Abstract
Recent advances in human genomics have made it possible to better understand the genetic basis of disease. In addition, genetic association studies can also elucidate the mechanisms by which ‘non-genetic’ exogenous and endogenous exposures influence the risk of disease. This is true both of studies that assess the marginal effect of a single gene and studies that look at the joint effect of genes and environmental exposures. For example, gene variants that are known to alter enzyme function or level can serve as surrogates for long-term biomarker levels that are impractical or impossible to measure on many subjects. Evidence that genetic variants modify the effect of an established risk factor may help specify the risk factor's biologically active components. We illustrate these ideas with several examples and discuss design and analysis challenges, particularly for studies of gene–environment interaction. We argue that to increase the power to detect interaction effects and limit the number of false positive results, large sample sizes will be needed, which are currently only available through planned collaborative efforts. Such collaborations also ensure a common approach to measuring variation at a genetic locus, avoiding a problem that has led to difficulties when comparing results from genetic association studies.
Keywords: genetic epidemiology, gene–environment interaction, study design, Mendelian randomization
1. Introduction
Tremendous advances in genomics, population genetics and genotyping technology over the last few years have dramatically improved our ability to test for associations between genes and disease in order to better understand how genetic variation correlates with variation in risk of disease. What is perhaps less immediately clear is that genetic association studies can also tell us something about traditional environmental risk factors such as exposure to carcinogens (e.g. smoking), lifestyle (e.g. physical activity) and physical characteristics (e.g. body mass index).
For example, if a particular exposure is difficult to measure accurately or too expensive to measure on a large number of subjects, we can study a gene that influences an intermediate phenotype that lies on the causal pathway between the exposure and disease. Finding an association between that gene and disease will help build the case for a causal role for the environmental exposure. We can also use what we know about genes to help dissect exposures that are complex mixtures of diverse components. There are many chemicals in cigarette smoke or in well-cooked red meat—which of them drives risk of colorectal cancer? By looking to see whether disease risk in exposure categories differs by genotype for a gene that encodes an enzyme that metabolizes a specific substrate, we can infer that that particular substrate plays a causal role in the risk of disease. Further, if we believe that individuals who carry a particular variant are more sensitive to the environmental exposure, we might focus on carriers to test whether that exposure is associated with disease risk (or focus on exposed subjects to test whether carriers of a particular genotype are at increased risk). Knowing how risk of disease varies across strata defined by genotype and exposure may also help suggest individualized treatment of disease. Pharmacogenetics is a particular example of this where ‘exposure’ is the drug dose. Finally, it has been suggested that detailed knowledge of the risks to particular gene-exposure strata could be used to provide personalized prevention, although genetic information may have poor predictive value for the modest effects anticipated in complex disease and the widespread use of genetic testing raises social and ethical concerns.
To further illustrate these ideas, we start with a brief review of the concept of ‘gene–environment interaction’, which we use loosely to mean the joint effect of genes and environment. We then comment briefly on statistical versus biological interaction modelling. We review available study designs for genetic association studies with particular emphasis on estimating joint gene–environment effects and close with a discussion of future trends.
2. Gene–environment interaction
The modern concept of gene–environment interaction dates back at least to the beginning of the twentieth century, before the discovery of DNA. In a 1902 landmark paper arguing that differences in chemical metabolism were inherited according to the principles of Mendelian genetics, Archibald Garrod noted that “such slight peculiarities of metabolism will necessarily be hard to trace by methods of direct analysis and will readily be masked by the influences of diet and of disease” (Garrod 1902, p.1620). He also suggested a genetic basis for “idiosyncrasies as regards drugs and the various degrees of natural immunity against infections” (Garrod 1902, p.1620).
