Abstract
Over the last two decades, identification of polymorphisms that influence human diseases has begun to have an impact on the provision of medical care. The promise of genetics lies in its ability to provide insights into an individual's susceptibility to disease, the likely nature of the disease and the most appropriate therapy. For much of its history, pharmacogenetics (PGx—the use of genetic information to impact drug choice) has been limited to comparatively simple phenotypes such as plasma drug levels. Progress in genetics technologies has broadened the scope of PGx efficacy and safety studies that can be implemented, impacting on a broad spectrum of drug discovery and development activities. Recent PGx data show the ability of this approach to generate information that can be applied to dose selection, efficacy determination and safety issues. This in turn will lead to significant opportunities to affect both the approach to clinical development and the probability of success—the latter being an important aspect for pharmaceutical companies and for the patients who will benefit from these new medicines.
Keywords: pharmacogenetics, pharmacogenomics, diagnosis, drug response, drug discovery, drug development
1. Introduction
The last twenty years have seen unprecedented international research programmes endeavouring to identify polymorphisms that are either causative of, or affect the susceptibility to, human disease. The programmes are beginning to have a visible impact on medical care in a growing number of ways. Firstly, in the late 80s and 90s, identification of causative mutations of monogenic diseases and the subsequent correlation studies linking phenotype and genotype have resulted in much greater definition of the sub-classification of these diseases. The resulting genetic sub-classification of many of these diseases has allowed clarification of their phenotypic heterogeneity and pathogenesis (Weatherall 2001). This knowledge has greatly facilitated implementation of effective prevention programmes around the world, significantly reducing the impact and burden of some of these diseases. This genetic knowledge has also provided scientific insight into possible therapies for diseases such as Huntington's disease, thalassaemias and muscular dystrophies. The more recent project to sequence the human genome has resulted in the identification of most of the human DNA sequence. This sequence information, coupled with the more recent publication of high density single nucleotide polymorphism (SNP) maps and other tools in genomics, bioinformatics, RNA expression and proteomics, has allowed the application of these new tools to more complex disease phenotypes, such as the more common disorders and inter-individually variable drug responses. This variability in drug response can take various forms but the new technologies allow insights into both variable efficacy and susceptibility to adverse events. The promise of these new technologies (the ‘new genetics’) lies in the potential to provide insight into an individual's genetic make-up and infer susceptibility to a given disease, as well as to understand and predict specific kinds of associated pathology (for example, vas deferens pathology in cystic fibrosis) and enable better therapeutic decision-making. Overall, this knowledge of DNA variations in an individual patient can—in combination with other clinical information—contribute to well-informed medical action.
Furthermore, other fields of medical science have also made significant progress in recent years, with considerable opportunity for synergistic benefits when they are combined with genetic information. For example, the recent availability of a number of imaging and molecular technologies allows for a more accurate diagnosis of the individual's physiological and pathological status. These technologies include structural imaging techniques that can now go far beyond crude anatomical structure to real-time visualization of biochemical and physiological processes in vivo, revealing correlations with behaviour, disease and treatment (such as functional MRI; Hariri & Weinberger 2003). What was recently remarkable is now routine—for example, pharmacological events can be observed in vivo using MR spectroscopy, positron emission tomography (PET) and single photon emission computed tomography (SPECT). Human biological processes—and their perturbation in disease—can be further examined by an increasingly sophisticated array of molecular tools exploring transcriptomic, proteomic and metabolomic phenomena (Plumb et al. 2003; Campbell & Ghazal 2004). All in all, recent technological advances have allowed previously undreamt of opportunities to explore the basis of human pathology.
The application of these molecular, genomic and imaging technologies will also provide new opportunities to explore the heterogeneity which patients clearly exhibit (which exists within the current diagnostic definitions) and provide a more accurate understanding of individual patients. This inter-patient variability is one of the key challenges in medicine and greater insight into this is critical to increasing the probability of successfully treating patients as they present to physicians.
It is important that the widely used term ‘personalized medicine’ is not misunderstood. While it could mean development of ‘tailored’ medicines to suit each patient—which is clearly impractical—it is more accurate to consider the term to represent a greater use of ‘personal’ medical information (arising from the application of emerging technologies) to increase the efficiency of both disease diagnosis and patient treatment. It has to be acknowledged that the contribution of confounders such as environmental variables (e.g. stress, diet, other medications) will to some degree limit the accuracy of the prediction; however, a major improvement in the probability of finding the right treatment for a given patient is eminently possible with the new information being generated today. The end result will be a reduction in ‘trial and error’ prescribing by the physician as a result of the new knowledge base and, of course, great benefit to the patients through improved efficacy and fewer side effects.
Among the panoply of new technologies described above, this paper will focus on the application of pharmacogenetics (PGx), currently the most advanced—and one of the most promising—of the emerging biomarker technologies for addressing the issues associated with variable response to medication. Unfortunately, nomenclature in this field is complex and confusing with many different definitions of the different terms. In this paper, we use the term ‘pharmacogenetics’ to refer to the study of inter-individual variations in the DNA sequence that are related to drug response, efficacy and toxicity (CPMP Position Paper on Terminology in Pharmacogenetics). (The related term ‘pharmacogenomics’, PGm, is usually employed in a broader sense that includes genome-wide variations and potential complex interactions as well as alterations in gene expression and post-translational modifications (e.g. proteomics) that correlate with drug response.) The development of PGx is associated with a number of issues that are relevant to the application of a range of other molecular or imaging technologies; hence PGx is used to show how the practice of medicine will change in the future. Pharmacogenomics—especially the use of gene expression patterns—is a rapidly developing technology with enormous potential that is only just being realized in the field of oncology, although it also has many potential applications elsewhere. In particular, the scope of this technology is enhanced by publications showing that changes in gene expression profiles in peripheral leucocytes may correlate with drug response in body systems not traditionally thought to be related to these immune-focused cells (e.g. Chon et al. 2004).
The science of PGx is approaching a major milestone—it is nearly 50 years since the word was first coined to describe the inheritance of an aberrant drug metabolism (Vogel 1959). Progress in the science in subsequent decades has been most impressive, particularly so in the last few years (Roses 2002; Goldstein et al. 2003): in fact, all aspects of genetics research—including PGx—have been transformed by major developments in the essential technologies, underpinning a qualitative leap in our ability to understand the basis of variable response to medicines.
