Abstract
Molecular marker data collected from natural populations allows information on genetic relationships to be established without referencing an exact pedigree. Numerous methods have been developed to exploit the marker data. These fall into two main categories: method of moment estimators and likelihood estimators. Method of moment estimators are essentially unbiased, but utilise weighting schemes that are only optimal if the analysed pair is unrelated. Thus, they differ in their efficiency at estimating parameters for different relationship categories. Likelihood estimators show smaller mean squared errors but are much more biased. Both types of estimator have been used in variance component analysis to estimate heritability. All marker-based heritability estimators require that adequate levels of the true relationship be present in the population of interest and that adequate amounts of informative marker data are available. I review the different approaches to relationship estimation, with particular attention to optimizing the use of this relationship information in subsequent variance component estimation.
Keywords: Markov Chain Monte Carlo, allele frequency, relatedness, pedigree reconstruction, likelihood
1. Introduction
Molecular marker data have been used in many areas of population biology and genetics, and are now finding an increasing role in the study of organisms in their natural environment. The marker data collected provides information on population structure, relatedness and inbreeding (Dobson et al. 1998; Surridge et al. 1999); all used in studies of, for example, mating systems (Avise 1994), paternities (Meagher 1986; Marshall et al. 1998) and isolation by distance (Barburjani 1987). In more recent times, more elaborate uses of molecular marker data have been exploited in the study of natural populations, with a study by Slate et al. (2002) combining sophisticated statistical methodology and an extensive bank of molecular markers to attempt to estimate quantitative trait loci (QTL) in wild red deer.
Over the last decade there has also been an increasing interest in using marker-based relationship information to enable the estimation of the phenotypic components of variance for a quantitative trait in populations of unknown pedigree. Knowledge of such variance components is important in both evolutionary and conservation studies. In evolutionary studies, they are important in the understanding of short-term evolution patterns, the reconstruction of historical patterns of natural selection, the prediction of genetic responses to selection and the study of clinical variation (Coyne & Beecham 1987). In conservation studies, estimates provide information on the number of individuals required in order to maintain a viable population and, thus, are required for the management of captive populations (Storfer 1996; Eding & Meuwissen 2001). Loss of genetic variation is a restricting factor in a species' ability to respond to natural selection and hence a limitation on its potential to evolve (Falconer & MacKay 1996; Lande & Shannon 1996). Maintaining variation is therefore critical for maintenance of species within a changing environment.
Several methodologies allowing molecular marker data to be combined with phenotypic information to estimate heritability have been developed (Ritland 1996b; Mousseau et al. 1998; Thomas & Hill 2000). Their properties have been explored through simulation, showing that, provided certain population criteria are met, tolerably accurate heritability estimates can be obtained. However, examples of their use in studying actual populations have been less convincing (Thomas et al. 2002; Wilson et al. 2003). The most fundamental problems with these methods are (i) how to make most efficient use of the molecular marker data to estimate relationship information and (ii) how to best account for the inherent uncertainty in this information in variance component analysis. It is the purpose of this review to discuss the different approaches to relationship estimation, with particular attention to optimizing its use in variance component (heritability) estimation.
2. Estimating relationship information
(a) Relationship coefficients
The most complete method of describing the genetic relationship between a pair of individuals uses the condensed coefficients of identity (Jacquard 1974). These describe nine possible patterns of alleles identical by descent that can be observed within, as well as between, two diploid individuals. In large random-mating populations allelic identity within individuals is effectively zero and so only two of the identities are required: the probability that a pair share two alleles identical by descent (Δ) and the probability they share one (Φ; Lynch & Ritland 1999). These are combined to calculate the coefficients of co-ancestry (θ) and relatedness (r):
| (2.1) |
The coefficient of co-ancestry describes the probability that two alleles, one randomly sampled from each individual, are identical by descent (Jacquard 1974). In an out-bred population the coefficients take on specific values for particular relationships (e.g. Φ=1 and Δ=0 for parent–offspring pairs, Φ=0.5 and Δ=0.25 for full-siblings, Φ=0.5 and Δ=0 for half-siblings and Φ=0 and Δ=0 for unrelated pairs). In the context of variance component estimation the most useful relationship parameters are Δ and r, because these are used to partition the genetic variance (σG2) into the additive (σA2) and dominance (σD2) components, with (Falconer & MacKay 1996; Lynch & Walsh 1998).
Two different approaches have been used to estimate the coefficients of a relationship from molecular marker data: (i) method of moments (MOM; e.g. Queller & Goodnight 1989) and (ii) maximum likelihood (ML; Thompson 1975; Milligan 2003). Conceptually, both work by partitioning the proportion of alleles within a pair that are identical in state into the proportion occurring due to chance and the proportion occurring due to a particular relationship. Unfortunately, individual parameter estimates are extremely noisy when taken on a pair by pair basis so that any joint analysis with quantitative trait data must include many pairs before it becomes useful (Ritland 1996a). In addition, the inherent uncertainty in the estimated relationship should be accounted for in the analysis of variance components in order to minimize subsequent error as well as bias. Since the two approaches to relationship estimation deal with the marker data in a fundamentally different manner, alternative approaches to variance component estimation have been developed for each.
(b) Method of moment relatedness estimators
Numerous MOM methods have been developed for estimating relationship coefficients from co-dominant marker loci data (Blouin 2003). These are divided into two groups: ‘correlation’ estimators, which regard the genetic similarity between the individuals jointly, and ‘regression’ estimators, which compare the genotype of one individual against that of another. These terms were adopted by Lynch & Ritland (1999) and are used to convey the concept that correlation estimators yield a single value for the relationship while the regression estimators can yield different results, depending on which individual is used as a reference. Most published estimators have fallen into the correlation category, since symmetrical statistics have an obvious innate appeal.
The estimators may then be divided into those estimating r and those estimating both Φ and Δ. Finally, they may be divided by whether the parameter estimate is based on the pattern of allelic similarity at each locus (i.e. is independent of allele frequencies) or the frequencies of the particular alleles shared. Divided in this manner, it is possible to see that for each correlation estimator there is an equivalent regression estimator (table 1). Any estimators highlighted by this categorization that have not been previously presented are derived and shown in appendix A.