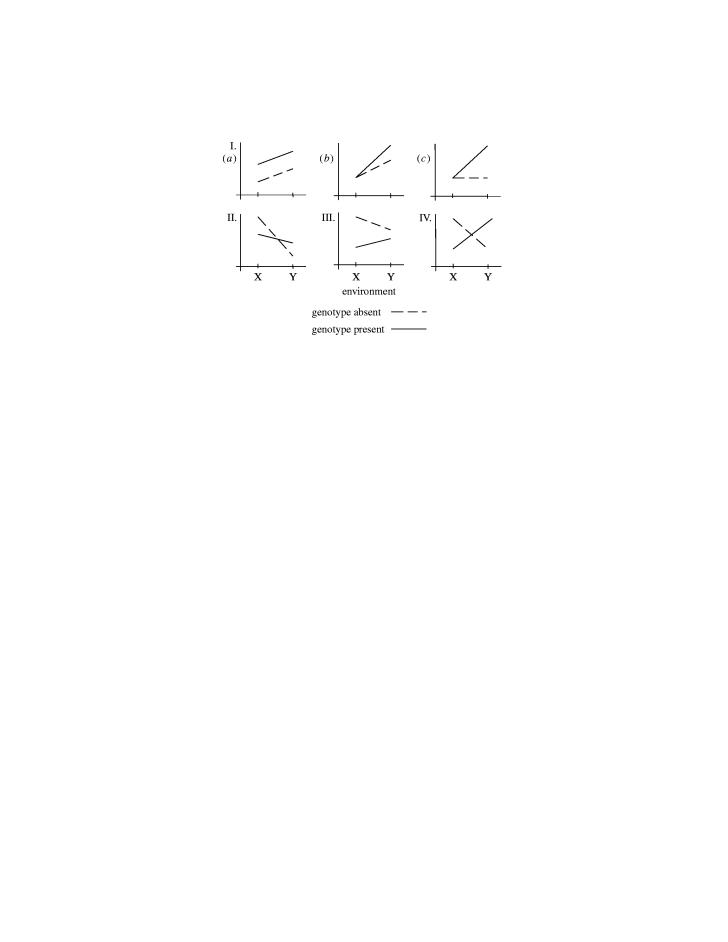
In his popular 1938 book debunking eugenic sterilization policies then in vogue in Western Europe and the United States, J. B. S. Haldane presented simple conceptual models for the ‘interaction of nature and nurture,’ shown in figure 1 (Haldane 1938). Under interaction model I, one of two genetically distinct populations (A and B) always has a higher trait value (mean height, weight, disease incidence, etc.) than the other, regardless of environment. Haldane gave the example of two breeds of dogs, mastiffs and dachshunds, under lean (X) and plentiful diets (Y)—the mastiff will be heavier than the dachshund in either environment. Under interaction model II, however, the relative position of the two populations changes depending on the environment. Haldane again turned to animal husbandry to illustrate. “Let A be Jersey cattle and B Highland cattle. Let X be a Wiltshire dairy meadow and Y a Scottish moor. On the English pasture the Jersey cow will give a great deal more milk than the highland cow. But on the Scottish pasture the order will probably be reversed. The Highland cow will give less milk than in England. But the Jersey cow will give still less. In fact, it is very likely that she will give none at all.”
Figure 1.
Four qualitative patterns of gene–environment interaction described by (and numbered after) Haldane (1938). The y-axis represents a trait value (e.g. mean height, disease prevalence or expected survival); the x-axis represents two environmental conditions.
Over the next fifty years many human diseases were discovered to follow the patterns outlined by Haldane, as reviewed in part by Khoury et al. (1988). For example (model I.b), exposure to sunlight increases the risk of skin cancer in all people, but the increase in risk is greater in xeroderma pigmentosa (XPD) patients. It is clear that gene–environment interaction is ubiquitous in the development of human traits, including disease, although most of the effects will be far more subtle than Haldane's farm animals or XPD and skin cancer.
3. Statistical measures and tests of interaction
For simple dichotomous genotypes (e.g. carrier versus non-carrier) and exposures (e.g. ever/never smoker) it is practical and useful to calculate stratum-specific trait summaries: for continuous traits, mean trait values for each gene–environment cross-classification (table 1); for binary traits, absolute incidence rates (if available) or relative measures such as relative risks or odds ratios (table 2). This presentation has the advantage of being ‘closest to the data’ (in the case of table 2 actually reporting the raw data) while allowing the reader to quickly assess the joint action of genes and environment (Botto & Khoury 2004). Formal statistical tests for gene–environment interaction—which are actually tests for departure from a specific statistical model for interaction—are less useful here, as (i) the test depends on the trait measurement scale (e.g. raw or log-transformed measurements for continuous traits; incidence or relative risks for binary traits; Greenland & Rothman 1998; Botto & Khoury 2004) and (ii) formal rejection or retention of a statistical model may yield little insight, as multiple (potentially contradictory) biological models can be consistent with the same statistical model for interaction (Thompson 1991; Cordell 2002).
Table 1.
Mean trait values by gene–environment stratum.
| exposure | ||
|---|---|---|
| genotype | 0 | 1 |
| 0 | (s.d.00) | (s.d.01) |
| 1 | (s.d.10) | (s.d.11) |
Dichotomous exposure (1, exposed) and genotype coding (1, carrier of minor allele); Y¯ is mean trait in gene–environment stratum ij; s.d.ij is the standard deviation of the trait.
Table 2.
