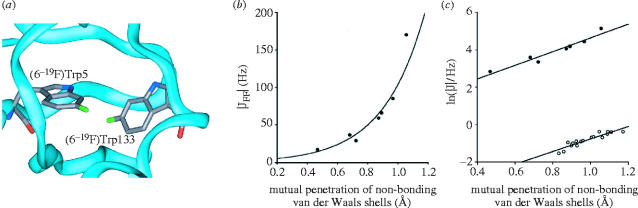
If 3hJNC′, does not arise from a partial covalent interaction, then what permits the intermolecular communication between nuclear spins? The theoretical results indicated that the magnitude of 3hJNC′, was clearly related to the mutual penetration of non-bonding van der Waals shells; however, there was no concentration of charge in the bonding region upon hydrogen bond formation. Thus, the electron density in the hydrogen bond is simply the sum of isolated donor and acceptor densities: the greater the van der Waals penetration, the greater the resulting summed electron density and the greater the experimentally observed J-coupling. This description of 3hJNC′ also provides a simple physical explanation for the existence of scalar couplings between nuclei that cannot possibly be covalently bonded to one another. For example, Kimber et al. (1978) observed a field-independent 19F–19F coupling of 17±2 Hz in a (dihydrofolate reductase) DHFR–NADPH–methotrexate complex which had been prepared biosynthetically with 6-fluorotryptophan residues. With no crystal structure of DHFR available, these researchers postulated that two out of the four 6-fluorotryptophan residues must be situated such that their 19F nuclei are ca 3 Å apart, based upon much earlier empirical observations of ‘through space’ 19F–19F J-couplings in small organic molecules (Kimber et al. 1978). Subsequently, it was found that the folded protein brings the 6-positions of Trp5 and Trp133 into extremely close proximity (figure 4a), and it was shown that sum-over-states (SOS)–DFT (Malkin et al. 1994) predicts a 19F–19F coupling of ca 30 Hz between two (model) fluoromethane molecules at this separation (Arnold et al. 2000b). An AIM atomic interaction line was shown to exist between the two 19F nuclei, and the non-bonding van der Waals shells of the monomers penetrate one another by Δr=0.47Å along this line. Long-range (>3JFF), intramolecular 19F–19F through-space J-couplings in a wide range of systems are also well described by SOS–DFT calculations, and small non-bonded dimer models such as (CH3F)2, give very similar results to covalently bonded models (Arnold et al. 2000b). The extent of the van der Waals penetration in several more fluoromethane dimers, whose F–C geometries were extracted from larger organic molecules possessing such long-range JFF couplings was then investigated, since it is well-established that the magnitudes of both JFF and 3hJNC′ increase exponentially with decreasing internuclear separation (Cornilescu et al. 1999; Mallory et al. 2000), and figure 4b,c shows that these magnitudes are related to the mutual penetration of non-bonding van der Waals shells in the same manner. Both are of the exponential form , where B is 3.36 for 3hJNC′ and 3.67 for JFF. The fact that In[|J|] versus Σ Δr has a very similar slope for both types of couplings strongly suggests that the through-space JFF coupling and the hydrogen bond 3hJNC′ coupling are subject to the same inductive mechanism, and require neither an attractive electrostatic bond nor a covalent bond, only that two atomic surfaces contact one another in the intervening space between coupled nuclei. Thus, both through bond as well as through space J-couplings can now be calculated using quantum chemical methods, with best results being obtained when dynamics is incorporated into the calculations (Case et al. 2000; Markwick et al. 2003).
Figure 4.
Through space J-couplings. (a) Structure of a [6-19F]Trp-labelled dihydrofolate reductase complex showing close approach between Trp6 and Trp133. (b) The exponential dependence of the magnitude of through-space 19F–19F J-couplings upon the mutual penetration of non-bonding van der Waals shells, Σ Δr. , R2=0.94. (c) ln(|J|) versus Σ Δr. Filled circles, JFF; open circles, 3hJNC′. Reprinted in part with permission from Arnold & Oldfield (2000) and Arnold et al. (2000b). Copyright 2000 American Chemical Society.
10. The chemical shift in NMR spectroscopy
In addition to these results, there has been much progress in predicting NMR chemical shifts in proteins. When a protein folds, there is a large range of chemical shift non-equivalence generated. These effects were first noted in the 1960s for 1H (Dwek 1973) and in the 1970s for 19F (Sykes & Weiner 1980) and 13C NMR (Allerhand et al. 1973). For the heavier elements (C, N, F), the effects can be very large—up to ca 10 p.p.m. for 13C, 35 p.p.m. for 15N, and ca 20 p.p.m. for 19F. However, the successful prediction of folding-induced chemical shifts has been reported only during the past decade. In early experimental work, it was found that 13Cα chemical shifts in helical versus random coil polypeptides were different, with the helical group resonating ca 3 p.p.m. downfield from the random coil shift (Allerhand & Oldfield 1973), but only with the development of 2D NMR techniques could clear patterns of Cα and Cβ helix-sheet differences be seen (Spera & Bax 1991), which set the stage for ab initio calculations of these properties, since they clearly appeared to be dependent on ϕ, ψ. There were, however, numerous questions or concerns at the outset as to whether such calculations would be possible, since it was unclear whether solvation, dynamics and electrostatics would also need to be incorporated into a calculation or set of calculations, to reproduce the experimental observations. By contrast, the observation that helices and sheets had generally well-separated chemical shifts and the observation that crystal and solution shifts were similar (Cole et al. 1988) made it unlikely that such effects would dominate the observed shifts.
In early test calculations, the 13C NMR chemical shifts in crystals of two amino acids: l-threonine and l-tyrosine were investigated (de Dios et al. 1994). The availability of tensor magnitude information from these crystalline samples provided an important test of what might be considered a ‘basic’ test of chemical shielding tensor predictions in model systems. It was found that both isotropic shifts as well as tensor magnitudes could be accurately predicted (de Dios et al. 1994) by using Hartree–Fock theory combined with the gauge including atomic orbitals (GIAO) technique (Ditchfield 1974). And in the case of l-threonine, where single crystal results were available, the tensor orientations could also be successfully predicted. An exception was found with the carboxyl tensors, but this could be corrected by including local electrostatic field effects using a lattice of point charges, the charge field perturbation (CFP)–GIAO approach (de Dios et al. 1994).