For much of its history, PGx has focused on studying the relation between DNA variants—especially in the genes coding for the body's drug absorption, metabolism and excretion system—and a medicine's pharmacokinetic profile. Pharmacokinetics refers to the analysis of how the active drug molecules are made available in the bloodstream, transported to the relevant target organ and subsequently metabolized and excreted. The effects of the drug molecule at its molecular target and the subsequent signalling or metabolic events that determine any therapeutic effect are facets of the medicine's pharmacodynamics. There are very real and practical reasons why pharmacokinetics has been the first drug response phenotype to be explored: this phenotype—concentration measurements of the drug and its metabolites in body fluids such as plasma or urine—is easily and accurately measured, the number of genes involved is somewhat limited (so the amount of genotyping required is small—certainly by today's standards) and the magnitude of the genetic variation and its penetrance is usually high (analogous to single gene disorders). Analysis of more complex phenotypes, such as the pharmacodynamic properties of a medicine or the basis for idiosyncratic toxicity not related to abnormal plasma levels, has only recently been possible as such studies required significantly more complex genetic analysis that was beyond the reach of these earlier technologies.
However, key technology improvements have been pivotal to making possible a wide range of pharmacogenetic efficacy and safety studies. Firstly, the completion of the human genome sequence has had a major impact: this key development has been central to the almost exponential increase in all human genetics activities, with PGx being no exception (Subramanian et al. 2001). Secondly, SNP-based high-throughput genotyping methods and platforms have led to a dramatic increase in both the capacity and speed of genotyping, together with a significant drop in the associated cost per genotype. The ability to genotype SNPs that are accurately positioned using reliable high-density maps (http://snp.cshl.org/) has now made genome wide scan (WGS) association studies a reality. These tools are critical in PGx, where family-based linkage analyses are highly impractical and rarely possible. In particular, the ability to carry out genome-wide association analyses allows PGx analyses to take advantage of the type of hypothesis-free genetics that family based linkage studies have been able to access for a number of years. The most recent—and still evolving—contribution to the technology progress is the HapMap, which will simplify genetic analysis by allowing SNP clustering according to their linkage disequilibrium relationships. This will facilitate selection of the most informative SNPs for genotyping, so that redundant or non-informative assays are avoided. It must be borne in mind, however, that these technology developments are now capable of generating such massive datasets that this creates new problems in data management and compensation for multiple testing, requiring modified statistical analysis techniques and the application of new data mining and pattern recognition methodologies.
(a) PGx applications: increased efficiency of pharmaceutical research and development
The last decade has seen remarkable technological developments in many sectors of biological and medical research. However, drug research has incorporated only a few novel developments, despite the commitment of considerable resources from pharmaceutical companies. This resource spending has had little impact on the productivity of the pharmaceutical industry: in fact, the industry has submitted 50% fewer new drug applications to the US Food and Drug Administration (FDA) in 2002–2003 compared with 1997–1998, whereas investment in research has increased over twofold in the same period (Lesko & Woodcock 2004). A critical challenge for the industry is to increase its productivity and focus on introducing novel strategies aimed at enhancing early stage discovery as well as the efficiency of decision making throughout the research process (Roses 2004).
(i) From gene to target to candidate selection
In the late 1990s, it was widely expected that the Human Genome Project would quickly lead to thousands of new potential pharmaceutical targets. This led to a host of genetics research ventures aimed at target identification for a variety of the most common diseases, mainly driven by the comparatively new (at the time) biotechnology sector. Most of these undertakings were based on the somewhat naïve extrapolation from successful gene discoveries in the rare, highly penetrant single gene diseases: it is clear that this somewhat simplistic approach significantly underestimated the inherent complexities of common diseases that increase the difficulty of identifying susceptibility genes. Furthermore, the difficulties in moving from a genome sequence derived from genetic research to disease-relevant, ‘tractable’ (i.e. amenable to chemical screening) targets which are conducive to subsequent pre-clinical drug discovery approaches were nearly always overlooked (Roses 2004). Tractable targets include members of families that have been demonstrated as being able to be modulated by suitable chemical ligands to produce beneficial therapeutic effects, such as the 7-transmembrane- or nuclear receptor-families. The number of known tractable targets has increased more than twofold in the last few years, from an approximate number of 500 to well over 1000. This was mainly the result of the availability of new sequence information from the Human Genome Project, together with the application of novel bioinformatic technologies that permit more accurate identification of coding sequences that bear similarities to gene classes that can be screened as well as more rapid exploration of gene pathways (Debouck & Metcalf 2000; DeFife & Wong-Staal 2002; Searls 2003). During the same period, our understanding of the molecular mechanisms underlying disease heterogeneity, gene–gene and gene–environment interactions is steadily increasing, as evidenced by the numbers of peer review publications in the international scientific literature and fuelled by the increasing armamentarium of high-throughput genotyping, data analysis and related technologies. Novel approaches for high-throughput experiments to discern associations between disease and disease traits with large numbers of tractable drug targets are now available (Roses et al. 2005).
(ii) From selecting a candidate to the clinic
The decision as to which molecules should progress from preclinical development into studies in humans is a pivotal point in the pharmaceutical development process that has tremendous implications in terms of financial costs, time and effort. This ‘critical path’ is defined as the path from candidate selection to an optimized drug. This firstly necessitates a series of (mostly animal) experiments to accumulate sufficient evidence of therapeutic effect and safety (preclinical phase), followed by (for a ‘successful’ molecule that manages to clear these hurdles) use in healthy and unhealthy human volunteers in clinical trials designed to determine the efficacy and safety of the medicine. The clinical testing prior to registration of a medicine is divided into different stages as experience of the molecule is gained during development (see table 1).
Table 1.
Phases of clinical development.