Table 1.
Classification of the method-of-moment estimators of relationship.
| correlation | regression | |
|---|---|---|
| estimators of r | ||
| pattern | SI_c: similarity index (Lynch 1988; Li et al. 1993) | SI_r: (appendix Aa) |
| allele specific—no weights | QG_c: unweighted estimator (eqn (6) in Ritland (1996) or eqn (10) in Lynch & Ritland (1999)) | QG_r: Queller & Goodnight (Queller & Goodnight 1989) |
| allele specific—weighted | R2_c: ‘two-gene’ correlation (Robertson & Hill 1984; Ritland 1996) | R2_r: (appendix Ab) |
| estimators of Φ and Δ | ||
| pattern | JW_c: Wang (Wang 2002) | JW_r: (appendix Ac) |
| allele and pattern specific—weighted | R4_c: ‘four-gene’ correlation (Ritland 1996) | R4_r: regression (Lynch & Ritland 1999) |
Note that the short codes defined for each pair are derived from the first member presented and are used for convenience rather than to indicate any inherent advantage of one estimator over another. The references given indicate the most comprehensive description of the estimator.
Several other estimators of relatedness exist that work in the presence of incomplete data (Broman & Weber 1998) or use information from loci containing dominant marker loci (Hardy 2003) or single nucleotide polymorphisms (SNPs, Glaubitz et al. 2003). However, when compared with using complete co-dominant microsatellite data these show relatively poor properties.
Usually, the allele frequencies required for relatedness estimation must be calculated from the sample under investigation, resulting in a positive covariance between the within-pair and within-sample allele frequencies and biasing relatedness estimates. For example, Wang (2002) demonstrated that, with samples of less than about 50 individuals, the R2_c estimator can have a 0.1 positive bias when used to estimate relationship information on unrelated individuals. To minimize the bias in allele frequencies, they should be calculated excluding the information from the pair currently under investigation (Queller & Goodnight 1989; Wang 2002). The sums of the powers of the estimated population allele frequencies are also biased and should be substituted with the exact formulae outlined by Crow & Kimura (1970; but see Wang 2002 for further details). A final source of bias in allele frequency estimation is due to the presence of relatives within the sample which introduces a positive covariance. In their study of co-ancestry in Iberian pigs, Toro et al. (2002) found large biases in the estimates of relatedness obtained when using molecular-based methodologies to calculate allele frequencies from the study sample, compared with those obtained using genealogy-based approaches. Furthermore, they found using simulation that the expected bias was greatly reduced when allele frequencies from the base population were used in place of sample estimates. It is much more difficult to address the bias introduced by relatives within the sample, because reducing it requires that estimates of relationships are available (Thomas et al. 2002; Wang 2004).
All the estimators may yield values of r or Φ and Δ outside their legitimate parameter spaces (0–1); a reflection of the large sample errors inherent in these methods (Ritland 1996a; Lynch & Ritland 1999; Wang 2002). Truncating the estimates so that they fall within the parameter space simply introduces bias (Milligan 2003) and offers few benefits.
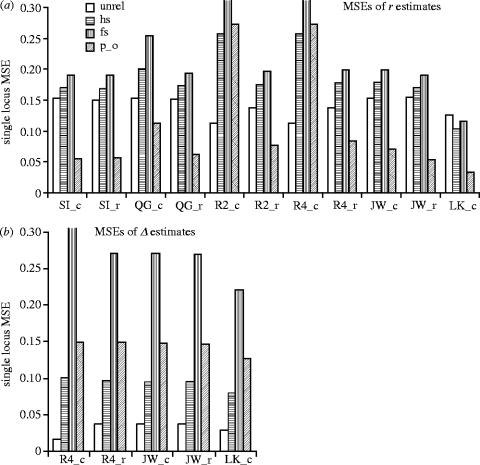
The main differences between the performances of the MOM estimators can be attributed to their different allele-specific weighting schemes. Optimal weights for any given pair are dependant upon their actual relationship, which is, obviously, not known in advance. Ritland (1996a) argued that, since the individuals of a pair, randomly selected from a sample, are most likely unrelated, it is reasonable to assume all pairs are unrelated and calculate weights accordingly. Although making such an assumption does not introduce any bias to an estimator, it does alter its efficiency. Since the weights are calculated under the assumption of no relationship, only measures of relatedness calculated from genuinely unrelated pairs are optimized. Relatedness measures calculated from full-sibling pairs, say, can be far from optimal (figure 1). The estimators may be organised with respect to the number of weights required for calculation as SI, QG, JW, R2 and R4, in ascending order. The greater the amount of weighting, the better the estimator performs for unrelated pairs, but the worse it performs for related pairs (reflected in the mean squared errors (MSE) of figure 1; see also results of Van De Casteele et al. 2001). The problem of decreased efficiency is compounded when there are rare alleles present due to the large weight placed on information from rare alleles (Ritland 1996a; Lynch & Ritland 1999); an effect most easily noted in full-sibling parameter estimation using Ritland's (1996a) estimators (R2_c & R4_c—which yield identical estimates of r). To minimize this error Ritland (1996a) suggested pooling alleles with a frequency of <0.05 into a single category. The problem of decreased weighting efficiency is further compounded in situations where weights are calculated using allele frequencies calculated from the sample of interest. In such situations it is even more important to follow the guidelines outlined above for dealing with allele frequency estimates. Similar problems, arising from the use of weights that are only optimal for unrelated pairs, are evident for the estimators of Φ and Δ (figure 1).
Figure 1.
Single locus mean squared errors (MSE) for estimates of (a) r and (b) Δ for the different relatedness estimators. Ten thousand pairs were simulated for each relationship class: unrelated (unrel), half-sibling (hs), full-sibling (fs) and parent–offspring (p_o). Marker information comprised 10 alleles in a triangular distribution. MSE value for r estimates from R2_c & R4_c estimators for fs=0.402. MSE value for Δ estimates from R4_c estimator for fs=1.04. Bias for likelihood estimation (LK_c): r-unrel=0.22, hs=0.14, fs=0.08 and p_o=0.06; Δ-unrel=0.03, hs=0.08, fs=0.07 and p_o=0.12.