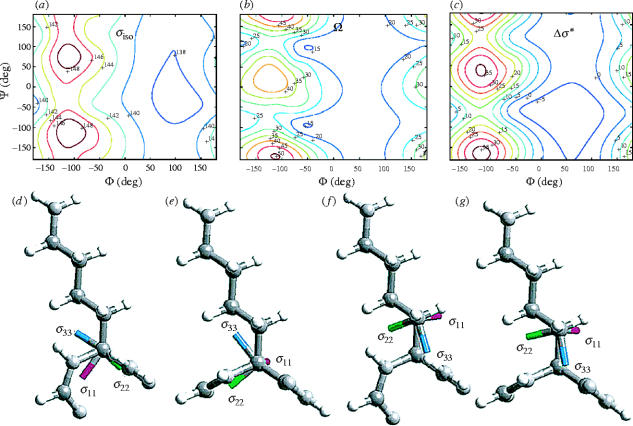
Hartree–Fock methods were then also used to predict 13C NMR chemical shifts, for Cα and Cβ, in proteins. An important first step here was to investigate how large a fragment was needed to predict these 13C shifts, since clearly the ϕ, ψ effects of neighbouring residues would need to be incorporated, based on the empirical observation that Cα, Cβ shifts were a function of these backbone torsion angles. After some test calculations, N-formyl-amino acid amides were selected, using a locally dense basis approach in which a larger number of basis functions is given to the atom of interest (Chesnut & Moore 1989). The coordinates of the atoms were based on protein X-ray crystallographic data. The first sets of calculations were uniform failures. Fortunately, it was pointed out by C. Jameson (private communication) that there might be significant errors in the crystallographic bond lengths and bond angles used, so the use of geometry optimization (using an EFG force field) was next investigated, to ‘regularize’ bond lengths and three-atom bond angles. This approach was successful, and using even these relatively small fragments and neglecting charge field and hydrogen bonding effects, it was found to be possible to predict 13Cα, 13Cβ NMR chemical shifts in a staphylococcal nuclease (SNase) protein with an r.m.s. error of ca 1 p.p.m. (de Dios et al. 1993). The same techniques were also found to permit 15N NMR chemical shift predictions in SNase, using a somewhat larger peptide fragment to take into account the conformational effects of the immediately preceding (i−1) residues which, based on empirical observations (Le & Oldfield 1994), were expected to make a significant contribution to shielding. The full ca 35 p.p.m. range of 15N shifts in valine residues in SNase was reproduced in the calculations, again with only a small error on the individual shifts. For valine, threonine and isoleucine, the 15N shift ranges are larger than with other residues, which can be attributed to a ‘γ-gauche’ effect on shielding (Cheney & Grant 1967), which varies with side-chain (χ1) conformation. The ability to predict isotropic 13Cα shifts in proteins and both isotropic and anisotropic shielding tensor information in amino acids together with 15N isotropic shifts in proteins (de Dios et al. 1993; Le & Oldfield 1996) then prompted a determination of complete isotropic and anisotropic shielding tensor information for all amino acids (except proline) on Ramachandran ϕ, ψ surfaces (Sun et al. 2002), as shown in, for example, figure 5a,b. The results obtained were interesting in that, for Cα, sheet residues were clearly predicted to resonate upfield (figure 5a), but the breadths of the tensors for both helical and sheet geometries were very similar, ca 34 p.p.m., for all amino acids except valine, threonine and isoleucine. For these residues, the tensor breadths were generally much smaller, ca 22 p.p.m., in helical geometries. This effect appears to be related to the presence of β-branched side-chains, reminiscent of the γ-gauche effect seen in the case of the 15N NMR shifts of these same three amino acids. The results of these calculations were then verified by using solid-state NMR techniques to deduce the magnitudes of the tensor elements in Ala, Val and Leu peptides (Havlin et al. 2001), and the Cα tensor orientations, shown in figure 5d,e, have now been confirmed experimentally in several amino acids and peptides by other groups (Yao & Hong 2002; Chekmenev et al. 2002).
Figure 5.
Computed isotropic shielding and anisotropic shielding tensors and their orientations for 13Cα in N-formyl alanine amide: (a) σiso; (b) Ω=σ33−σ11; (c) Δσ*. (d) Helical 13Cα tensor orientation in lysine. (e) Sheet 13Cα tensor orientation in lysine. (f) Helical 13Cβ tensor orientation. (g) Sheet Cβ tensor orientation. Reprinted in part with permission from Sun et al. (2002). Copyright 2002 American Chemical Society.
These computational results were at first somewhat surprising in that, in solution NMR, it had been found that a chemical shift anisotropy (CSA), Δσ*, had been found to vary very considerably between helical and sheet geometries, with Δσ*≈6 p.p.m. in helices and Δσ*≈27 p.p.m. in sheets (Tjandra & Bax 1997). However, this CSA, determined by dipole–dipole/CSA interference techniques, is not the same as that determined theoretically or by solid-state NMR since it is convoluted with tensor orientation information. In particular, the σ22 element of the 13Cα tensor is oriented approximately along the C–H bond vector in helical geometries (figure 5d) while σ11 is oriented approximately along the C–H bond vector in sheet geometries (Walling et al. 1997; figure 5e). It is, however, a simple matter to effect the appropriate transformation, and typical Δσ*(ϕ, ψ) surface results are shown in figure 5c. There was found to be generally good agreement between experimental and theoretical Δσ* values (Case 1998; Sun et al. 2002), with Δσ*≈0 p.p.m. being found for ideal helical residues and Δσ* of ca 25 p.p.m. being predicted for ideal sheet residues. Interestingly, for Cβ, there is essentially no change in tensor orientation between helices and sheets, as shown in figure 5f,g.