| Phase I trials | These comprise the first exposure of humans to a putative medicine and are intended to explore pharmacokinetic parameters and ensure that there are no grossly unacceptable safety or tolerability issues. Typically up to 100 individuals may take part in a series of studies aimed at generating information on pharmacokinetic parameters such as bioavailability, the rate and route of clearance and any signs of drug–drug interactions. |
| Phase II studies | These are conducted in patients and look to establish an initial indication that the compound is effective. The studies (which may involve up to 1000 individuals) are sufficiently large that some safety signals may be apparent, especially so-called ‘nuisance’ or quality of life (QOL) side effects such as reversible changes to liver function tests. Establishing key efficacy (and safety) parameters is a crucial part of the phase II studies. |
| Phase III studies | These large trials, costing tens or even hundreds of millions of pounds, provide the most convincing evidence of efficacy and safety to support a regulatory submission. To minimize bias and variability, they are usually randomized controlled trials (RCT). For example, the double-blind RCT model, where neither the physician nor the research subjects know which arm of the study (active drug or placebo/standard care) the patient is assigned to, is the phase III ‘gold standard’. Owing to the size of these studies, less common adverse events may become apparent. |
| Phase IV | Once a medicine is registered, it can be used by a much wider population of patients. At this stage (phase IV), rare adverse events may be identified that could not be discovered during clinical development: even the large pivotal phase III studies lack the power to detect adverse events occurring at rates less than 0.1% |
Clinical studies are based on a statistical model of frequency, aiming at compiling evidence of efficacy and safety through the use of the drug in large numbers of patients. These numbers are needed to overcome many issues such as disease heterogeneity, partial understanding of underlying disease mechanisms, variability in drug responses, placebo effects and so on. Currently, the failure rate of potential products in development is more than 90%: the main causes of this being poor efficacy and sub-optimal safety. This ‘pipeline attrition’ has an enormous cost—both financially and time-wise—as the average cost of developing a market product is estimated to be in excess of $800 million (DiMasi et al. 2003) and the average time from first screening to marketing varies from between 8 and 15 years. The large-scale phase III trials consume the majority of these resources and also represent up to 50% of the overall attrition (Gilbert 2003). There is a burgeoning recognition that novel approaches and tools such as PGx may enable exploration of the pathophysiological mechanisms underlying differences in drug response and reduce attrition (Lesko et al. 2003). Pharmacogenetics can be applied either retrospectively or prospectively. Retrospectively PGx looks back over results of clinical trials, using genotype data to generate insights into issues such as the kinetic and dynamic properties of drugs, efficacy and adverse events. Prospective PGx allows proactive identification of patient subgroups (e.g. disease subtypes or poor metabolizers) that would be predictive of either positive or negative responses to a drug. If such data were available before or between phase IIa and IIb trials, this would significantly shorten and simplify phase III and increase the probability of success (Lesko & Woodcock 2004). Furthermore, the ability to prospectively identify subgroups of patients by therapeutic response during early phase II development would permit the progression of multiple compounds that can treat overlapping groups of patients with the same disease sub-component (Roses 2004).
It is important to differentiate between the application of PGx for safety and efficacy outcomes. It is undoubtedly the case that the efficacy of a medicine is strongly impacted by genetic variation but it is also affected by factors such as environmental influences (such as compliance, diet or other medicines) and placebo response. This means that PGx efficacy is unlikely to be completely deterministic at an individual level but will have a critical role in significantly increasing the probability of effective response for identified subgroups of patients. In contrast, safety PGx is focused on the individual, with specific decisions about whether or not to prescribe a medicine based on genetic information predicting rare dangerous events and common adverse effects (AE).
(iii) PGx and drug pharmacokinetics
As mentioned earlier, the first use of PGx from the 1950s onwards was to explore genetic variants that affected pharmacokinetics—especially the metabolism of drugs as this appears to limit the rate at which many medicines are cleared from the body (Daly 2003). The first PGx phenotype studied was toxicity in patients that was due to excessive exposure to medicines and a severely reduced rate at which medicine was cleared from the body.
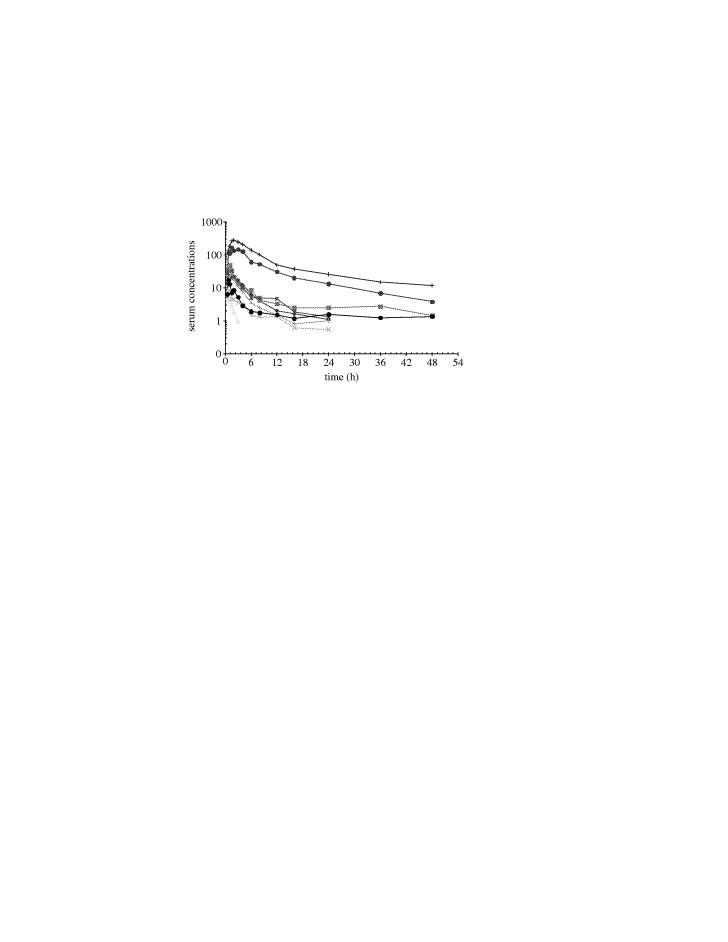
Figure 1 gives an example of the type of variable pharmacokinetics that can be seen in phase I studies. Each line represents the serum concentration over time for an individual subject. While most subjects cluster with similar peak concentrations and time course, there are unusually high exposures in a couple of patients and low exposure in one patient. There are a number of reasons for such variability, (e.g. diet etc but phase I studies attempt to minimize variation in these environmental variables); however, polymorphic variation in the drug metabolising enzymes is a fundamental variable to consider.
Figure 1.
A typical graph of serum concentration versus time for individual study subjects.
The principles underpinning the design of such PGx experiments have remained essentially the same for many years, although the current analyses are inevitably more sophisticated reflecting recent progress in genetic technology and understanding. For example, we are much more aware of the range of enzymes and transporters involved in drug disposition and clearance. Absorption and distribution (not just metabolism of drugs) are better understood and the relevant alleles are well characterized, as well as their distribution in different ethnic groups (Lin et al. 1996, 2001).
Studies of drug clearance in different individuals with known genotypes have established that at least 40% of drug metabolism is via polymorphic CYP450 enzymes (Ingelman-Sundberg 2004). While these data may appear straightforward, we need to be aware of complexities that may impact the interpretation. For example, environmental confounders (such as smoking) can affect CYP expression and change the metabolic route. The ability of grapefruit juice to impact the clearance of drugs such as anti-hypertensive calcium channel blockers is well described. The whole area of drug–drug interactions shows the complexities that can be observed. It has been estimated that 6% of patients on two medications experience adverse drug reactions (ADRs), whereas ADRs are reported by 50% of those on five medications and nearly 100% of those on 10 medications. A major cause of these reactions is changes produced by one drug in the metabolism of another through P450 pathways. Guzey et al. (2002) describe a case wherein a CYP2D6 extensive metabolizer became a phenotypically poor metabolizer during treatment with the potent 2D6 inhibitor bupropion, demonstrating the complexity of assessing the exact causes of ADRs.