In general, the regression forms of the estimators perform better than their correlation counterparts. Again, this is because there are relatively fewer weights incorporated into the regression estimators. Take, for example, a locus containing 10 alleles. The R2_c estimator requires 10 different weights optimized at r=0, while R2_r requires, at most, two weights.
In practice, it is unclear which of the estimators is best. To minimize the error over all possible pairs in most populations would require the use of R2_c or R4_c, since the vast majority of randomly chosen pairs are likely to be unrelated. However, if we are interested in estimating information from full-sibling pairs then the use of the SI estimator might be preferred. Simulation methods should be adopted in order to determine the most appropriate estimator for the proposed study population and allele distribution (Van De Castelle 2001).
(c) Likelihood-based relatedness measures
The likelihood of observing the genetic data of a given pair can be expressed in terms of Φ and Δ
| (2.2) |
(modified from Thompson 1975), where P(gl|no), P(gl|one) and P(gl|two)are the probabilities of the observed genotype at locus l, given that the pair share zero, one and two alleles that are identical by descent, respectively (table 2). Standard ML techniques may then be used to estimate values for Φ and Δ, given the observed marker data and the population allele frequencies. Generally speaking, ML methods show lower MSE than the MOM approaches but small datasets (i.e. few marker loci) yield very biased estimates (figure 1; see also results of Milligan (2003)). This observation is due to the fact that ML estimates, unlike MOM estimates, are constrained to lie within the legitimate parameter space. The bias approaches that of the MOM estimators with higher numbers of multi-allelic marker loci (e.g. 20+).
Table 2.
The probability of observing a pair of genotypes given the two (Φ) and four (Δ) gene coefficients.
| coefficient | |||
|---|---|---|---|
| pairwise genotype | 1−Φ−Δ | Φ | Δ |
| ii–ii | pi4 | pi3 | pi2 |
| ii–ij | 4pi3pj | 2pi2pj | 0 |
| ii–jj | 2pi2pj2 | 0 | 0 |
| ij–ij | 4pi2pj2 | pipj(pi+pj) | 2pipj |
| ii–jk | 4pi2pjpk | 0 | 0 |
| ij–ik | 8pi2pjpk | 2pipjpk | 0 |
| ij–kl | 0 | 0 | |
p represents a population allele frequency and i, j, k and l index mutually exclusive alleles.
The likelihood approach has mainly been used in a restricted manner to determine the probability of a pair falling into each of a number of different relationship categories (Mousseau et al. 1998). Subsequent analysis may then incorporate the probability information for each category or may assume that the pair belongs to the most likely category only and ignore uncertainty (Thomas & Hill 2000). In practice, relationships are assigned using likelihood ratio tests to test the support for the pair falling into one pre-specified category versus another. Statistical significance for the assigned relationship may be obtained using simulation techniques (Marshall et al. 1998; Goodnight & Queller 1999; Slate et al. 2000; García et al. 2002). Similar techniques may be adopted to assign specific relationships using the MOM estimates (Blouin et al. 1996), although these techniques are less intuitive than using ML.
Methods based on pairs are less efficient than those based on larger groups (Thomas & Hill 2002). For example, when analysing full-sibling groups, exclusion of incompatible sibling relationships is not possible on a strictly pairwise basis but is on a triplet-wise and greater basis. Of academic interest is the fact that one does not need to analyse groups larger than triplets in full-sibling group analyses because there is no excluded group of four that does not also contain at least one excluded triplet group. Hence, all exclusion information is contained within triplets (unpublished result). A great advantage of likelihood techniques is that they can be readily extended to jointly estimate a number of relationships, as well as account for errors in the marker data (Sieberts et al. 2002).
With increasing numbers of individuals, a full likelihood analysis of all the possible population structures becomes impractical and Markov chain Monte Carlo techniques (MCMC) need to be adopted to optimize structure (Painter 1997). These have been based on either a combination of multiple pairwise likelihoods (Smith et al. 2001), parent–offspring pairs and triplets (Almudevar 2003) or on the reconstruction of half or full-sibling relationships (Thomas et al. 2000; Smith et al. 2001; Thomas & Hill 2002; Wang 2004). Under limiting circumstances, for example, when examining a single generation of a population, the MCMC methodologies are superior to other methods (Wilson et al. 2003). Unfortunately, there is the inbuilt assumption that reconstructed pedigrees are true, since no indication of their accuracy is carried forward into subsequent analysis. Almudevar (2001) attempted to use bootstrap procedures to assign an accuracy statistic to reconstructed pedigrees but it is difficult to see how these statistics could then be used in subsequent analysis. For some purposes, such as variance component analysis, little bias is introduced by the assumption of pedigree accuracy, provided that pedigree errors are low (Thomas & Hill 2000). Such an assumption might be inappropriate for other analyses where sibling relationship sizes must be accurate (Chapman 2003).
Most of the MCMC methods also assume that the marker data is accurate; a potentially serious limitation since microsatellite data can contain a high percentage of errors and thereby severely reduce the accuracy of the reconstructed pedigree (Butler et al. 2004). More recent developments by Wang (2004) have shown how the MCMC methodology can be extended to account for marker errors, significantly improving sibling relationship accuracy. The MCMC approaches also require that allele frequency estimates from the parental generation be known (Thomas & Hill 2000). These can be estimated simultaneously with the pedigree through one of several suggested methodologies (Thomas & Hill 2000; Smith 2001; Wang 2004). Assuming a sibling-relationship analysis only and that the marker data contains errors, then Wang's (2004) method based on the likelihood of putative parental genotypes is the most appropriate, although the method of Thomas & Hill (2000) is general for all relationship structures.
An alternative method for pedigree reconstruction in a single generation was developed by Almudevar & Field (1999). This was based on using exclusions to sequentially build up putative sibling relationships, scoring them based on a pre-defined statistic. The major appeal of this method is that it is independent of allele frequency estimates. In comparison to other MCMC analyses, it proved equally effective at estimating a few large sibling relationship groups but less effective when estimating small groups where exclusion information was lower (Butler et al. 2004).