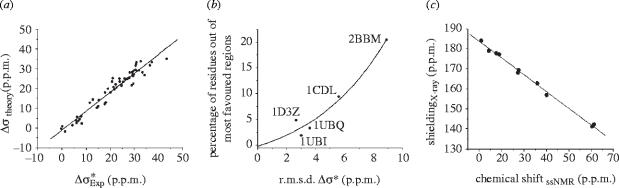
The experimental Δσ* results for Cα and the theoretical predictions were all found to be highly correlated, as shown in figure 6a. Moreover, the errors that were observed were found to be a function of the quality of the (crystallographic) structure used to set the ϕ,ψ values in the calculations, as can be seen in figure 6b. Future work on geometry optimization, as outlined above, may reduce these errors. The GIAO approach was also found to be capable of accurately predicting 13C side-chain shifts, as shown in figure 6c for Cβ, Cγ1, Cγ2, Cδ (and Cα) for isoleucine side-chains, where again (γ-gauche) torsional effects make major contributions to shielding (Sun et al. 2002).
Figure 6.
Experimental versus theoretical shift/shielding results in proteins. (a) Correlations between experimental and computed 13C Δσ* values in ubiquitin using PDB structure 1D3Z and individual amino acid Δσ* surfaces; (b) Relation between Δσ* error (r.m.s. error between the experimental Δσ* and that computed from the structure indicated) and the structure quality (percentage residues outside the most favoured regions of the protein structure). Structure used: ubiquitin, 1UBI, 1UBQ and 1D3Z; calmodulin, 1CDL and 2BBM. (c) Correlation between experimental isotropic chemical shifts, δ, in p.p.m. (from TMS) and the computed absolute shieldings, σ, for Cα, Cβ, Cγ1, Cγ2 and Cδ in Ile-18 and Ile-19 in the bovine pancreatic trypsin inhibitor. Experimental solid state shift/X-ray predicted shieldings; slope: −0.84, y-intercept: 192.1 p.p.m., R2=1.00, r.m.s.d.=1.1 p.p.m. Reprinted in part with permission from Sun et al. (2002). Copyright 2002 American Chemical Society.
In most of the work described above, chemical shifts were calculated using individual or specific molecules. A much faster method is to use a database approach, which relies on additivity. Here, Case and coworkers (Sitkoff & Case 1997; Xu & Case 2001, 2002) have been very successful in producing a fast algorithm for quite accurately predicting isotropic 13C and 15N shifts, based on a large database of chemical shifts computed using DFT. This prediction algorithm (‘SHIFTS’) is expected to be of great use in NMR peak assignments, as well as in structure refinement.
Finally, in the area of solution 13C, 15N NMR chemical shift calculations, there have been important advances made by Vila et al. (2003) in predicting ‘random coil’ shifts, or more precisely Boltzmann-averaged values of the chemical shifts of a series of tetra-peptides. This is an exciting new development since it potentially opens up the use of NMR chemical shifts and quantum chemistry for the investigation of less well-structured regions in proteins (Vila et al. 2003).
11. Fluorine NMR chemical shifts and electrostatics
A rather challenging task is posed by 19F NMR chemical shifts in proteins. These had been investigated experimentally for many years, but unlike the situation with 13C and 15N NMR shifts, it seemed unlikely that the 19F NMR chemical shifts of, for example, a fluorophenylalanine residue, would be related to ϕ, ψ or χ1, χ2 conformational effects. It was found to be possible to accurately predict 19F chemical shifts in many chemically different fluorobenzenes (de Dios & Oldfield 1994), but what would cause a large range of chemical shift non-equivalence in a protein, due to folding?
One likely possibility appeared to be that 19F NMR chemical shifts in proteins might be dominated by electrostatic field effects, an idea which was based on earlier observations that 13C and 17O NMR shifts of CO bound to different haem proteins, the C17O EFG and the CO infrared vibrational frequencies were all related (Park et al. 1991). Moreover, they could be predicted by using an electrical polarization model (Augspurger et al. 1991, 1993a). To test this idea, we investigated how the 19F NMR chemical shifts in fluorobenzene (a phenylalanine model) might be perturbed by an electrostatic field, generated by clusters of HF molecules or point charges/point dipoles (the protein), using supermolecule cluster calculations, and the CFP method outlined above for the amino acids threonine and tyrosine (de Dios & Oldfield 1993). The shift effects were found to be large at plausible intermolecular distances, and the results of the supermolecule and point charge/point dipole calculations agreed. In addition, a third method was developed, in which the derivatives of the shielding tensor with respect to the field and field gradient were evaluated, by using derivative Hartree–Fock theory (Augspurger & Dykstra 1991; Augspurger et al. 1992). For example, the response of the shift to a uniform field is given by
where is called the dipole shielding polarizability tensor. That is, is the formal derivative of σ with respect to a uniform field, while is the formal derivative of σ with respect to the non-uniform field or field gradient components. Hyperpolarizabilities (∂2/∂F2) and mixed derivatives (∂2/∂F∂V) were also evaluated (Augspurger et al. 1993b), but based on the results of model calculations, only the and tensors were found to be significant. The results of these ‘multipole shielding polarizability’ calculations on the fluorobenzene–HF clusters were found to agree with those obtained from supermolecule or CFP techniques (Augspurger et al. 1993b; de Dios & Oldfield 1993).
What this means for proteins is that the electrostatic contributions to the 19F NMR chemical shifts can be calculated by simply multiplying the and tensors by the electrostatic fields and field gradients in the protein. To evaluate the fields in a protein, the Enzymix program was used. This involves use of a point dipole–Langevin dipole method to evaluate the uniform field component, F, and by finite difference, the field gradient, V (Pearson et al. 1993). This is computationally lengthy, since solvation as well as dynamical effects need to be taken into account, at least if solvent-exposed residues are to be considered, which necessitates a molecular dynamics (MD) approach. That is, the fields (F) and field gradients (V) are calculated over an MD trajectory, then the uniform and non-uniform electrostatic field contributions to shielding can be evaluated given knowledge of the dipole and quadrupole shielding polarizability. Perhaps surprisingly, the method was found to give an excellent description of the 19F NMR chemical shifts of the five [5-19F]Trp residues in a galactose binding protein (Pearson et al. 1993) as well as quite good accord with the experimental [4-19F]Phe residue shifts in myoglobin and haemoglobin (Pearson et al. 1997). In the case of the galactose binding protein, good accord was also found with the CFP method for buried residues, but the slope of the correlation was worse since the dielectric constant (in the protein) was not considered in the calculations (de Dios et al. 1993). The effects of electrostatic interactions can also be extremely well modelled by use of hybrid QM–MM techniques, as demonstrated by Cui & Karplus (2000), who applied this approach to a wide number of systems, including haem protein models. It seems likely that the QM–MM technique will be of great future importance, especially when combined with the Car–Parrinello MD technique (Gervasio et al. 2003), enabling the incorporation of both inter-residue interactions as well as dynamics, into property calculations.