However, despite almost 50 years of research, PGx applications have yet to have a significant impact on clinical practice. For example, it is well established that CYP2D6 poor metabolizers have increased biological exposure and increased risk of adverse events to a variety of commonly prescribed medicines. However, these observations have not as yet resulted in CYP2D6 testing being a routine part of the prescribing practice or even being recommended as such. Despite the extensive research literature, there is a shortage of clinical outcome studies that demonstrate the clinical benefit of such interventions. This lack of clinical utility data makes it difficult for physicians to apply this information in their prescribing decisions. Suitable prospective studies (e.g. looking at the effect of genotyping CYP2C9 on warfarin dosing and bleeding events) are now being initiated, and the results from such studies—although not available for several years—will be critical if PGx is to make significant inroads into medical practice.
Although prescribing practices have not yet been significantly altered, PGx information is starting to appear in the label information provided when the drug is dispensed. In the USA, labels for atomoxetine and 6-mercaptopurine provide the physician with information regarding the metabolism of these medicines by polymorphic enzymes (CYP2D6 and thiopurinemethyl transferase, respectively). Physicians are alerted in the label that tests are available to identify poor metabolizers but the physicians are not required to carry out these tests prior to prescribing these medicines (Lesko et al. 2003).
The application of PGx to study pharmacokinetics is nonetheless a strong focus of attention at the moment, as drug regulatory authorities are actively exploring PGx tools to better understand drug exposure (and, in particular, toxicity), utilize this information more widely in selecting drug doses for development and make this information available to physicians. For example, the US FDA and Japanese Ministry of Health, Labour and Welfare have already released draft guidelines relating to genomic data submission (Lesko et al. 2003) and the Committee for Proprietary Medicinal Products in Europe has a PGx expert group to address these issues.
(iv) PGx of drug efficacy
In addition to its direct clinical impact on patients and healthcare systems alike, variable efficacy is also an important issue for drug development. Failure to show efficacy in phase II studies is the most common reason for terminating the development of medicines. As has been noted above, the large phase III studies required to establish safety and efficacy are very expensive; therefore, if more information could be extracted from earlier, smaller phase II studies to more clearly define the efficacy of a candidate medicine, then valuable time and resources would be saved during phase III. In fact, the variable efficacy of medicines, even in apparently homogeneous patient groups recruited in phase II studies, can obscure true and significant efficacy in a subset of patients, thus leading to inappropriate termination of the compound. A major challenge in PGx efficacy is that, in contrast to the more ‘single gene’ character of the pharmacokinetic PGx described above, the efficacy phenotype is likely to be multigenic and potentially confounded by environmental factors, and thus require more research to be fully clinically applicable.
Nonetheless, efficacy prediction is a hugely promising area for PGx (see table 2). By using genetic and other biomarkers to identify appropriately responding subgroups in phase II studies, compounds that are effective in patient subgroups may be developed further, significantly increasing the delivery of new medicines to meet unmet patient needs and increasing the productivity of pharmaceutical research and development. The critical issue is whether the phase II studies are appropriate to generate robust PGx data that can be reliably used to support the further development of compounds. Although published data are scarce, some initial findings seem promising.
Table 2.
Pharmacodynamic (drug target) polymorphisms associated with variation in medication response.
| gene | medication | phenotype change | references |
|---|---|---|---|
| angiotensin converting enzyme (ACE) | ACE inhibitors (imidapril, enalapril) | blood pressure, kidney damage reduction, left ventricular hypertrophy reduction, blood vessel stenosis | Ohmichi et al. (1997), Jacobsen et al. (1998), Penno et al. (1998), Kohno et al. (1999) and Okamura et al. (1999) |
| arachidonate 5 lipoxygenase (ALOX) | anti-asthmatics (leukotriene inhibitors) | forced expiratory volume (FEV-1) improvement | Drazen et al. (2003) |
| beta 2 adrenergic receptor (ADRBR2) | beta-2 agonists (albuterol) | vascular reactivity, bronchodilation | Lima et al. (1999), Martinez et al. (1999), Dishy et al. (2001), Israel et al. (2001) and Cockcroft et al. (2000) |
| corticotrophin releasing hormone receptor 1 | inhaled coricosteroids | improved lung function (FEV-1) | Tantisira et al. (2004) |
| dopamine D3 | traditional antipsychotics (chlorpromazine, haloperidol) | abnormal involuntary muscle movements (tardive dyskinesia), akathisia | Steen et al. (1997), Basile et al. (1999) and Lerer et al. (2002) |
| dopamine D2 | risperidone (antipsychotic) | response of schizophrenia symptoms | Yamanouchi et al. (2003) |
| growth hormone receptor | growth hormone | increased responsiveness to growth hormone | Dos Santos et al. (2004) |
| serotonin transporter | antidepressants | mood improvement, side effects | Smeraldi et al. (1998), Serretti et al. (2000), Murphy et al. (2004) and Mundo et al. (2001) |
The example of Herceptin (trastuzumab) highlights how a pharmacogenetic test can be used to progress medicines through the research and development pipeline. Over-expression of the ErbB2 gene is associated with increased tumour aggressiveness and poorer prognosis. Herceptin—a humanized monoclonal antibody against the ErbB2 receptor—is now approved for the treatment of breast cancer (Vogel & Franco 2003; Noble et al. 2004). Retrospective examination of the clinical trials of Herceptin showed that a positive response was more likely in patients with tumours over-expressing ErbB2. So the measurement of ErbB2 over-expression can be used to assess whether treatment with Herceptin is appropriate. The availability of a test for a subgroup with a higher probability of responding to treatment with Herceptin allowed this drug to progress through further studies to approval. The same paradigm has recently been applied to understand the response of lung cancer patients to Iressa (gefitinib), where a positive response is closely associated with the presence of activating mutations in the drug target (EGFR) in the tumour (Lynch et al. 2004). These striking results have had an immediate effect on the way clinicians assess the role of gefitinib in cancer treatment. They will also have an impact on the questions that regulatory authorities require to be answered during drug development.