Odds ratios from an unmatched case–control study by gene–environment stratum.
| case | control | OR | |
|---|---|---|---|
| G=0, E=0 | n00 | m00 | 1 (ref) |
| G=0, E=1 | n01 | m01 | OR01 (CI01) |
| G=1, E=0 | n10 | m10 | OR10 (CI10) |
| G=1, E=1 | n11 | m11 | OR11 (CI11) |
After Botto & Khoury (2004). Odds ratios are calculated relative to unexposed non-carriers (E=0, G=0). For example, the usual cross product estimate for OR10 is n10m00/(n00m10). CIij is the confidence interval for ORij.
It is impractical to fit such stratified models for more finely cross-classified data (multiple exposure categories, multi-allelic markers such as haplotypes of linked single nucleotide polymorphisms (SNPs) or multiple genes) as many strata will have few observations, leading to highly variable (or inestimable) strata (Robins & Greenland 1986; Botto & Khoury 2004). Here it seems some sort of statistical modelling will be indispensable. For example, a hierarchical model, which treats the ‘first-level’ stratum-specific parameters as random variables and then regresses these on ‘second-stage’ variables (e.g. groupings of genes based on function or decompositions of environmental exposures into their biologically active components) could be fit (Aragaki et al. 1997; Hung et al. 2004). Various levels of detail can be included in the second-stage model, improving the model fit if the details are accurate but harming the model fit if they are not. Alternatively, a space of potential working models based on simple ‘main effects plus cross-product interaction’ parameterizations can be explored and summarized using Bayesian model-selection and model-averaging techniques (Conti et al. 2003). The principal aim of both these approaches is to minimize prediction error (i.e. to reduce over-fitting) while producing parsimonious and useful summaries of the data; they do not necessarily aim to estimate parameters with direct biological meaning. Another approach (dubbed ‘toxikinetic modelling’) builds very detailed models for the joint action of genes and environmental exposures (perhaps including external information on substrate-specific kinetics for different enzyme isoforms; Conti et al. 2003; Cortessis & Thomas 2004); parameter estimates have an immediate biological interpretation but are necessarily model-specific.
If the primary scientific question is not ‘how do this gene and this environmental exposure jointly affect trait distribution?’ but simply ‘does this gene or this environmental exposure affect trait distribution?’ then it will often suffice to test the marginal association between a gene and disease—even if the principal focus is on the environmental factor. This is the idea behind ‘Mendelian randomization’, illustrated in figure 2a (Clayton & McKeigue 2001; Brennan 2004; Thomas & Conti 2004). If an environmental exposure influences the risk of disease through an internal phenotype that is itself influenced by variation in a known gene, then the association between the exposure and disease can be tested by examining the association between the gene and the disease. This approach—apparently first proposed to test the causal relationship between serum cholesterol and cancer by studying variants in the apolipoprotein A (APOE) gene (Katan 1986; Keavney 2004)—has several potential advantages: accurate measurements of the environmental exposure may be unavailable or prohibitively expensive, and genotypes are not susceptible to recall bias and other forms of confounding seen in case–control studies. However, other sources of bias are possible in genetic association studies (Thomas & Conti 2004). If differences in allele frequencies and disease rates are correlated across subpopulations, a significant association need not imply a causal relationship between the gene (or the environmental exposure) and disease (this is known as population stratification bias; Thomas & Witte 2002; Wacholder et al. 2002). Further, the gene may influence several internal phenotypes, so the association between variation in the gene and disease may not be due to the same internal phenotype being affected by the environmental exposure (see figure 2b); again, a significant association between the gene and the trait may not indicate a causal role for the exposure (mediated through the internal phenotype).
Figure 2.
A cartoon depiction of ‘Mendelian randomization’ after Thomas & Conti (2004). In scenario (a), finding an (induced) association between the gene (G) and disease (D) supports the hypothesis for a causal relationship between environmental exposure (E) and disease. In scenario (b), an association between the gene and disease gives no information about the causality of the exposure. IP, internal phenotype.
4. Available study designs
Until recently, many studies of genetic susceptibility to disease have collected limited (if any) information on environmental exposures. Similarly, traditional epidemiologic studies have collected detailed information on exposure but have not collected blood samples or other sources of DNA that would allow joint study of genes and environment. Now, with a greater focus on multifactoral and common complex diseases, there is increased awareness of the need to collect high-quality information on both genes and environmental exposures in a population-based context (Thomas 2000).