3. Combining with quantitiative data
(a) Method of moments heritability estimator
The first of the molecular-based methods of variance component estimation was developed by Ritland (1996b), and was based upon the regression of pairwise phenotypic similarity against the relationship (Grimes & Harvey 1980). The additive genetic variance is calculated as:
| (3.1) |
where is the variance of pairwise relationship and CZr is the covariance of pairwise relationship and phenotypic similarity (, with y1 and y2 being the phenotypic values for the individuals in the pair). The environmental variance (σE2) may then be estimated from the sample variance and heritability calculated accordingly (Falconer & MacKay 1996).
Replacing the actual value of the relationship with an estimate requires an additional step be added to the estimation procedure to account for the noise introduced through the use of estimated relationship parameters. Straightforward calculation of would include this noise, resulting in the parameter being overestimated. Ritland (1996b) therefore proposed an ANOVA to partition the variance of the relatedness into between and within locus components; the intraclass covariance providing an estimate of the actual variance of the relationship for use in equation (3.1).
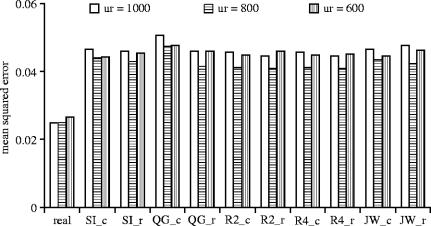
Obviously, any MOM estimators of relationship may be used in the regression framework outlined above. The additional error introduced through using estimated relationship information instead of known information outstrips any differences in the amount of error due to the choice of estimator (figure 2). In general though, the Queller & Goodnight (1989; QG_r) estimator performs least well overall and the Lynch & Ritland (1999; R4_r) estimator performs best. As the percentage of unrelated pairs increase and the amount of allele information decreases, the Ritland (1996a) estimators (R2_c or R4_c) perform best. ML may also be used to generate relatedness information, although its inherent bias can seriously bias subsequent heritability estimates. For example, with ML, relatedness is overestimated for both unrelated and full-sibling individuals, with unrelated individuals being more biased. Hence the phenotypic similarity of full-siblings is attributed to a smaller genetic similarity, biasing heritability upwards. In addition, the clever ANOVA, described by Ritland (1996b) to minimize the error introduced through estimating relatedness, cannot be used to analyse ML estimates because of the large biases shown by single-locus ML estimates. For the regression framework to be useful, estimates of relatedness must be either unbiased or all be biased by the same amount.
Figure 2.
MSE for heritability estimates made using relationship information derived from the different method of moment relatedness estimators. Two hundred replicates were simulated for each relationship structure. Different structures comprised 200 full-sibling pairs and varying numbers of unrelated pairs (ur=600, 800 and 1000). Marker information comprised 10 marker loci each with 10 alleles in a triangular distribution.
Ritland (1996b) extended his regression estimator to include the estimation of the dominance variance (requiring unbiased estimates of Δ) and the genetic covariance between traits. Curiously, his method for estimating the covariance may be further developed so that no marker data need be used at all (Lynch 2000). Unfortunately, the estimates produced are extremely noisy and require that unrealistic numbers of related pairs be present in the sample.
(b) Likelihood-based heritability estimation
The likelihood approach to variance component estimation was based on the assumption that the distribution of relationships within the population was known and the fact that the pair's phenotypic information also provides information on the relationship—the distribution of a pair's phenotypes is dependant on their relationship (Mouseau et al. 1998). The molecular marker and phenotypic information are combined to give the joint likelihood of the observed data:
| (3.2) |
where t indexes a particular pair, r indexes a particular class of relationship (e.g. full-sibling, half-sibling, unrelated), ar is the prior probability of a random pair sharing relationship r, mt|r is the likelihood of the molecular data given r (table 2) and zt|r is the probability density of the phenotypic data given r and the population parameters, such as the additive genetic variance, to be estimated. Several phenotypic distribution functions have been suggested but the one yielding the least biased results is based on the phenotypic difference of the pair (Thomas et al. 2000). Alternatively, the joint distribution of the pair's observed phenotypes can be regarded as following a multivariate normal distribution
| (3.3) |
where is the vector of trait means and A, D and E are the additive, dominance and environmental covariance matrices, respectively. This model allows parameters with more than one trait to be estimated simultaneously. As each pairwise likelihood is dependant upon the phenotypic parameters being estimated, a covariance is introduced that results in biases (Thomas et al. 2000). The size of the bias is dependant upon the relative contribution of the phenotypic information versus the genotype information, and the number of traits being analysed.
Since equation (3.2) does not represent the full likelihood of the population but only the product of pairwise likelihoods, standard methods of error estimation (from the matrix of second derivatives) are inappropriate. The best alternative is the bootstrap (Efron & Tibshirani 1993), using repeat sampling at the level of individuals (Thomas et al. 2002). In analyses where all pairwise combinations of individuals are being considered, repeat sampling of pairs leads to a gross underestimate of the standard error. The bootstrap may also be used to estimate standard errors for the above MOM approach.
A problem of pairwise approaches is their relatively poor weighting of family information and information from different relationship classes (Thomas & Hill 2002). Families are weighted by the number of pairs within which they are represented, thus, inappropriately large weights are placed on larger families. It is unclear exactly how different relationship classes are weighted under the pairwise methodologies. These problems helped motivate the use of MCMC reconstructed sibling relationships to estimate variance components. Under the assumption that MCMC reconstructed sibling relationships are accurate, standard methods for variance component analysis that weight family sizes and relationships appropriately may be adopted (e.g. restricted maximum likelihood methods; Lynch & Walsh 1998).
An appeal of the MCMC approaches is that they appear to be conservative in nature, favouring the non-assignment of a relationship (or assigning a lower relationship) to a related pair—type 2 errors—over assigning a relationship to a genuinely unrelated pair—type 1 errors (Thomas et al. 2000; Thomas & Hill 2002). Errors of type 2 do not greatly bias subsequent variance component estimates unless present in large numbers, while errors of type 1 can rapidly bias estimates. Simulations have shown that, even though biased, variance components estimated in this manner are, in general, more precise than components estimated using the pairwise approaches. However, the methodology has currently only been applied to single generation cohorts. Standard errors of variance components may be estimated in the traditional manner, but must be acknowledged to be underestimates because they ignore the uncertainty in the pedigree.