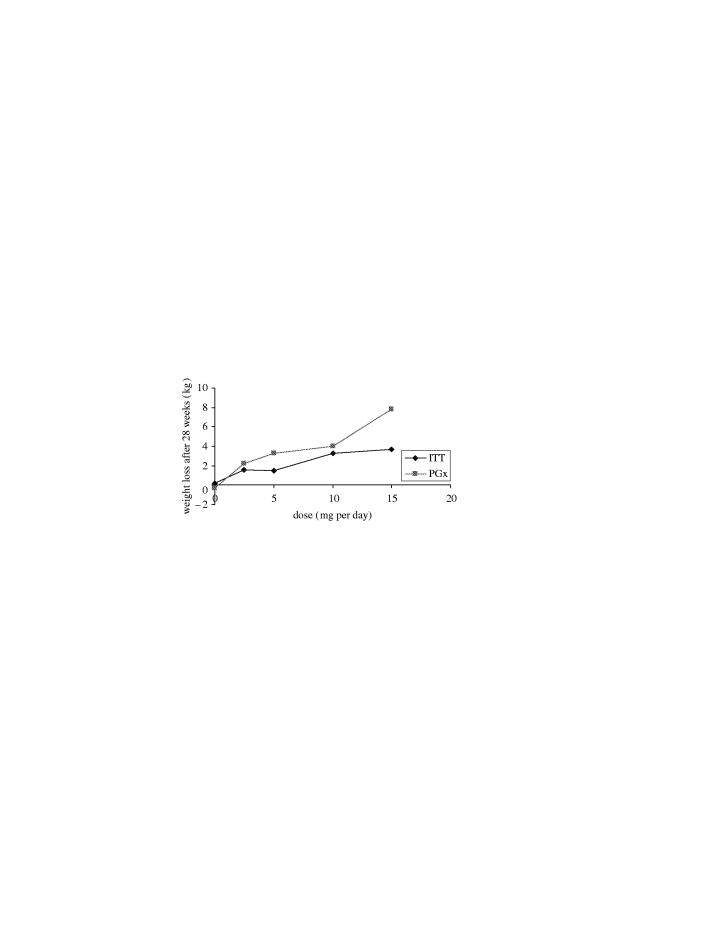
In another example in a recent phase II study of a GlaxoSmithKline anti-obesity compound, analysis of all of the patients treated showed less efficacy than that reported in the literature for the current ‘gold standard’ therapies. However, PGx analysis using candidate genes based on the compound's target and putative mechanism of action showed association between three PGx markers and weight loss during the study. Using the presence of any one of these alleles to identify a subgroup, 36% of the patients could be clustered to show significantly greater weight loss. The dose response in the whole (‘ITT’) patient group and the PGx-defined subgroup (‘PGx’) is shown in figure 2.
Figure 2.
Graph of weight loss in the whole (ITT) study group versus weight loss in the genotype-selected (PGx) group.
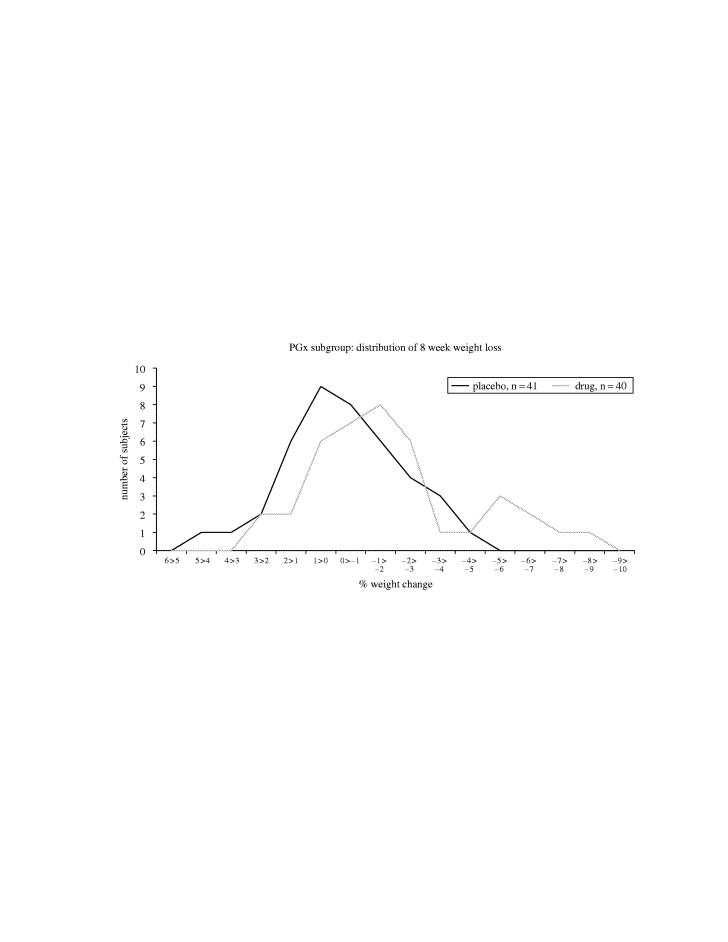
Analysis of a subsequent phase II study with a different anti-obesity compound showed a revealing histogram of patient numbers versus response (figure 3). Although the average response in the placebo group was 0% change, there was considerable variation even in eight weeks. Most of the treated patients showed some benefit, while there were some who showed much greater effects (‘super-responders’). However, there were also some that showed very little benefit, gaining weight during the study. The visualization of this data clearly indicates opportunities for PGx to provide insights into the variable response seen in different patient subgroups.
Figure 3.
Histogram showing weight change during an eight week obesity study.
Data such as these show that phase II studies are sufficiently sized to generate PGx hypotheses that can be used to influence subsequent development approaches for candidate medicines. The data can be replicated and further refined in subsequent phase IIb or phase III studies.
(v) PGx of drug safety
While there is ample evidence that medicines provide significant benefit in terms of mortality, morbidity and cost-effectiveness, it is unfortunately inevitable that ADRs are observed. Lazarou et al. (1998) estimated that ADRs caused approximately 106 000 deaths each year in the US. There is increasing evidence to show that genetic variations can predispose individuals to such ADRs. For example, Rau et al. (2004) demonstrated that when patients were treated with antidepressants, nearly 30% of those with ADRs were CYP2D6 poor metabolizers, a condition that is normally present in only 7% of Caucasians. In addition to establishing efficacy, a key objective of clinical development is to define the potential safety issues that might be associated with a new medicine, so that the risk/benefit of the medicine can be assessed.
Mitigating risk in development. Although the full safety profile of a medicine cannot be established until it is in widespread use after launch, potential safety signals can be apparent in early phase II studies, and can have a significant (negative) impact on the risk for further development. For example, if reversible changes in liver function tests are seen in a small subset of patients in a phase II study, it can be difficult to assess the importance of this. Many valuable and effective medicines have a small impact on liver function but, on the other hand, a number of medicines have failed either in late development or after launch due to a subset of patients who exhibit these liver function changes and subsequently develop severe liver failure. If high risk patients could be identified before starting the drug (e.g. with inexpensive genetic screening), the overall safety of a medicine in use would increase considerably and the abrupt abandonment of drugs at later stages of development could be avoided.