Table 3 summarizes the three main genetic association designs—retrospective case–control, prospective cohort and family-based—in terms of characteristics relevant to the study of gene–environment interaction: their susceptibility to population stratification bias, recall bias, survivor bias, the availability of prospectively collected plasma phenotypes (or other relevant biomarkers) and the required sample sizes. We briefly summarize these designs and their characteristics here; more detailed comparisons have been provided by Caparaso et al. (1999), Langholz et al. (1999) and Garcia-Closas et al. (2004).
Table 3.
Select characteristics of well-established designs for gene–environment interaction.
| characteristic | study design | ||
|---|---|---|---|
| family-based | case–control | cohort | |
| potential for population stratification bias | nil if appropriately analysed | varies; able to be minimized via good design, genomic control | varies but generally less than retrospective case–control study; able to be minimized via good design, genomic control |
| potential for recall bias | moderate to high | moderate to high | nil |
| potential for survivor bias | moderate to high | moderate to high | nil to moderate if DNA is not obtained on all cases and controls at base-line |
| ability to use plasma phenotypes in cases | no | no | yes |
| required sample sizes achievable? | common disease: yes | common disease: yes | common disease: yes with adequate follow up |
| rare disease: yes | rare disease: yes | rare disease: no, unless multiple studies are pooled | |
(a) Family-based designs
Family-based association designs use the assumed Mendelian transmission of alleles from parents to offspring to test and estimate the association between genes and traits (Laird et al. 2000; Weinberg & Umbach 2000). Because they condition on observed parental genotypes (or an appropriate sufficient statistic in case the parental genotypes are missing, as in case–sibling control studies) and rely on departures from Mendelian transmission, they are immune to population stratification bias (if appropriately analysed). In some realistic situations, family-based tests of gene–environment interaction may be more powerful than analogous tests in population-based studies (Gauderman 2001). However, it may be more difficult to collect genetic information on parents (especially for late-onset disease) or find appropriate sibling controls (Witte et al. 1999; Weinberg & Umbach 2000), and family-based tests generally have less power for genetic main effects than a population-based case–control study with the same number of genotyped subjects.
Since information on environmental exposures (and genotypes) is usually collected retrospectively, family-based studies share problems of recall bias and survivor bias with retrospective case–control studies.
(b) Case–control designs
In retrospective case–control studies, data on environmental exposures and samples for DNA and biomarker studies are obtained after diagnosis of disease in the cases. Selection bias occurs when controls do not represent the population in which the cases occurred (e.g. hospital-based controls); survival bias occurs when the cases that can be interviewed or genotyped differ systematically from those who cannot (e.g. cases with a particularly lethal genetic form of the disease die before they can be enrolled in the study). Population stratification bias can arise if the ethnicity of the controls is substantially different from that of the cases and the allele frequencies of the variants being tested also vary by ethnicity. In many cases, large bias due to population stratification can be eliminated by following basic principles of good study design and matching on self-reported ethnicity (Wacholder et al. 2000, 2002; Cardon & Palmer 2003; Reiner et al. 2005). However, this may not suffice for recently mixed populations, such as African or Hispanic Americans (Kittles et al. 2002; Thomas & Witte 2002), and even in relatively homogeneous populations such as non-Hispanic European Americans small biases cannot be ruled out (Freedman et al. 2004)—which is relevant as the effects of many genes underlying complex traits may themselves be small. ‘Genomic control’ methods that test and adjust for population stratification are available, although they require subjects be genotyped on a second panel of putatively anonymous markers (Devlin & Roeder 1999; Pritchard et al. 2000; Reich & Goldstein 2001; Tang et al. 2005)—preferably ‘ancestry informative markers,’ markers that are known to have different allele frequency in ancestral populations, for example, Spanish, Native Americans and Africans from Hispanic Americans (Bonilla et al. 2004).
The major problem with respect to gene–environment interactions is likely to be misclassified information on environmental exposures. ‘Recall bias’ can arise if cases report their pre-diagnosis exposure histories differently after their diagnosis relative to what they would have reported prior to diagnosis. Although this form of misclassification may not bias the estimates of certain gene–environment interaction parameters (Garcia-Closas et al. 1998), it will certainly bias the estimates of environmental ‘main effects’ and reduce the power to detect interactions.
Finally, it is difficult to assess the effect of such biomarkers (which might in principle represent better measurements of long-term environmental exposure, e.g. plasma nutrient levels) on disease risk, or the effect of genes on biomarker levels in cases because any biomarkers are collected after disease diagnosis and temporality is impossible to establish—did altered biomarker levels lead to disease or vice versa?