ML techniques may also be adopted to estimate variance components in hybrid situations when only part of the pedigree is unknown. If there is uncertainty in only a small part of the pedigree, it is feasible to attach a likelihood to each of the different possible pedigrees and maximize with respect to the parameters of interest (Foulley et al. 1990) using methods analogous to Mousseau et al. (1998). Another approach useful in situations where only maternal identities are known is to use paternity assignment to help reconstruct pedigrees (Kruuk et al. 2000; Milner et al. 2000). In both these studies, pedigrees were reconstructed using Cervus with paternities assigned different confidence levels (Marshall et al. 1998). The lower the set confidence level, the larger the number of relationships reconstructed and the larger the number of informative phenotypic contrasts upon which to base variance component estimates. However, a lower confidence interval also means a larger number of incorrect assignments and a larger bias (Thomas et al. 2002).
(c) Real data examples
There is a distinct lack of real-data examples of variance component estimation based solely upon marker-based information on relatedness. Ritland's (1996b) regression approach was used to estimate heritability in a wild plant population, Mimulus guttatus (Ritland & Ritland 1996). Resulting estimates were larger than those determined under more controlled conditions, a result contrary to expectation since environmental variance might be expected to be lower under these conditions (Coyne & Beecham 1987). Alternatively, the result may simply reflect the large sampling variance associated with this approach. The pairwise likelihood technique was applied to a captive salmon population (Oncorhynchus tshawytscha), resulting in heritability estimates that were similar to previously derived estimates (Mousseau et al. 1998). However, the salmon population was set up under rather specific conditions to allow prior information about the population structure to be determined.
Thomas et al. (2002) compared the three marker-based approaches when estimating the heritability of body weight in Soay sheep (Ovis aries). In addition, they contrasted these against estimates made when maternities were assumed known, but other relationships required estimation, and estimates derived from a pedigree determined using known maternal data and paternity inference. Results showed that the two pairwise approaches yielded inaccurate heritability estimates that were not significantly different from zero, even when ‘known’ relationships were used in the pairwise framework (arguably a reflection of the poor family and relationship-specific weights). The MCMC-based approaches estimated levels of heritability that were significantly greater than zero. However, the greater the number of relationships assigned the lower the estimate, which perhaps reflects the bias introduced through misassigned relationships. A second example study compared the regression and MCMC approaches using the rainbow trout (Oncorhynchus mykiss; Wilson et al. 2003). The population had a much higher average level of relatedness than the sheep population examined by Thomas et al. (2002) and, in addition, comprised a half-sibling/full-sibling structure that was more suited to MCMC analysis. Despite the advantage of a more appropriate population, results were qualitatively similar to Thomas et al. with MCMC analysis out-performing the regression estimator, which was biased and unreliable.
Understandably, variance components determined from pedigrees made up of a combination of known and marker-based relationships display the most convincing properties (Kruuk et al. 2000; Milner et al. 2000; Thomas et al. 2002). At present, it is likely that the role of marker-based techniques will be as a supplement to, rather than the replacement of, known (observed) relationships.
4. Discussion
A number of statistical tricks are available that help maximize the relationship information gained from marker-data, for example, weighting and minimizing errors in allele frequency estimates and their functions. For the purposes of variance component estimation, the regression-based relatedness estimator of Lynch & Ritland (1999) shows the most desirable properties over the widest range of marker-data. Ideally however, simulation should be used to check that this holds true for the particular population being studied. If information on population structure is known in advance, then likelihood approaches improve estimation by restricting relationship estimation to those classes defined by the structure. If data are known to come from a single generation, for example, a captive population of fishes, then the MCMC-based approaches improve estimation.
With expanding amounts of marker-data becoming readily available (e.g. with SNPs), it is conceivable that MCMC approaches will be expanded to account for more general population structures. This may take the form of reconstruction using triplet-wise likelihoods, which would probably offer a reasonable compromise between capturing useful full-sibling and parent–offspring exclusions and losing indirect information from the extended families, while still leaving the tractable problem.
With a further expansion to the amount of marker data available for analysis, two additional, but linked, concepts become important: the role of linkage between markers and the transition from estimating the expected relationship value, given the pedigree of expectation, to estimating the realized (actual) relationship value.
The statistical methods discussed here have all required that there is no linkage between the marker loci and, hence, each can be regarded as an independent estimate of the relatedness. In truth, loci may be linked and hence provide non-independent information. Accounting for linkage in adjacent loci using an ML framework is conceptually straightforward, requiring the inclusion of a function describing the probability of jointly inheriting a linked pair given the distance (recombination rate) between them (Boehnke & Cox 1997). Recently, a study by Leutenegger et al. (2003) demonstrated the use of a probability-based model to examine the inbreeding of an individual scored for a number of markers across their genome, given a known marker map. Their methodology allowed them to determine a distribution for inbreeding status at each locus in the individual. An equivalent method could be applied to a pair of individuals to examine the distribution of their relatedness. This approach would provide estimates of the relatedness specific to each genomic region. In addition, the distribution of the size of each region would also provide information on the degree of relationship, with more distant relationships tending to share smaller segments then closer relationship (Browning 1998; Zhao & Liang 2001).
Categorical ML and MCMC estimators of relationship assign a specific relationship and, hence, fixed values for Φ, Δ and r (e.g. Φ=0.5, Δ=0.25 and r=0.5 for full-siblings). It has long been recognized that these values are not fixed for a given class of relationship (except parent–offspring) but are merely expectations (Suarez et al. 1979; Donnelly 1983; Hill 1993; Bickeböller & Thompson 1996). ML and MCMC approaches actually estimate this expected relationship value, not the realized value for the relationship. Alternatively, MOM and continuous ML estimators determine a continuous measure of relatedness. At their limit, with full genotype data, they estimate the actual relatedness between two individuals. Consequently, once a certain level of marker information is attained, more accurate relationships will be estimated using the approaches that are least reliable with little marker-data.