PGx is expected to greatly illuminate the basis of safety concerns such as liver function changes seen during drug development. For example, in recent clinical studies on tranilast (a product under development for reducing restenosis after coronary angioplasty), some 8% of individuals showed an increase in unconjugated serum bilirubin, although this dissipated on termination of drug treatment. This phenotype showed some similarity to Gilbert's syndrome, a well-recognized condition characterized by episodic increases in unconjugated bilirubin but not associated with any long-term impact on liver functioning.
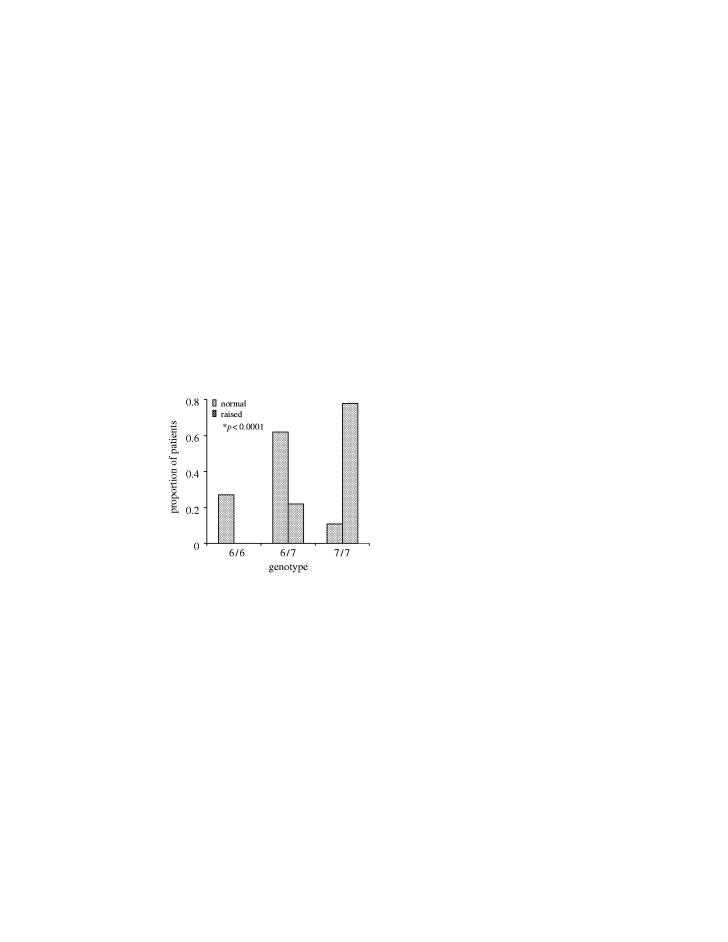
Genetic analysis of Gilbert's syndrome patients has established a strong genetic susceptibility marker in the promoter of the gene UGT-1A, where a variable TA repeat is located. ‘Wild-type’ activity is associated with six copies of the TA repeat, whereas seven copies of the TA repeat is associated with reduced expression of UGT-1A and increased propensity to Gilbert's syndrome.
Figure 4 shows the result of an association analysis studying bilirubin levels as phenotype in the tranilast-treated subjects. There is a highly significant association between the TA repeat genotype and the likelihood of developing increased levels of bilirubin after treatment with tranilast. This strongly supports the hypothesis that the observed hyperbilirubinaemia can be described as a tranilast-induced Gilbert's syndrome.
Figure 4.
Association analysis of bilirubin levels versus phenotype in tranilast treated subjects.
As the elevated bilirubin could be ascribed to a well-known, benign syndrome, with no evidence for progression to serious liver disease, the likelihood that this safety signal with tranilast could lead to serious liver complications was significantly reduced. A similar clinical observation (i.e. elevated unconjugated bilirubin in a subset of patients) has also been seen in studies with atazanavir (BMS) and the phenotype in this example was also strongly associated with the UGT-1A promoter polymorphism, suggesting a similar basis for the clinical observation.
In summary, these results not only show how PGx can provide insights into safety signals that may become apparent at phase II but also underscore the role of PGx in providing information to help research and development decision-making. This use of PGx data will be critical in facilitating the rational development of new medicines by pharmaceutical companies.
Significant numbers of samples were available from phase III studies with tranilast (where elevated bilirubin was also seen in a small percentage of subjects) and this dataset has provided significant information on the power of genetic data to identify PGx signals and to provide insights into other aspects of experimental design. For example, perhaps not surprisingly, greater power can be obtained by increasing the number of controls, while keeping the number of cases constant. Perhaps more surprising is that epidemiological controls from an unrelated (although ethnically analogous) population can be as effective as the matched controls from the phase III study sites. This is particularly true if genomic control measurements (Devlin & Roeder 1999; Pritchard & Rosenberg 1999) are performed to document and correct for stratification in the samples. Remarkably, it is clear in this example that, using a sufficient number of controls, as few as 10 cases of hyperbilirubinaemia would be needed to establish a positive association with the UGT-1A promoter polymorphism—findings which will help immensely in the design of future PGx experiments (Danoff et al. 2004).
(vi) Post-launch PGx and adverse events
Much PGx discussion has focused on the contribution of PGx to understanding rare AEs. Because there are clearly fewer cases of such AEs, it is unlikely that the approaches described above, that is, the selection of cases from a clinical trial population using the remainder of the cohort as controls, is viable; sufficient cases may only be reported once a medicine is registered and more widely available. In this case, retrospective collection of both cases and controls may be required and various approaches are being investigated to expedite efficient collection of such cases and controls to allow PGx analysis.
The studies on hypersensitivity reaction (HSR) to abacavir (ABC) have been used as proof of (i) the feasibility of retrospective case/control approaches where the number of cases (rare AEs) is comparatively small and (ii) the feasibility of genome-wide association studies. HSR affects approximately 4% of subjects with HIV who are treated with ABC and, while the clinical sequelae can be serious, rigorous clinical management of HIV subjects has prevented significant mortality due to this AE. DNA was collected from subjects reporting HSR after exposure to abacavir and from matched controls with no HSR symptoms after abacavir treatment. Genotyping included both candidate gene and genome-wide SNP association approaches.
Candidate gene analysis identified a position in the human leucocyte antigen (HLA) region on chromosome 6 with a strong association with HSR (Hetherington et al. 2002). This finding has been confirmed by at least two other laboratories. The association between markers in the HLA region is strong in Caucasians, weaker in Hispanics and cannot be detected in Blacks. Work to analyse the genome-wide association data is being finalized and will be reported in the near future.
Studies have also reported on genetic associations with Stevens Johnson Syndrome (SJS), a rare, serious cutaneous reaction that is occasionally seen in response to a range of medicines (Chung et al. 2004). The finding that the marker HLA-B*1502 is strongly associated with carbamazepine-induced SJS in the Taiwanese population is another indication that PGx technology can provide significant new insights into the basis of such rare adverse events.