(c) Cohort designs
For prospective cohort studies, information on environmental exposures is collected at the base-line (and ideally at repeated subsequent follow-up intervals) on a large number of disease-free subjects. DNA and biomarker information should also be collected at base-line, although DNA for nested case–control studies could be obtained from cases (and matched controls) as soon as possible after diagnosis for existing prospective studies without banked samples (although this creates the potential for survivor bias). Effective follow-up and the prospective collection of data on genes and environmental exposures should minimize or eliminate selection, survivor and recall biases. If follow-up and participation in nested case–control studies does not differ by ethnicity, then the potential for population stratification bias is reduced relative to retrospective case–control studies, although it may still be a concern in some populations and require the use of appropriate genomic control methods.
The principal difficulty with prospective studies is the large number of subjects who must be enrolled at the base-line in order to ensure an adequate number of cases for reliable analysis. This limits cohorts to the study of relatively common diseases such as myocardial infarction or the more common cancers. Prospective studies would not yield sufficient power to study rare diseases. Furthermore, prospective studies may be less practical when the focus is on a particular subtype of disease, for example, tumours with similar gene expression profiles (Carr et al. 2004). Not only are these subtypes by definition less common, but it may also be difficult to obtain the fresh tissue necessary to classify cases.
(d) General design concerns
Whether formally testing for gene–environment interaction, estimating stratum-specific parameters or using statistical learning machinery to explore unsuspected gene–environment combinations, sample size is a major limiting factor in the study of gene–environment interaction. A rule of thumb says that the sample size necessary to depart from a multiplicative model for the joint effect of two variables (on the odds ratio or relative risk scale) is at least four times the sample size needed to evaluate the main effect of either of the variables (Smith & Day 1984). Given that environmental exposures are almost certainly measured with some error, the necessary sample sizes will be even larger (Garcia-Closas et al. 1999; Wong et al. 2003, 2004). Lack of power is already a key reason why so many studies of main genetic effects fail to replicate results and are probably false positives (Hirschhorn & Altshuler 2002; Wacholder et al. 2004); with current sample sizes in the order of a few hundred, only the strongest interaction effects are likely to be replicable and most ‘significant’ interactions will be false positives.
As ongoing prospective cohort studies will not be able to accrue sufficient numbers of cases for rare diseases, well-designed case–control studies remain the only option for the assessment of gene–environment interaction for rare diseases; case–control studies are also a cost-effective option for common diseases. One way to increase the power of cohort studies is to pool data across several studies. For example, the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) is currently examining gene–environment interactions in over 6000 cases of breast cancer and 8000 cases of prostate cancer, pooled across 10 prospective studies with over 800 000 people under follow-up and over 7 million person-years of follow-up already accrued (http://epi.grants.cancer.gov/BPC3/cohorts.html). An additional benefit of such an approach is the increased coordination among participating studies, across disciplinary lines (linking genomics and epidemiology) and among the epidemiologic community in general. To ensure across-study comparability of the genetic variants measured, participating studies will have to choose a common set to genotype. This is particularly important given the interest in ‘tagging’ SNPs, as results from equally efficient but distinct sets of tag SNPs may be difficult to synthesize. Further, to the extent that the tag SNPs and the data used to choose them are made public (as the BPC3 has; http://www.uscnorris.com/MECGenetics/), other researchers can use this information, saving time and resources and ensuring greater comparability. Combining information across several ongoing cohorts can mitigate the main weakness of prospective studies (lack of incident cases) while capitalizing on the methodological strengths of the prospective design.
5. An application: assessing complex mixtures
An important problem in environmental epidemiology is deciding which components of ‘complex mixtures’ (air pollution, diet, cigarette smoke, etc.) are causally related to disease. This is difficult to study observationally, as the components in their most relevant form are almost always found together and are highly correlated, making it difficult to statistically separate their effects. If, however, the effect of exposure changes according to variation in a particular gene that lies on the exposure-disease pathway, then an argument could be made that those components affected by that gene's function are causally related to disease (see figure 3). For example, cooking protein at high heat forms heterocyclic amines, which are carcinogenic in animal models. Heterocyclic amines are sometimes present in grilled and pan-fried meats (Sinha et al. 1998). While red meat intake has been quite consistently associated with the risk of colorectal cancer, red meat is a complex mixture of fatty acids, haem iron, and protein; which of these components underlies the increased risk is unknown. Exposure to heterocyclic amines is one hypothesis but obtaining detailed information on meat preparation is difficult in epidemiologic studies. Some (Roberts-Thomson et al. 1996; Chen et al. 1998; Kampman et al. 1999; Le Marchand et al. 2002) but not all (Barrett et al. 2003) studies have found the association of red meat intake with colorectal neoplasia is stronger in carriers of the ‘rapid’ NAT2 alleles. These alleles are associated with the faster metabolism of a variety of substrates, including heterocyclic amines. Aragaki et al. (1997) went further and attempted to explicitly estimate the effects of various heterocyclic amines, using external information about the chemical composition of different meats and the substrate-specific kinetics of various NAT2 alleles in a hierarchical model. Thus, an interaction between a complex environmental exposure and a gene with a relatively well-characterized function can ‘point the finger’ at the causal component(s) of the exposure—perhaps leading to the identification of unsuspected causal components (Rothman et al. 2001).