12. Metal ion NMR
Another NMR challenge for quantum chemistry was to try to predict the NMR shifts and shift (or shielding) tensors for a transition metal in a protein, a difficult problem given the large number of electrons in such metals. In early work, several groups investigated experimentally the 57Fe (I=1/2) NMR chemical shifts in the proteins myoglobin (Mb) and ferrocytochrome c (Baltzer et al. 1985; Baltzer 1987; Chung et al. 1990), and there was found to be an extremely large (ca 3000 p.p.m.) chemical shift range. In addition, relaxation measurements enabled the determination of the tensor spans, Δσ, for MbCO, MbRNC and cyt c. Early investigations of Rh(I) NMR chemical shifts in organometallic compounds using Hartree–Fock methods were unsuccessful, but good correlations were later found when using DFT techniques, especially when using the hybrid exchange/correlation functional B3LYP (Donkervoort et al. 1999). This approach was thus adapted to investigate 57Fe and 59Co shifts and shift tensors in a series of model systems and in metalloproteins themselves (Godbout & Oldfield 1997; Godbout et al. 1998). Again, perhaps surprisingly, it was found to be possible to predict both the isotropic and anisotropic shift (shielding) tensor properties, when using the DFT/B3LYP approach, and suitably large basis sets. Initial results with a protein, carbonmonoxymyoglobin, were very poor, but this was found to be the result of using an early crystallographic structure in which the CO ligand bound to Fe appeared to adopt a bent geometry. Use of an upright geometry, based on crystallographic investigations of a small haem–CO complex, corrected this problem and led to the idea, as described below, that NMR shifts might be used to accurately determine at least some aspects of protein structure.
13. Towards structure determination
The results outlined above indicate that it is now possible to predict many spectroscopic observables in proteins and model systems; δFe and ΔEQ in Mössbauer spectroscopy; δ(13C, 15N, 19F, 57Fe) and Δδ in proteins, peptides and other model systems; δNMR hyperfine shifts in proteins and model systems; and ∇E (the EFG) in amino acids and haems. There is, therefore, a correlation between ‘structure’ and ‘spectra’. In what we have described so far, we have typically gone in the ‘forward’ direction: structure to spectra. But what might be more interesting would be to go in the reverse direction: spectra to structure. At least at the empirical level, this is not new since, for example, J-couplings have been used for many years as structure restraints just using empirical parameterizations of the Karplus relation. However, with quantum chemistry there might be more possibilities using, for example, chemical shifts deduced from Ramachandran shielding surfaces or indeed, ab initio structure determinations based in part on NMR shifts, or chemical shift assisted approaches to ab initio folding.
Two simple examples of de novo approaches to structure determination are given in which the structure to spectral relation is inverted analytically. Assume a property, P, is a function of two independent variables, . For example, P might be a Cα chemical shift and a=ψ, b=ϕ, the backbone torsion angles. For a given a, b, the value of the property can be derived from the property surface P=P(a, b) and the probability, Z, that a, b represent the actual experimental values will be
where Pexpt is the experimental value of the property, P(a, b) is the calculated value of the property for the given a, b values and W is a search width parameter related to the r.m.s. error in the property prediction. When Pexpt=P(a, b), Z=1 and the a, b values chosen are possible solutions. Of course, the actual form of P(a, b) may yield multiple a, b solutions yielding Z=1, in which case it is necessary to use more than one property to deduce unique values for a and b. In this case, it is possible to use conditional probability such that
where there are N observables. In an early application of this idea, isotropic and anisotropic 13Cα NMR shift information was used to predict the backbone ϕ, ψ torsion angles in a tripeptide with an error of between 5° and 10° (Heller et al. 1997), and in another application, δ(13C), Δδ(13C), δ(17O), ∇E(17O), δ(57Fe) and ΔEQ (57Fe) Z surface information was used to predict the Fe–CO ligand tilt and bend angles for CO bound to Fe(II) in myoglobin (McMahon et al. 1998). The nature of this ligand bonding had been controversial for many years, with early crystallographic and indeed other results suggesting a highly bent (ca 120°) ligand-binding geometry. Using the combination of NMR and Mössbauer spectroscopic property information, a ligand tilt (τ)=4° and a bend (β)=7° were found (McMahon et al. 1998). After these results were published, the X-ray structure was re-determined using synchrotron radiation (Kachalova et al. 1999). The new results obtained showed only a ca 0.60° variance between the Z-surface predictions, based on quantum chemistry, and the new crystal structure. In a similar vein, we have recently investigated the geometry of NO bound to haemoglobin and myoglobin. These results again vary considerably from some crystallographic structures, but enable the prediction of both δFe and ΔEQ in the Mössbauer spectra, as well as the electron paramagnetic resonance observables, and are essentially the same as those obtained by using quantum chemical geometry optimization on a large molecular cluster (Y. Zhang, W. Gossman and E. Oldfield, unpublished data).