This section has listed an increasing number of studies—HSR to abacavir, SJS in response to carbamazepine, hyperbilirubinaemia in response to tranilast—that have established robust PGx associations with only a few tens of adverse event cases. While this may reflect the genetic ‘truism’ that the rarer an event, the more likely there is to be a strong genetic component, it underlines the power of this technology to provide insights that were previously difficult—if not impossible—to obtain with other methods.
These studies have led to several findings. Firstly, retrospective case/control PGx studies on AEs are certainly feasible, although care must be taken to define the phenotype with great accuracy. Phenotype collection is arguably the most critical part of any genetics study and it may be difficult to collect sufficient detail for AEs observed away from the controlled, monitored clinical trial environment. Secondly, findings in one ethnic group are not necessarily applicable to others. It is unclear at this stage whether this is due to different genetic architecture at the same locus or different aetiology of the AE, but resolution of this issue is important for moving forward.
Taking the physicians' point of view, they repeatedly face the same dilemma each day: what is the right drug and what is the right starting dose for this patient? Medical training and clinical experience has taught the physician some of the important parameters that influence this decision, including the patient's age, sex, race, compliance and level of anxiety. However, as every physician knows, prescribing medications is a combination of clinical arts and inexact sciences, leading to an often bewildering variety of outcomes across a group of patients. At times, physicians may be overwhelmed by the complexity and unpredictability of a patient's response to a medication. The solution to this trial and error prescribing lies in increasing our knowledge on the causes of variable responses and side effects. A paradigm shift will need to occur in the way physicians prescribe medicines. In the coming years, automated—and progressively more inexpensive—genotyping will provide the physician with increasing amounts of information regarding how a given patient will react to a particular medication. However, most of this information will arrive as probabilities, particularly in predictions of efficacy, because of the interactions between multiple genetic and environmental determinants. This type of information may be new to physicians more used to the (admittedly spurious) binary nature of most diagnostic decisions. Physician training may need to progress to include new topics on pharmacogenetics, with input from not only pharmacology and genetics, but also from statistics, epidemiology and genetic counselling. Building on the experience of other parts of medicine—such as cardiovascular risk assessment—where estimation and communication of risk has been central for a number of years will be critical to avoid re-inventing too many wheels.
The examples given above show the potential of PGx to influence the development of new medicines and their use. When the technology is fully mature, integrated into drug development processes and incorporated into clinical practice we would expect that:
The rate of delivery of new medicines through pharmaceutical research and development will increase, utilizing a greater understanding of safety issues and defining genetically defined patient groups in which a compound is effective.
The effectiveness of these medicines in clinical use will be similarly enhanced.
High-risk individuals can be directed towards alternative therapies, reducing the mortality and morbidity associated with AEs.
The development of PGx and other technologies applicable to ‘personalization’ of medicines is proceeding at such a pace that it is not a matter of ‘if’ these technologies impact on medicine prescription, but ‘when’—and to what extent (Lesko & Woodcock 2004). Although these transformations will make better use of resources, both by healthcare providers and pharmaceutical companies, it is the lives of patients that will see the most significant differences resulting from these developments.
Footnotes
One contribution of 12 to a Discussion Meeting Issue ‘Genetic variation and human health’.
References
- Basile V.S, Masellis M, Badri F, et al. Association of the MscI polymorphism of the dopamine D3 receptor gene with tardive dyskinesia in schizophrenia. Neuropsychopharmacology. 1999;21:17–27. doi: 10.1016/S0893-133X(98)00114-6. [DOI] [PubMed] [Google Scholar]
- Campbell C.J, Ghazal P. Molecular signatures for diagnosis of infection: application of microarray technology. J. Appl. Microbiol. 2004;96:18–23. doi: 10.1046/j.1365-2672.2003.02112.x. [DOI] [PubMed] [Google Scholar]
- Chon H, et al. Broadly altered gene expression in blood leukocytes in essential hypertension is absent during treatment. Hypertension. 2004;43:947–951. doi: 10.1161/01.HYP.0000123071.35142.72. [DOI] [PubMed] [Google Scholar]
- Chung W.H, Hung S.I, Hong H.S, Hsih M.S, Yang L.C, Ho H.C, Wu J.Y, Chen Y.T. Medical genetics: a marker for Stevens–Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- Cockcroft J.R, Gazis A.G, Cross D.J, Wheatley A, Dewar J, Hall I.P, Noon J.P. Beta(2)-adrenoceptor polymorphism determines vascular reactivity in humans. Hypertension. 2000;36:371–375. doi: 10.1161/01.hyp.36.3.371. [DOI] [PubMed] [Google Scholar]
- Daly A.K. Pharmacogenetics of the major polymorphic metabolizing enzymes. Fundam. Clin. Pharmacol. 2003;17:27–41. doi: 10.1046/j.1472-8206.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- Danoff T.M, et al. A Gilbert's syndrome UGT1A1 variant confers susceptibility to tranilast-induced hyperbilirubinemia. Pharmacogenomics J. 2004;4:49–53. doi: 10.1038/sj.tpj.6500221. [DOI] [PubMed] [Google Scholar]
- Debouck C, Metcalf B. The impact of genomics on drug discovery. Annu. Rev. Pharmacol. Toxicol. 2000;40:193–207. doi: 10.1146/annurev.pharmtox.40.1.193. [DOI] [PubMed] [Google Scholar]
- DeFife K.M, Wong-Staal F. Integrated approaches to therapeutic target gene discovery. Curr. Opin. Drug Discov. Dev. 2002;5:683–689. [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- DiMasi J, Hansen R.W, Grabowski H.G. The price of innovation: new estimates of drug development costs. J. Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Dishy V, Sofowora G.G, Xie H.G, Kim R.B, Byrne D.W, Stein C.M, Wood A.J. The effect of common polymorphisms of the beta2-adrenergic receptor on agonist-mediated vascular desensitization. N. Engl. J. Med. 2001;345:1030–1035. doi: 10.1056/NEJMoa010819. [DOI] [PubMed] [Google Scholar]