Currently ‘actual’ relationship information derived from marker-loci is only being used in the context of variance-component estimation to search for QTL (Williams et al. 1997; George et al. 2000). By partitioning the phenotypic variance into components explained by (i) the expected relationship matrix and (ii) the realized relationship matrix derived from locus-specific identities between the individuals, an area thought to closely linked to a QTL can be determined. This approach has been recently applied to a natural population of red deer (Slate et al. 2002) whose pedigree had been originally determined through observation and marker-based paternity assignment. With complete marker information, theoretically similar partitioning models that explore phenotypes in terms of blocks of actual genome identity could be described, without requiring the expected relationship matrix.
The increased use of markers in the study of natural populations has made it feasible to detect heritability estimates in certain types of natural population. Comparison of real data and simulated studies, however, clearly indicate that not all populations are suitable for marker-based analysis. The most important single feature that allows for marker-based analysis is having adequate numbers of relations within the sample (Ritland 1996b). Clearly, the more individuals collected the greater the number of related pairs. Unfortunately, as sample size increases so too must the amount of marker-data typed per individual, because the probability of sampling two individuals of similar marker structure due to chance alone also increases. Thus, more marker-data is required to distinguish true relatives from pairs that are genetically similar at a few loci due to chance. Establishing the limits of the trade-off between marker number and sample size requires further theoretical and practical work.
Acknowledgments
I would like to thank Peter Visscher for his help and encouragement during the completion of this manuscript. S.C.T. works as a full-time medical doctor and is self-financed.
Appendix A
(a) Similarity index regression form
The regression form of the similarity index is derived in a similar manner to the correlation form (see Li et al. 1993). For this estimator, relationship estimates are defined as a ratio of the observed similarity of a pair corrected for similarity due to chance, and the expected similarity of an identical pair (i.e. 1), also corrected for similarity due to chance (Rousset 2002). Two forms of the regression form are available, depending upon whether the reference individual is homozygous or heterozygous (table 3). Since either individual of a pair may be the reference, unique locus-specific estimates are calculated as the arithmetic mean of both estimates.
Table 3.
Derivation of the regression form of the similarity index (SIr).
| pair-wise pattern | pattern frequency-given base (FF) | similarity (S) | FF×S |
|---|---|---|---|
| homozygous reference individual | |||
| ii–ii | pi2 | 1 | pi2 |
| ik–ii | 2pi(1-pi) | 3/4 | 3pi(1−pi)/2 |
| kk–ii | (1−pi)2 | 0 | 0 |
| similarity due to chance | |||
| SIr estimator: homozygous reference | |||
| heterozygous reference individual | |||
| ii–ij | pi2+pj2 | 3/4 | 3(pi2+pj2)/4 |
| ij-ij | 2pipj | 1 | 2pipj |
| il–ij | 1/2 | ||
| ll–ij | 0 | 0 | |
| similarity due to chance | |||
| SIr estimator: heterozygous reference |
Similarity is the pre-defined value for the similarity of a given genotype, k represents any allele that is not of type i and l represents any allele that is not of type i or j.
(b) Weighted allele specific estimator of r regression form
The weighted allele specific estimator of r is derived from a single locus form of the Queller & Goodnight (1989) estimator. By moving the summation terms to outside the division and including allele specific weights, the locus-specific estimator becomes
| (A1) |
where a indexes the different alleles observable at the locus in the reference individual, pa1 and pa2 are the allele frequency of a in the proband and the reference individual, respectively, and is the population frequency of a. When the reference individual is homozygous wa=1. In the heterozygous case, allele-specific weights can be derived by assuming that r=0 using the method outlined in Ritland (1996a), and are equal to
| (A2) |
where and are the allele frequencies of the first and second alleles in the reference individual and b indexes the other reference allele to a (i.e. b=3−a).
(c) Wang joint estimator of Φ and Δ regression form
The regression form of Wang's (2002) joint estimator is derived in a similar manner to the correlation form. With a homozygous base individual three alternative pairwise genotype patterns are observable (table 4). The probability of observing these patterns are
| (A3) |
| (A4) |
and
| (A5) |
(table 4). By setting the equation corresponding to an observed pairwise genotype pattern to one, and the other equations to zero, a locus-specific estimate of Φ and Δ can be derived by solving any two of the equations (since A 3+A 4+A 5=1).
Table 4.
Probabilities of observing the different pairwise genotype patterns given the two- and four-gene relatedness coefficients. (, where p indicates an allele frequency, n indicates the number of alleles at the locus, k represents any allele that is not of type i and l represents any allele that is not of type i or j. Entries are derived from table 2.)
| pairwise pattern | probability of reference individual | probability of observed pattern coefficient | ||
|---|---|---|---|---|
| 1−Φ−Δ | Φ | Δ | ||
| homozygous reference individual | ||||
| ii–ii | a2 | a4 | a3 | a2 |
| ik–ii | a2 | 2(a3−a4) | a2−a3 | 0 |
| kk–ii | a2 | a2−2a3+a4 | 0 | 0 |
| heterozygous reference individual | ||||
| ii–ij | 1−a2 | 2(a3−a4) | a2−a3 | 0 |
| ij–ij | 1−a2 | 2(a22−a4) | a2−a3 | 1−a2 |
| il–ij | 1−a2 | 0 | ||
| ll–ij | 1−a2 | 0 | 0 | |
When considering a heterozygous reference individual, four alternative pairwise genotypes are observable (table 4). Hence, no closed solution is obtainable since there are more independent probability equations than parameters. A notable exception to this is for biallelic loci where there is only one ‘independent’ equation and no solution is available. The weighting approach of Ritland (1996a) can be adopted to solve more than two independent equations, with weights being derived numerically under the assumption Φ=Δ=0.
Footnotes
One contribution of 16 to a Theme Issue ‘Population genetics, quantitative genetics and animal improvement: papers in honour of William (Bill) Hill’.