14. Conclusions and future prospects
The results we have described above cover developments in predicting a wide variety of spectroscopic and other observable properties in proteins and model systems, which have been carried out over the past decade. At perhaps the most basic level, ab initio SCF energy calculations have been successful in reproducing the fine details of protein structure seen in very-high-resolution crystal structures. In the future, it therefore seems likely that it will be possible to directly incorporate quantum chemical results in structure refinement. From the associated ground state wave functions, most electrostatic properties (ρ(r), λi, ∇2ρ(r), Hessian-of-ρ(r), Φ(r), ∇E) can be derived. All these properties should be accessible in proteins. From a spectroscopic standpoint, it is now also possible to accurately predict the Mössbauer observables as follows: δFe, the isomer shift (from ρ(0)), and ΔEQ, the quadrupole splitting (from VZZ, the z-component of the EFG tensor). This opens the way to using these observables to determine local geometries, and also gives more confidence in using MO methods to probe details of metal–ligand bonding. In the area of NMR spectroscopy, it is now possible to predict 13C, 15N, 19F and even 57Fe chemical shifts and shift (shielding) tensors, as well as their orientations, in addition to enabling detailed studies of J-couplings, of interest in the context of structure refinement and hydrogen bonding. And for paramagnetic systems, it has been found to be possible to quite accurately predict many NMR contact shifts ab initio, which for haem proteins encompass a ca 6000 p.p.m. shift range. Already, the structure to spectra nexus is being used to make structure refinements, and in the future it seems likely that NMR chemical shifts may play an even more important role in structure determination, especially in non-crystalline proteins. The success of quantum chemical methods in predicting such a wide range of properties also gives added confidence in using quantum chemical methods to investigate reaction mechanisms (Gogonea et al. 2001; Gao & Truhlar 2002) which, since they involve bond rearrangements, must necessarily be treated, at least in part, quantum mechanically. Hopefully, over the next few years, these two rather distinct areas: structural/spectroscopic (large basis/small fragment/property calculations) and enzyme mechanism (small basis—QM–MM, entire protein) of quantum chemistry will merge, and it will be possible to use one method to determine or refine structures, to make property calculations, and to investigate reaction mechanisms and their inhibition. For example, at the level of using NMR chemical shift information, it should be possible to probe drug–protein interactions—a potentially ‘useful’ application of quantum chemical technology. It should also be possible to use QM methods in a more direct manner in drug design, rather than simply using force-field-based methods to make structure–activity correlations, by more accurately calculating interaction energies, and by using AIM theory (O'Brien & Popelier 2002). As with most things computational, it is just a matter of time.
Acknowledgments
The author thanks A. C. de Dios and C. Jameson for their early contributions to this work and Y. Zhang and H. Sun for providing the figures. This work was supported by the United States Public Health Service (National Institutes of Health grants EB00271 and GM50694).
References
- Alderman D.W, Sherwood M.H, Grant D.M. Comparing, modeling, and assigning chemical-shift tensors in the Cartesian, irreducible spherical, and icosahedral representations. J. Magn. Res. Ser. A. 1993;101:188–197. [Google Scholar]
- Allerhand A, Oldfield E. Determination of rotational mobilities of backbone and side chain carbons of poly(γ-benzyl l-glutamate) in the helical and random-coil states from measurements of carbon-13 relaxation times and nuclear Overhauser enhancements. Biochemistry. 1973;12:3428–3433. doi: 10.1021/bi00742a011. [DOI] [PubMed] [Google Scholar]
- Allerhand A, Childers R.F, Oldfield E. Natural-abundance carbon-13 nuclear magnetic resonance studies in 20 min sample tubes. Observation of numerous single-carbon resonances of hen egg-white lysozyme. Biochemistry. 1973;12:1335–1341. doi: 10.1021/bi00731a013. [DOI] [PubMed] [Google Scholar]
- Arnold W.D, Oldfield E. The chemical nature of hydrogen bonding in proteins via NMR: J-couplings, chemical shifts, and AIM theory. J. Am. Chem. Soc. 2000;122:12 835–12 841. [Google Scholar]
- Arnold W.D, Sanders L.K, McMahon M.T, Volkov R.V, Wu G, Coppens P, Wilson S.R, Godbout N, Oldfield E. Experimental Hartree–Fock, and density functional theory investigations of the charge density, dipole moment, electrostatic potential, and electric field gradients in l-asparagine monohydrate. J. Am. Chem. Soc. 2000a;122:4708–4717. [Google Scholar]
- Arnold W.D, Mao J.H, Sun H.H, Oldfield E. Computation of through-space F-19–F-19 scalar couplings via density functional theory. J. Am. Chem. Soc. 2000b;122:12 164–12 168. [Google Scholar]
- Augspurger J.D, Dykstra C.E. Electromagnetic properties of molecules from a uniform procedure for differentiation of molecular wave-functions to high-order. J. Phys. Chem. 1991;95:9230–9238. [Google Scholar]
- Augspurger J.D, Dykstra C.E, Oldfield E. Correlation of carbon-13 and oxygen-17 chemical shifts and the vibrational frequency of electrically perturbed carbon monoxide: a possible model for distal ligand effects in carbonmonoxyheme proteins. J. Am. Chem. Soc. 1991;113:2447–2451. [Google Scholar]