Figure 3.
Using gene–environment interactions to dissect complex mixtures (e.g. smoking, air pollution and diet). In panel (a), an association between the exposure and disease is observed but which of the many components of exposure (Xi) play causal roles is unknown. In panel (b), an interaction between the exposure and a polymorphism (G) known to metabolize one of the components of the exposure is observed. In the example cited in the text, the exposure is dietary red meat, the disease is colorectal cancer, X3 is heterocyclic amines and G is NAT2.
6. Future prospects
Investment in studies of the joint and independent action of genes and environmental exposures is likely to pay off in terms of increased knowledge about disease biology, which in turn will lead to better treatments (e.g. by suggesting drug targets) or preventive measures (e.g. by discovering the causal components of an environmental exposure). Beyond these indirect benefits, it has been suggested that studies of gene–environment interaction may directly yield targeted ‘personalized prevention’ strategies. According to one scenario that has made it into the popular press, a patient will soon hand his or her personal physician a card that contains information on hundreds or thousands of genetic variants and the physician will then base treatment or prevention advice on a combination of this genetic information, the patient's clinical history, lifestyle, occupational exposures and so on. This scenario assumes that the relevant gene–environment interactions will have been proposed, replicated and validated—and all that very soon—so that this advice is evidence-based and efficacious. However, as sketched in this article, we are only beginning to understand gene–environment interactions for a few diseases and the challenges to understanding these interactions in the context of common, complex disease are formidable. Affordably sequencing an individual's genome may be the easy part.
Furthermore, the concept of personalized prevention may also conflict with the view that population-wide interventions are usually more effective in reducing the incidence of common diseases than interventions targeting high-risk individuals (Rose 1985). The idea that inherited susceptibility is a major determinant of disease risk could increase latent feelings of genetic determinism and actually undermine support for ‘broad brush’ preventive recommendations that are the cornerstone of many public health campaigns or investment in general public health infrastructure (in much the same way discussions of potentially real but quite subtle differences in patterns of standardized test performance or brain function between men and women can undermine support for policies that remove historical barriers to women's participation in scientific research). Extending genetic testing beyond counselling-intensive, high-penetrance disorders raises complex issues, and past experiences, such as screening programmes for sickle cell anaemia in southern U.S. states (Scott & Castro 1979), urge caution. The psychological and social consequences of genotyping individuals in order to make preventive recommendations are still uncertain and require research—to say nothing of the financial costs and benefits. At a minimum, a DNA-based screening test should not become widely used until there is a proven intervention that takes advantage of the genetic information.
Integrating information on inherited genetic variation into epidemiologic studies promises to be a powerful tool for improving the study of disease aetiology. The ramifications of this information for public health policy and clinical decision-making are uncertain and will require a measured, evidenced-based approach.
Footnotes
One contribution of 12 to a Discussion Meeting Issue ‘Genetic variation and human health’.