References
- Almudevar A. A bootstrap assessment of variability in pedigree reconstruction based on genetic markers. Biometrics. 2001;57:757–763. doi: 10.1111/j.0006-341x.2001.00757.x. [DOI] [PubMed] [Google Scholar]
- Almudevar A. A simulated annealing algorithm for maximum likelihood pedigree reconstruction. Theor. Popul. Biol. 2003;63:63–75. doi: 10.1016/s0040-5809(02)00048-5. [DOI] [PubMed] [Google Scholar]
- Almudevar A, Field C. Estimation of single generation sibling relationships based on DNA markers. J. Agric. Biol. Environ. Stat. 1999;4:136–165. [Google Scholar]
- Avise J.C.Molecular markers, natural history and evolution1st edn.1994Chapman & Hall; New York [Google Scholar]
- Barburjani J. Autocorrelation of gene frequencies under isolation by distance. Genetics. 1987;117:777–782. doi: 10.1093/genetics/117.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickeböller H, Thompson E.A. Distribution of genome shared IBD by half-sibs: approximation by the poisson clumping heuristic. Theor. Popul. Biol. 1996;50:66–90. doi: 10.1006/tpbi.1996.0023. [DOI] [PubMed] [Google Scholar]
- Blouin M.S. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol. Evol. 2003;18:503–511. [Google Scholar]
- Blouin M.S, Parsons M, Lacaille V, Lotz S. Use of microsatellite loci to classify individuals by relatedness. Mol. Ecol. 1996;5:393–401. doi: 10.1111/j.1365-294x.1996.tb00329.x. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox N.J. Accurate inference of relationships in sib-pair linkage studies. Am. J. Hum. Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W, Weber J.L. Estimation of pairwise relationships in the presence of genotyping error. Am. J. Hum. Genet. 1998;63:1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. Relationship information contained in gamete identity by descent data. J. Comput. Biol. 1998;5:323–334. doi: 10.1089/cmb.1998.5.323. [DOI] [PubMed] [Google Scholar]
- Butler K, Field C, Herbinger C.M, Smith B.R. Accuracy, efficiency and robustness of four algorithms allowing full sibship reconstruction from DNA marker data. Mol. Ecol. 2004;13:1589–1600. doi: 10.1111/j.1365-294X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- Chapman R.E, Wang J, Bourke A.F.G. Genetic analysis of spatial foraging patterns and resource sharing in bumble bee pollinators. Mol. Ecol. 2003;12:2801–2808. doi: 10.1046/j.1365-294x.2003.01957.x. [DOI] [PubMed] [Google Scholar]
- Coyne J.A, Beecham E. Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics. 1987;117:727–737. doi: 10.1093/genetics/117.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J, Kimura M.An introduction to population genetics1970Burgess; Minneapolis [Google Scholar]
- Dobson F.S, Chesser R.K, Hoogland J.L, Dugg D, Foltz D.W. Breeding groups and gene dynamics in a socially structured population of prairie dogs. J. Mamm. 1998;79:671–680. [Google Scholar]
- Donnelly K.P. The probability that related individuals share some section of the genome identical by descent. Theor. Popul. Biol. 1983;23:34–64. doi: 10.1016/0040-5809(83)90004-7. [DOI] [PubMed] [Google Scholar]
- Eding H, Meuwissen T.H.E. Marker-based estimates of between and within population kinships for the conservation of genetic diversity. J. Anim. Breed. Genet. 2001;118:141–159. [Google Scholar]
- Efron B, Tibshirani R.J.An introduction to the bootstrap1993Chapman & Hall; New York [Google Scholar]
- Falconer D.S, MacKay T.F.C. An introduction to quantitative genetics. 4th edn. Longman; Harlow: 1996. [Google Scholar]
- Foulley J.L, Thompson R, Gianola D. On sire evaluation with uncertain paternity. Genet. Sel. Evol. 1990;22:373–376. doi: 10.1186/1297-9686-19-1-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García D, Carleos C, Parra D, Cañón J. Sib-parentage testing using molecular markers when parents are unknown. Anim. Genet. 2002;33:364–371. doi: 10.1046/j.1365-2052.2002.00882.x. [DOI] [PubMed] [Google Scholar]
- George A.W, Visscher P.M, Haley C.S. Mapping quantitative trait loci in complex pedigrees: a two step variance components approach. Genetics. 2000;156:2081–2092. doi: 10.1093/genetics/156.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz J.A, Rhodes E.J.R, Dewoody J.A. Prospects for inferring pairwise relationships with single nucleotide polymorphisms. Mol. Ecol. 2003;12:1039–1047. doi: 10.1046/j.1365-294x.2003.01790.x. [DOI] [PubMed] [Google Scholar]
- Goodnight K, Queller D.C. Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol. Ecol. 1999;8:1231–1234. doi: 10.1046/j.1365-294x.1999.00664.x. [DOI] [PubMed] [Google Scholar]
- Grimes L.W, Harvey W.R. Estimation of genetic variances and covariances using symmetric differences squared. J. Anim. Sci. 1980;50:634–644. [Google Scholar]
- Hardy O.J. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol. Ecol. 2003;12:1577–1588. doi: 10.1046/j.1365-294x.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Hill W.G. Variation in genetic composition in back-crossing programs. J. Hered. 1993;84:212–213. [Google Scholar]
- Jacquard A.The genetic structure of populations1974Springer; New York [Google Scholar]
- Kruuk L.E.B, Clutton-Brock T.H, Slate J, Pemberton J.M, Botherstone S, Guinness F.E. Heritability of fitness in a wild mammal population. Proc. Natl Acad. Sci. USA. 2000;97:698–703. doi: 10.1073/pnas.97.2.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Shannon S. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution. 1996;50:434–437. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- Leuteneggar A, Prum B, Génin E, Verny C, Lemainque A, Clerget-Darpoux F, Thompson E.A. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.C, Weeks D.E, Chakravarti A. Similarity of DNA fingerprints due to chance and relatedness. Hum. Hered. 1993;43:45–52. doi: 10.1159/000154113. [DOI] [PubMed] [Google Scholar]
- Lynch M. Estimation of relatedness by DNA fingerprinting. Mol. Biol. Evol. 1988;5:584–599. doi: 10.1093/oxfordjournals.molbev.a040518. [DOI] [PubMed] [Google Scholar]