- Augspurger J.D, Pearson J.G, Oldfield E, Dykstra C.E, Park K.D, Schwartz D. Chemical-shift ranges in proteins. J. Magn. Reson. 1992;100:342–357. [Google Scholar]
- Augspurger J.D, Dykstra C.E, Oldfield E, Pearson J.G. Intra- and intermolecular electrical effects on nuclear magnetic resonance, nuclear quadrupole resonance and infra-red spectroscopic parameters from ab initio calculation and experiment: from CO to proteins. In: Tossell J.A, editor. Nuclear magnetic shieldings and molecular structure. NATO ASI Series C. vol. 386. Kluwer; Boston, MA: 1993. pp. 75–94. [Google Scholar]
- Augspurger J.D, de Dios A.C, Oldfield E, Dykstra C.E. Computing nuclear magnetic resonance chemical shielding in large systems via multipole shielding polarizabilities. Chem. Phys. Lett. 1993;213:211–216. [Google Scholar]
- Bader R.F.W. Oxford University Press; 1990. Atoms in molecules—a quantum theory. [Google Scholar]
- Baltzer L. 57Fe NMR of heme proteins: the chemical shift anisotropy of ferrocytochrome-c. J. Am. Chem. Soc. 1987;109:3479–3481. [Google Scholar]
- Baltzer L, Becker E.D, Tschudin R.G, Gansow O.A. 57Fe NMR spectroscopy of heme proteins: chemical shift anisotropy and relaxation parameters of carbonylmyoglobin. J. Chem. Soc. Chem. Commun. 1985:1040–1041. [Google Scholar]
- Bernal J.D, Crowfoot D. X-ray photographs of crystalline pepsin. Nature. 1934;794:133–134. [Google Scholar]
- Case D.A. The use of chemical shifts and their anisotropies in biomolecule structure determination. Curr. Opin. Struct. Biol. 1998;8:624–630. doi: 10.1016/s0959-440x(98)80155-3. [DOI] [PubMed] [Google Scholar]
- Case D.A, Scheurer C, Bruschweiler R. Static and dynamic effects on vicinal scalar J-couplings in proteins and peptides: a MD/DFT analysis. J. Am. Chem. Soc. 2000;122:10 390–10 397. [Google Scholar]
- Challacombe M, Schwegler E. Linear scaling computation of the Fock matrix. J. Chem. Phys. 1997;106:5526–5536. doi: 10.1063/1.1869470. [DOI] [PubMed] [Google Scholar]
- Chekmenev E.Y, Xu R.Z, Mashuta M.S, Wittebort R.J. Glycyl Cα chemical shielding in tripeptides: measurement by solid-state NMR and correlation with X-ray structure and theory. J. Am. Chem. Soc. 2002;124:11 894–11 899. doi: 10.1021/ja026700g. [DOI] [PubMed] [Google Scholar]
- Cheney B.V, Grant D.M. Carbon-13 magnetic resonance. VIII. The theory of carbon-13 chemical shifts applied to saturated hydrocarbons. J. Am. Chem. Soc. 1967;89:5319–5327. [Google Scholar]
- Chesnut D.B, Moore K.D. Locally dense basis-sets for chemical-shift calculations. J. Comput. Chem. 1989;10:648–659. [Google Scholar]
- Chung J, Lee H.C, Oldfield E. Iron-57 nuclear magnetic resonance spectroscopic study of alkyl isocyanide myoglobins and a comparison of the 57Fe chemical shift anisotropies of alkyl isocyanide myoglobins, carbonmonoxy-myoglobin, ferrocytochrome c and [57Fe(bipy)3]X2·5H2O (x=Cl, Br, I) J. Magn. Res. 1990;90:148–157. [Google Scholar]
- Cole H.B.R, Sparks S.W, Torchia D.A. Comparison of the solution and crystal structures of staphylococcal nuclease with 13C and 15N chemical shifts used as structural fingerprints. Proc. Natl Acad. Sci. USA. 1988;85:6362–6365. doi: 10.1073/pnas.85.17.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Hu J.S, Bax A. Identification of the hydrogen bonding network in a protein by scalar couplings. J. Am. Chem. Soc. 1999;121:2949–2950. [Google Scholar]
- Cremer D, Kraka E. A description of the chemical bond in terms of local properties of electron density and energy. Croat. Chem. Acta. 1984;57:1259–1281. [Google Scholar]
- Cui Q, Karplus M. Molecular properties from combined QM/MM methods. 2. Chemical shifts in large molecules. J. Phys. Chem. B. 2000;104:3721–3743. [Google Scholar]
- de Dios A.C, Oldfield E. Methods for computing nuclear magnetic resonance chemical shielding in large systems. Multiple cluster and charge field approaches. Chem. Phys. Lett. 1993;205:108–116. [Google Scholar]
- de Dios A.C, Oldfield E. Evaluating 19F chemical shielding in fluorobenzenes: implications for chemical shifts in proteins. J. Am. Chem. Soc. 1994;116:7453–7454. [Google Scholar]
- de Dios A.C, Pearson J.G, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- de Dios A.C, Laws D.D, Oldfield E. Predicting carbon-13 nuclear magnetic resonance chemical shielding tensors in zwitterionic l-threonine and dl-tyrosine via quantum chemistry. J. Am. Chem. Soc. 1994;116:7784–7786. [Google Scholar]
- Ditchfield R. Self-consistent perturbation theory of diamagnetism. I. A gauge-invariant LCAO method for N.M.R. chemical shifts. Mol. Phys. 1974;27:789–807. [Google Scholar]
- Dittrich B, Flaig R, Koritsanszky T, Krane H.G, Morgenroth W, Luger P. Topological properties of the peptide bond in glycyl-l-threonine dihydrate based on a fast synchrotron/CCD-diffraction experiment at 100 K. Chem. A Eur. J. 2000;6:2582–2589. doi: 10.1002/1521-3765(20000717)6:14<2582::aid-chem2582>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Donkervoort J.G, Buhl M, Ernsting J.M, Elsevier C.J. Steric and electronic effects on the Rh-103 NMR chemical shifts of Rh-I(cyclooctadiene) compounds bearing N-donor ligands. Chem. Eur. J. 1999:27–33. [Google Scholar]
- Dwek R.A. Clarendon Press; Oxford: 1973. Nuclear magnetic resonance (N.M.R.) in bio-chemistry. [Google Scholar]
- Gao J, Truhlar D.G. Quantum mechanical methods for enzyme kinetics. Annu. Rev. Phys. Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- Gervasio F.L, Schettino V, Mangani S, Krack M, Carloni P, Parrinello M. Influence of outer-shell metal ligands on the structural and electronic properties of horse liver alcohol dehydrogenase zinc active site. J. Phys. Chem. B. 2003;107:6886–6892. [Google Scholar]