References
- Aragaki C.C, Greenland S, Probst-Hensch N, Haile R.W. Hierarchical modeling of gene–environment interactions: estimating NAT2 genotype-specific dietary effects on adenomatous polyps. Cancer Epidemiol. Biomarkers Prev. 1997;6:307–314. [PubMed] [Google Scholar]
- Barrett J.H, et al. Investigation of interaction between N-acetyltransferase 2 and heterocyclic amines as potential risk factors for colorectal cancer. Carcinogenesis. 2003;24:275–282. doi: 10.1093/carcin/24.2.275. [DOI] [PubMed] [Google Scholar]
- Bonilla C, et al. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann. Hum. Genet. 2004;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- Botto L, Khoury M. Facing the challenge of complex genotypes and gene–environment interaction: the basic epidemiologic units in case–control and case-only designs. In: Khoury M, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. Oxford University Press; Oxford: 2004. [Google Scholar]
- Brennan P. Commentary: Mendelian randomization and gene–environment interaction. Int. J. Epidemiol. 2004;33:17–21. doi: 10.1093/ije/dyh033. [DOI] [PubMed] [Google Scholar]
- Caparaso N, Rothman N, Wacholder W. Case–control studies of common alleles and environmental factors. Monogr. Natl Cancer Inst. 1999;26:25–30. doi: 10.1093/oxfordjournals.jncimonographs.a024222. [DOI] [PubMed] [Google Scholar]
- Cardon L.R, Palmer L.J. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Carr K.M, Rosenblatt K, Petricoin E.F, Liotta L.A. Genomic and proteomic approaches for studying human cancer: prospects for true patient-tailored therapy. Hum. Genomics. 2004;1:134–140. doi: 10.1186/1479-7364-1-2-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Stampfer M.J, Hough H.L, Garcia-Closas M, Willett W.C, Hennekens C.H, Kelsey K.T, Hunter D.J. A prospective study of N-acetyltransferase genotype, red meat intake, and risk of colorectal cancer. Cancer Res. 1998;58:3307–3311. [PubMed] [Google Scholar]
- Clayton D, McKeigue P.M. Epidemiological methods for studying genes and environmental factors in complex diseases. Lancet. 2001;358:1356–1360. doi: 10.1016/S0140-6736(01)06418-2. [DOI] [PubMed] [Google Scholar]
- Conti D.V, Cortessis V, Molitor J, Thomas D.C. Bayesian modeling of complex metabolic pathways. Hum. Hered. 2003;56:83–93. doi: 10.1159/000073736. [DOI] [PubMed] [Google Scholar]
- Cordell H.J. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Cortessis V, Thomas D.C. Toxicokinetic genetics: an approach to gene–environment and gene–gene interactions in complex metabolic pathways. IARC Sci. Publ. 2004;157:127–150. [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Freedman M.L, et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Thompson W.D, Robins J.M. Differential misclassification and the assessment of gene–environment interactions in case–control studies. Am. J. Epidemiol. 1998;147:426–433. doi: 10.1093/oxfordjournals.aje.a009467. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Rothman N, Lubin J. Misclassification in case–control studies of gene–environment interactions: assessment of bias and sample size. Cancer Epidemiol. Biomarkers Prev. 1999;8:1043–1050. [PubMed] [Google Scholar]
- Garcia-Closas M, Wacholder S, Caporaso N, Rothman N. Inference issues in cohort and case–control studies of genetic effects and gene–environment interactions. In: Khoury M, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. Oxford University Press; Oxford: 2004. [Google Scholar]
- Garrod A. The incidence of alkaptonuria: a study in chemical individuality. Lancet. 1902;2:1616–1620. [PMC free article] [PubMed] [Google Scholar]
- Gauderman W. Sample size requirements for matched case–control studies of gene–environment interaction. Stat. Med. 2001;15:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Greenland S, Rothman K. Concepts of interaction. In: Rothman K, Greenland S, editors. Modern epidemiology. Lippincott Williams & Wilkins; Philadelphia: 1998. [Google Scholar]
- Haldane J. W.W Norton, Company; New York: 1938. Heredity and politics. [Google Scholar]
- Hirschhorn J.N, Altshuler D. Once and again—issues surrounding replication in genetic association studies. J. Clin. Endocrinol. Metab. 2002;87:4438–4441. doi: 10.1210/jc.2002-021329. [DOI] [PubMed] [Google Scholar]
- Hung R.J, Brennan P, Malaveille C, Porru S, Donato F, Boffetta P, Witte J.S. Using hierarchical modeling in genetic association studies with multiple markers: application to a case–control study of bladder cancer. Cancer Epidemiol. Biomarkers Prev. 2004;13:1013–1021. [PubMed] [Google Scholar]
- Kampman E, Slattery M.L, Bigler J, Leppert M, Samowitz W, Caan B.J, Potter J.D. Meat consumption, genetic susceptibility, and colon cancer risk: a United States multicenter case–control study. Cancer Epidemiol. Biomarkers Prev. 1999;8:15–24. [PubMed] [Google Scholar]
- Katan M.B. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- Keavney B. Commentary: Katan's remarkable foresight: genes and causality 18 years on. Int. J. Epidemiol. 2004;33:11–14. doi: 10.1093/ije/dyh056. [DOI] [PubMed] [Google Scholar]