- Lynch M. Estimating genetic correlations in natural populations. Genet. Res. 2000;7:255–264. doi: 10.1017/s0016672399004243. [DOI] [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B.Genetics and analysis of quantitative traits1998Sinauer; Sunderland, MA [Google Scholar]
- Marshall T.C, Slate J, Kruuk L.E.B, Pemberton J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Meagher T.R. Analysis of paternity within a natural population of Chamaelirium luteum. 1. Identification of most-likely male parents. Am. Nat. 1986;128:199–215. [Google Scholar]
- Milligan B.G. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J, Pemberton J.M, Botherstone S, Albon S.D. Estimating variance components and heritabilities in the wild: a case study using the ‘animal model’ approach. J. Evol. Biol. 2000;13:804–813. [Google Scholar]
- Mousseau T.A, Ritland K, Heath D.D. A novel method for estimating heritability using molecular markers. Heredity. 1998;80:218–224. [Google Scholar]
- Painter I. Sibship reconstruction without parental information. J. Agric. Biol. Environ. Stat. 1997;2:212–229. [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Ritland K. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 1996a;67:175–185. [Google Scholar]
- Ritland K. A marker-based method for inferences about quantitiative inheritance in natural populations. Evolution. 1996b;50:1062–1073. doi: 10.1111/j.1558-5646.1996.tb02347.x. [DOI] [PubMed] [Google Scholar]
- Ritland K, Ritland C. Inferences about quantitative inheritance based on natural population structure in the yellow monkey flower, Mimulus guttatus. Evolution. 1996;50:1074–1082. doi: 10.1111/j.1558-5646.1996.tb02348.x. [DOI] [PubMed] [Google Scholar]
- Robertson A, Hill W.G. Deviations from Hardy–Weinberg proportions: sampling variances and use in estimation of inbreeding coefficients. Genetics. 1984;l07:703–718. doi: 10.1093/genetics/107.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Inbreeding and relatedness coefficients: what do they measure? Heredity. 2002;88:371–380. doi: 10.1038/sj.hdy.6800065. [DOI] [PubMed] [Google Scholar]
- Sieberts S.K, Wijsman E.M, Thompson E.A. Relationship inference from trios of individuals in the presence of typing error. Am. J. Hum. Genet. 2002;20:170–180. doi: 10.1086/338444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J, Marshall T.C, Pemberton J.M. A retrospective assessment of the paternity inference program CERVUS. Mol. Ecol. 2000;9:801–808. doi: 10.1046/j.1365-294x.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Slate J, Visscher P.M, MacGregor S, Stevens D, Tate M.L, Pemberon J.M. A genome scan for quantitative trait loci in a wild population of red deer (Cervus elaphus) Genetics. 2002;162:1863–1873. doi: 10.1093/genetics/162.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.R, Herbinger C.M, Merry H.R. Accurate partition of individuals into full-sib families from genetic data without parental information. Genetics. 2001;158:1329–1338. doi: 10.1093/genetics/158.3.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storfer A. Quantitative genetics: a promising approach for the assessment of genetic variation in endangered species. Trends Ecol. Evol. 1996;11:343–348. doi: 10.1016/0169-5347(96)20051-5. [DOI] [PubMed] [Google Scholar]
- Suarez B.K, Reich T, Fishman P.M. Variability in sib pair identity. Hum. Hered. 1979;29:37–41. doi: 10.1159/000153013. [DOI] [PubMed] [Google Scholar]
- Surridge A.K, Ibrahim K.M, Bell D.J, Webb N.J, Rico C, Hewitt G.M. Fine-scale genetic structuring in a natural population of European wild rabbits (Oryctolagus cuniculus) Mol. Ecol. 1999;8:299–307. doi: 10.1046/j.1365-294x.1999.00570.x. [DOI] [PubMed] [Google Scholar]
- Thomas S.C, Hill W.G. Estimating quantitative genetic parameters using sibships reconstructed from marker data. Genetics. 2000;155:1961–1972. doi: 10.1093/genetics/155.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.C, Hill W.G. Sibship reconstruction in hierarchical population structures using Markov chain Monte Carlo techniques. Genet. Res. 2002;79:227–234. doi: 10.1017/s0016672302005669. [DOI] [PubMed] [Google Scholar]
- Thomas S.C, Pemberton J.M, Hill W.G. Estimating variance components in natural populations using inferred relationships. Heredity. 2000;84:427–436. doi: 10.1046/j.1365-2540.2000.00681.x. [DOI] [PubMed] [Google Scholar]
- Thomas S.C, Coltman D.W, Pemberton J.M. The use of marker-based relationship information to estimate the heritability of body weight in a natural population: a cautionary tale. J. Evol. Biol. 2002;15:92–99. [Google Scholar]
- Thompson E.A. The estimation of pairwise relationships. Ann. Hum. Genet. 1975;39:173–188. doi: 10.1111/j.1469-1809.1975.tb00120.x. [DOI] [PubMed] [Google Scholar]
- Toro M, Barragán C, Óvilo C, Rodrigañez J, Rodriguez C, Silió L. Estimation of coancestry in Iberian pigs using molecular markers. Conserv. Genet. 2002;3:309–320. [Google Scholar]
- Van De Casteele T, Galbusera P, Matthysen E. A Comparison of microsatellite-based pairwise relatedness estimators. Mol. Ecol. 2001;10:1539–1549. doi: 10.1046/j.1365-294x.2001.01288.x. [DOI] [PubMed] [Google Scholar]
- Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.T, Duggirala R, Blangero J. Statistical properties of a variance components method for quantitative trait linkage analysis in nuclear families and extended pedigrees. Genet. Epidemiol. 1997;14:1065–1070. doi: 10.1002/(SICI)1098-2272(1997)14:6<1065::AID-GEPI84>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Wilson A.J, McDonald G, Moghadam H.K, Herbinger C.M, Ferguson M. Marker-assisted estimation of quantitative genetic parameters in rainbow trout Oncorhynchus mykiss. Genet. Res. 2003;81:145–156. doi: 10.1017/s0016672302006055. [DOI] [PubMed] [Google Scholar]
- Zhao H, Liang F. On relationship inference using gamete identity by descent data. J. Comput. Biol. 2001;8:191–200. doi: 10.1089/106652701300312940. [DOI] [PubMed] [Google Scholar]