- Godbout N, Oldfield E. Density functional theory study of cobalt-59 nuclear magnetic resonance chemical shifts and shielding tensor elements in Co(III) complexes. J. Am. Chem. Soc. 1997;119:8065–8069. [Google Scholar]
- Godbout N, Havlin R, Salzmann R, Debrunner P.G, Oldfield E. Iron-57 NMR chemical shifts and Mössbauer quadrupole splittings in metalloporphyrins, ferrocytochrome c, and myoglobins: a density functional theory investigation. J. Phys. Chem. A. 1998;102:2342–2350. [Google Scholar]
- Gogonea V, Suarez D, van der Vaart A, Metz K.W. New developments in applying quantum mechanics to proteins. Curr. Opin. Struct. Biol. 2001;11:217–223. doi: 10.1016/s0959-440x(00)00193-7. [DOI] [PubMed] [Google Scholar]
- Havlin R.H, Laws D.D, Bitter H.-M.L, Sanders L.K, Sun H, Grimley J.S, Wemmer D.E, Pines A, Old-field E. An experimental and theoretical investigation of the chemical shielding tensors of 13Cα of alanine, valine, and leucine residues in solid peptides and in proteins in solution. J. Am. Chem. Soc. 2001;123:10 362–10 369. doi: 10.1021/ja0115060. [DOI] [PubMed] [Google Scholar]
- Hehre W.J, Radom L, von Schleyer P.R, Pople J.A. Wiley; New York: 1986. Ab initio molecular orbital theory. [Google Scholar]
- Heisenberg W. Mehrköperproblem und resonanz in der quantenmechanik. Zeitschr. Phys. 1926;38:411–426. [Google Scholar]
- Heitler W, London F. Wechselwirkung neutraler atome und homöopolare bindung nach der quantenmechanik. Zeitschr. Phys. 1927;44:455–472. [Google Scholar]
- Helgaker T, Watson M, Handy N.C. Analytical calculation of nuclear magnetic resonance indirect spin–spin coupling constants at the generalized gradient approximation and hybrid levels of density-functional theory. J. Chem. Phys. 2000;113:9402–9409. [Google Scholar]
- Heller J, Laws D.D, Tomaselli M, King D.S, Wemmer D.E, Pines A, Havlin R.H, Oldfield E. Determination of dihedral angles in peptides through experimental and theoretical studies of α-carbon chemical shielding tensors. J. Am. Chem. Soc. 1997;119:7827–7831. [Google Scholar]
- Kachalova G.S, Popov A.N, Bartunik H.D. A steric mechanism for inhibition of CO binding to heme proteins. Science. 1999;284:473–476. doi: 10.1126/science.284.5413.473. [DOI] [PubMed] [Google Scholar]
- Karplus M. Theory of vicinal coupling. In: Grant D, Harris R, editors. Encyclopedia of nuclear magnetic resonance. Wiley; New York: 1996. pp. 420–422. [Google Scholar]
- Kimber B.J, Feeney J, Roberts G.C.K, Birdsall B, Griffiths D.V, Burgen A.S.V, Sykes B.D. Proximity of two tryptophan residues in dihydrofolate reductase determined by fluorine-19 NMR. Nature. 1978;271:184–185. doi: 10.1038/271184a0. [DOI] [PubMed] [Google Scholar]
- Kohn W, Becke A.D, Parr R.G. Density functional theory of electronic structure. J. Phys. Chem. 1996;100:12 974–12 980. [Google Scholar]
- Le H.B, Oldfield E. Correlation between 15N NMR chemical shifts in proteins and secondary structure. J. Biomol. NMR. 1994;4:341–348. doi: 10.1007/BF00179345. [DOI] [PubMed] [Google Scholar]
- Le H.B, Oldfield E. Ab initio studies of amide 15N chemical shifts in dipeptides: applications to protein NMR spectroscopy. J. Phys. Chem. 1996;100:16 423–16 428. [Google Scholar]
- Li X, Wu G, Abramov Y.A, Volkov A.V, Coppens P. Application of charge density methods to a protein model compound: calculation of Coulombic intermolecular interaction energies from the experimental charge density. Proc. Natl Acad. Sci. USA. 2002;99:12 132–12 137. doi: 10.1073/pnas.192438999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiWang A.C, Bax A. Solution NMR characterization of hydrogen bonds in a protein by indirect measurement of deuterium quadrupole couplings. J. Magn. Res. 1997;127:54–64. doi: 10.1006/jmre.1997.1187. [DOI] [PubMed] [Google Scholar]
- McMahon M.T, de Dios A.C, Godbout N, Salzmann R, Laws D.D, Le H, Havlin R.H, Oldfield E. An experimental and quantum chemical investigation of CO binding to heme proteins and model systems: a unified model based on 13C, 17O, and 57Fe nuclear magnetic resonance and 57Fe Mössbauer and infrared spectroscopies. J. Am. Chem. Soc. 1998;120:4784–4797. [Google Scholar]
- Malkin V.G, Malkina O.L, Casida M.E, Salahub D.R. Nuclear magnetic resonance shielding tensors calculated with a sum-over-states density functional perturbation theory. J. Am. Chem. Soc. 1994;116:5898–5908. [Google Scholar]
- Mallory F.B, et al. Nuclear spin–spin coupling via nonbonded interactions. 8. The distance dependence of through-space fluorine–fluorine coupling. J. Am. Chem. Soc. 2000;122:4108–4116. [Google Scholar]
- Mao J, Zhang Y, Oldfield E. Nuclear magnetic resonance shifts in paramagnetic metalloporphyrins and metalloproteins. J. Am. Chem. Soc. 2002;124:13 911–13 920. doi: 10.1021/ja020297w. [DOI] [PubMed] [Google Scholar]
- Markwick P.R.L, Sprangers R, Sattler M. Dynamic effects on J-couplings across hydrogen bonds in proteins. J. Am. Chem. Soc. 2003;125:644–645. doi: 10.1021/ja028875w. [DOI] [PubMed] [Google Scholar]
- Naito A, McDowell C.A. Determination of the 14N quadrupole coupling tensors and the 13C chemical shielding tensors in a single-crystal of l-asparagine monohydrate. J. Chem. Phys. 1984;81:4795–4803. [Google Scholar]
- O'Brien S.E, Popelier P.L.A. Quantum topological molecular similarity. Part 4. A QSAR study of cell growth inhibitory properties of substituted (E)-1-phenylbut-1-en-3-ones. J. Chem. Soc. Perkin Trans. 2002;2:478–483. [Google Scholar]