- Khoury M.J, Adams M.J, Jr, Flanders W.D. An epidemiologic approach to ecogenetics. Am. J. Hum. Genet. 1988;42:89–95. [PMC free article] [PubMed] [Google Scholar]
- Kittles R.A, et al. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum. Genet. 2002;110:553–560. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- Laird N, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19(Suppl. 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Langholz B, Rothman N, Wacholder S, Thomas D. Cohort studies for characterizing measured genes. Monogr. Natl Cancer Inst. 1999;26:39–42. doi: 10.1093/oxfordjournals.jncimonographs.a024224. [DOI] [PubMed] [Google Scholar]
- Le Marchand L, et al. Well-done red meat, metabolic phenotypes and colorectal cancer in Hawaii. Mutat. Res. 2002;506–507:205–214. doi: 10.1016/s0027-5107(02)00167-7. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Rosenberg N, Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Goldstein D. Detecting association in a case–control study while correcting for population stratification. Genet. Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Reiner A.P, et al. Population structure, admixture, and aging-related phenotypes in African American adults: the cardiovascular health study. Am. J. Hum. Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Thomson I.C, Ryan P, Khoo K.K, Hart W.J, McMichael A.J, Butler R.N. Diet, acetylator phenotype, and risk of colorectal neoplasia. Lancet. 1996;347:1372–1374. doi: 10.1016/s0140-6736(96)91012-0. [DOI] [PubMed] [Google Scholar]
- Robins J, Greenland S. The role of model selection in causal inference from nonexperimental data. Am. J. Epidemiol. 1986;123:392–402. doi: 10.1093/oxfordjournals.aje.a114254. [DOI] [PubMed] [Google Scholar]
- Rose G. Sick individuals and sick populations. Int. J. Epidemiol. 1985;14:32–38. doi: 10.1093/ije/14.1.32. [DOI] [PubMed] [Google Scholar]
- Rothman N, Wacholder S, Caporaso N.E, Garcia-Closas M, Buetow K, Fraumeni J.F., Jr The use of common genetic polymorphisms to enhance the epidemiologic study of environmental carcinogens. Biochim. Biophys. Acta. 2001;1471:C1–C10. doi: 10.1016/s0304-419x(00)00021-4. [DOI] [PubMed] [Google Scholar]
- Scott R.B, Castro O. Screening for sickle cell hemoglobinopathies. J. Am. Med. Assoc. 1979;241:1145–1147. [PubMed] [Google Scholar]
- Sinha R, et al. Heterocyclic amine content in beef cooked by different methods to varying degrees of doneness and gravy made from meat drippings. Food Chem. Toxicol. 1998;36:279–287. doi: 10.1016/s0278-6915(97)00162-2. [DOI] [PubMed] [Google Scholar]
- Smith P.G, Day N.E. The design of case–control studies: the influence of confounding and interaction effects. Int. J. Epidemiol. 1984;13:356–365. doi: 10.1093/ije/13.3.356. [DOI] [PubMed] [Google Scholar]
- Tang H, et al. Genetic structure, self-identified race/ethnicity, and confounding in case–control association studies. Am. J. Hum. Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Genetic epidemiology with a capital “E”. Genet. Epidemiol. 2000;19:289–300. doi: 10.1002/1098-2272(200012)19:4<289::AID-GEPI2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Thomas D, Conti D. Commentary: the concept of ‘mendelian randomization’. Int. J. Epidemiol. 2004;33:21–25. doi: 10.1093/ije/dyh048. [DOI] [PubMed] [Google Scholar]
- Thomas D, Witte J. Point: population stratification: a problem for case–control studies of candidate gene associations? Cancer Epidemiol. Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- Thompson W. Effect modification and the limits of biological inference from epidemiologic data. J. Clin. Epidemiol. 1991;44:221–232. doi: 10.1016/0895-4356(91)90033-6. [DOI] [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J. Natl Cancer Inst. 2000;92:1151–1158. doi: 10.1093/jnci/92.14.1151. [DOI] [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol. Biomarkers Prev. 2002;11:513–520. [PubMed] [Google Scholar]
- Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg C, Umbach D. Choosing a retrospective design to assess joint genetic and environmental contributions to risk. Am. J. Epidemiol. 2000;152:197–203. doi: 10.1093/aje/152.3.197. [DOI] [PubMed] [Google Scholar]
- Witte J.S, Gauderman W.J, Thomas D.C. Asymptotic bias and efficiency in case–control studies of candidate genes and gene–environment interactions: basic family designs. Am. J. Epidemiol. 1999;148:693–705. doi: 10.1093/oxfordjournals.aje.a009877. [DOI] [PubMed] [Google Scholar]
- Wong M.Y, Day N.E, Luan J.A, Chan K.P, Wareham N.J. The detection of gene–environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int. J. Epidemiol. 2003;32:51–57. doi: 10.1093/ije/dyg002. [DOI] [PubMed] [Google Scholar]
- Wong M.Y, Day N.E, Luan J.A, Wareham N.J. Estimation of magnitude in gene–environment interactions in the presence of measurement error. Stat. Med. 2004;23:987–998. doi: 10.1002/sim.1662. [DOI] [PubMed] [Google Scholar]