- Park K.D, Guo K, Adebodun F, Chiu M.L, Sligar S.G, Oldfield E. Distal and proximal ligand interactions in heme–proteins: correlations between C–O and Fe–C vibrational frequencies, O-17 and C-13 nuclear magnetic resonance chemical shifts and O-17 nuclear quadrupole coupling constants in C17O-labeled and 13CO-labeled species. Biochemistry. 1991;30:2333–2347. doi: 10.1021/bi00223a007. [DOI] [PubMed] [Google Scholar]
- Pauling L. The application of the quantum mechanics to the structure of the hydrogen molecule and hydrogen molecule-ion and to related problems. Chem. Rev. 1928a;5:173–213. [Google Scholar]
- Pauling L. The shared-electron chemical bond. Proc. Natl Acad. Sci. USA. 1928b;14:359–362. doi: 10.1073/pnas.14.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L. Interatomic distances in covalent molecules and resonance between two or more Lewis electronic structures. Proc. Natl Acad. Sci. USA. 1932;18:293–297. doi: 10.1073/pnas.18.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L, Corey R.B. Two hydrogen-bonded spiral configurations of the polypeptide chain. J. Am. Chem. Soc. 1950;72:5349. [Google Scholar]
- Pauling L, Corey R.B. Configurations of polypeptide chains with favored orientations around single bonds: two new pleated sheets. Proc. Natl Acad. Sci. USA. 1951;37:729–740. doi: 10.1073/pnas.37.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.G, Oldfield E, Lee F.S, Warshel A. Chemical shifts in proteins: a shielding trajectory analysis of the fluorine nuclear magnetic resonance spectrum of the Escherichia coli galactose binding protein using a multipole shielding polarizability-local reaction field-molecular dynamics approach. J. Am. Chem. Soc. 1993;115:6851–6862. [Google Scholar]
- Pearson J.G, Montez B, Le H.B, Oldfield E, Chien E.Y.T, Sligar S.G. Assignment and analysis of fluorine nuclear magnetic resonance spectra of 4-fluorotryptophan myoglobins and hemoglobins. Biochemistry. 1997;36:3590–3599. doi: 10.1021/bi961664h. [DOI] [PubMed] [Google Scholar]
- Ramachandran G.N, Kartha G. Structure of collagen. Nature. 1954;174:269–270. doi: 10.1038/174269c0. [DOI] [PubMed] [Google Scholar]
- Ramachandran G.N, Kartha G. Structure of collagen. Nature. 1955;176:593–595. doi: 10.1038/176593a0. [DOI] [PubMed] [Google Scholar]
- Ramachandran G.N, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan C. Remembrances of Professor G. N. Ramachandran. Protein Sci. 2001;10:1689–1693. doi: 10.1002/pro.101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A, Crick F.H.C. The structure of collagen. Nature. 1955;176:915–916. doi: 10.1038/176915a0. [DOI] [PubMed] [Google Scholar]
- Sitkoff D, Case D.A. Density functional calculations of proton chemical shifts in model peptidcs. J. Am. Chem. Soc. 1997;119:12 262–12 273. [Google Scholar]
- Spera S, Bax A. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- Sun H.H, Sanders L.K, Oldfield E. Carbon-13 NMR shielding in the twenty common amino acids: comparisons with experimental results. J. Am. Chem. Soc. 2002;124:5486–5495. doi: 10.1021/ja011863a. [DOI] [PubMed] [Google Scholar]
- Svedberg T, et al. A discussion on the protein molecule. Proc. R. Soc. B. 1939;846:1–40. [Google Scholar]
- Sykes B.D, Weiner J.H. Biosynthesis and 19F NMR characterization of fluoroamino acid containing proteins. In: Cohen J.S, editor. Magnetic resonance in biology. Wiley; New York: 1980. pp. 171–194. [Google Scholar]
- Tjandra N, Bax A. Large variations of 13Cα chemical shift anisotropy in proteins correlate with secondary structure. J. Am. Chem. Soc. 1997;119:9576–9577. [Google Scholar]
- Van Alsenoy C, Yu C.-H, Peeters A, Martin J.M.L, Schäfer L. Ab initio geometry determinations of proteins. 1. Crambin. J. Phys. Chem. 1998;102:2246–2251. [Google Scholar]
- Vila J.A, Baldoni H.A, Ripoll D.R, Scheraga H.A. Unblocked statistical-coil tetrapeptides in aqueous solution: quantum-chemical computation of the 13C NMR chemical shifts. J. Biomol. NMR. 2003;26:113–130. doi: 10.1023/a:1023524727484. [DOI] [PubMed] [Google Scholar]
- Walling A.E, Pargas R.E, de Dios A.C. Chemical shift tensors in peptides: a quantum mechanical study. J. Phys. Chem. A. 1997;101:7299–7303. [Google Scholar]
- Wilkens S.J, Xia B, Weinhold F, Markley J.L, Westler W.M. NMR investigations of Clostridium pasteurianum rubredoxin. Origin of hyperfine H-1, H-2, C-13 and N-15 NMR chemical shifts in iron–sulfur proteins as determined by comparison of experimental data with hybrid density functional calculations. J. Am. Chem. Soc. 1998;120:4806–4814. [Google Scholar]
- Xu X.P, Case D.A. Automated prediction of 15N, 13Cα, Cβ and 13C′ chemical shifts in proteins using a density functional database. J. Biomol. NMR. 2001;21:321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- Xu X.P, Case D.A. Probing multiple effects on 15N, 13Cα, 13Cβ, and 13C′ chemical shifts in peptides using density functional theory. Biopolymers. 2002;65:408–423. doi: 10.1002/bip.10276. [DOI] [PubMed] [Google Scholar]
- Yao X.L, Hong M. Determination of Cα chemical shift tensor orientation in peptides by dipolar-modulated chemical shift recoupling NMR spectroscopy. J. Am. Chem. Soc. 2002;124:2730–2738. doi: 10.1021/ja017137p. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mao J.H, Oldfield E. Fe-57 Mössbauer isomer shifts of heme protein model systems: electronic structure calculations. J. Am. Chem. Soc. 2002a;124:7829–7839. doi: 10.1021/ja011583v. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mao J.H, Godbout N. Mössbauer quadrupole splittings and electronic structure in heme proteins and model systems: a density functional theory investigation. J. Am. Chem. Soc. 2002;124:13 921–13 930. doi: 10.1021/ja020298o. [DOI] [PubMed] [Google Scholar]