- Dos Santos C, et al. A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nature Genet. 2004;36:720–724. doi: 10.1038/ng1379. [DOI] [PubMed] [Google Scholar]
- Drazen J.M, et al. Pharmacogenetic association between ALOEX5 promoter genotype and the response to anti-asthma treatment. Nature Genet. 2003;22:168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- Gilbert J. Rebuilding big PhRMA's business model. In vivo. 2003:73–80. [Google Scholar]
- Goldstein D.B, Tate S.K, Sisodiya S.M. Pharmacogenetics goes genomic. Nature Rev. Genet. 2003;4:937–947. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- Guzey C, Norstrom A, Spigset O. Change from the CYP2D6 extensive metabolizer to the poor metabolizer phenotype during treatment with bupropion. Ther. Drug Monit. 2002;24:436–437. doi: 10.1097/00007691-200206000-00018. [DOI] [PubMed] [Google Scholar]
- Hariri A.R, Weinberger D.R. Functional neuroimaging of genetic variation in serotonergic neurotransmission. Genes Brain Behav. 2003;2:341–349. doi: 10.1046/j.1601-1848.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- Hetherington S, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359:1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn-Schmiedebergs Arch. Pharmacol. 2004;369:89–104. doi: 10.1007/s00210-003-0819-z. [DOI] [PubMed] [Google Scholar]
- Israel E, et al. National Heart, Lung, and Blood Institute's Asthma Clinical Research Network. Effect of polymorphism of the beta(2)-adrenergic receptor on response to regular use of albuterol in asthma. Int. Arch. Allergy Immunol. 2001;124:183–186. doi: 10.1159/000053705. [DOI] [PubMed] [Google Scholar]
- Jacobsen P, Rossing K, Rossing P, Tarnow L, Mallet C, Poirier O, Cambien F, Parving H.H. Angiotensin converting enzyme gene polymorphism and ACE inhibition in diabetic nephropathy. Kidney Int. 1998;53:1002–1006. doi: 10.1111/j.1523-1755.1998.00847.x. [DOI] [PubMed] [Google Scholar]
- Kohno M, Yokokawa K, Minami M, Kano H, Yasunari K, Hanehira T, Yoshikawa J. Association between angiotensin-converting enzyme gene polymorphisms and regression of left ventricular hypertrophy in patients treated with angiotensin-converting enzyme inhibitors. Am. J. Med. 1999;106:544–549. doi: 10.1016/s0002-9343(99)00067-4. [DOI] [PubMed] [Google Scholar]
- Lazarou J, Pomeranz B.H, Corey P.N. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. J. Am. Med. Assoc. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- Lerer B, et al. Pharmacogenetics of tardive dyskinesia: combined analysis of 780 patients support association with dopamine D3 receptor gene Ser9Gly polymorphism (a multi-center study) Neuropsychopharmacology. 2002;27:105–119. doi: 10.1016/S0893-133X(02)00293-2. [DOI] [PubMed] [Google Scholar]
- Lesko L.J, Woodcock J. Translation of pharmacogenomics and pharmacogenetics: a regulatory perspective. Nature Rev. Drug Discov. 2004;3:763–769. doi: 10.1038/nrd1499. [DOI] [PubMed] [Google Scholar]
- Lesko L.J, et al. Pharmacogenetics and pharmacogenomics in drug development and regulatory decision making: report of the first FDA-PWG-PhRMA-DruSafe Workshop. J. Clin. Pharmacol. 2003;43:342–358. doi: 10.1177/0091270003252244. [DOI] [PubMed] [Google Scholar]
- Lima J.J, Thomason D.B, Mohamed M.H, Eberle L.V, Self T.H, Johnson J.A. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin. Pharmacol. Ther. 1999;65:519–525. doi: 10.1016/S0009-9236(99)70071-8. [DOI] [PubMed] [Google Scholar]
- Lin K.M, Poland R.E, Wan Y.Y, Smith M, Lesser I.M. The evolving science of pharmacogenetics: clinical and ethnic perspectives. Psychopharmacol. Bull. 1996;32:205–217. [PubMed] [Google Scholar]
- Lin K.M, Smith M.W, Ortiz V. Culture and psychopharmacology. Psychiatr. Clin. North Am. 2001;24:523–537. doi: 10.1016/s0193-953x(05)70245-8. [DOI] [PubMed] [Google Scholar]
- Lynch T.J, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Martinez F.D, Graves P.E, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. Clin. Invest. 1997;100:3184–3188. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundo E, Walker M, Cate T, Macciardi F, Kennedy J.L. The role of serotonin transporter protein gene in antidepressant-induced mania in bipolar disorder. Arch. Gen. Psychiatry. 2001;58:539–544. doi: 10.1001/archpsyc.58.6.539. [DOI] [PubMed] [Google Scholar]
- Murphy D.L, Lerner A, Rudnick G, Lesch K.P. Serotonin transporter: gene, genetic disorders, and pharmacogenetics. Mol. Interv. 2004;4:109–123. doi: 10.1124/mi.4.2.8. [DOI] [PubMed] [Google Scholar]
- Noble M.E, Endicott J.A, Johnson L.N. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- Ohmichi N, Iwai N, Uchida Y, Shichiri G, Nakamura Y, Kinoshita M. Relationship between the response to the angiotensin converting enzyme inhibitor imidapril and the angiotensin converting enzyme genotype. Am. J. Hypertens. 1997;10:951–955. doi: 10.1016/s0895-7061(97)00121-0. [DOI] [PubMed] [Google Scholar]
- Okamura A, et al. Pharmacogenetic analysis of the effect of angiotensin-converting enzyme inhibitor on restenosis after percutaneous transluminal coronary angioplasty. Angiology. 1999;50:811–822. doi: 10.1177/000331979905001005. [DOI] [PubMed] [Google Scholar]
- Penno G, Chaturvedi N, Talmud P.J, Cotroneo P, Manto A, Nannipieri M, Luong L.A, Fuller J.H. Effect of angiotensin-converting enzyme (ACE) gene polymorphism on progression of renal disease and the influence of ACE inhibition in IDDM patients: findings from the EUCLID Randomized Controlled Trial. EURODIAB Controlled Trial of Lisinopril in IDDM. Diabetes. 1998;47:1507–1511. doi: 10.2337/diabetes.47.9.1507. [DOI] [PubMed] [Google Scholar]
- Plumb R.S, Stumpf C.L, Granger J.H, Castro-Perez J, Haselden J.N, Dear G.J. Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid Commun. Mass Spectrom. 2003;17:2632–2638. doi: 10.1002/rcm.1250. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Rosenberg N.A. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau T, Wohlleben G, Wuttke H, Thuerauf N, Lunkenheimer J, Lanczik M, Eschenhagen T. CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants—a pilot study. Clin. Pharmacol. Ther. 2004;75:386–393. doi: 10.1016/j.clpt.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Roses A.D. Genome-based pharmacogenetics and the pharmaceutical industry. Nature Rev. Drug Discov. 2002;1:541–554. doi: 10.1038/nrd840. [DOI] [PubMed] [Google Scholar]
- Roses A.D. Pharmacogenetics and drug development: the path to safer and more effective drugs. Nature Rev. Drug Discov. 2004;3:645–656. doi: 10.1038/nrg1432. [DOI] [PubMed] [Google Scholar]
- Roses A.D, Burns D.K, Chissoe S, Middleton L, St Jean P. Disease-specific target selection: a critical first step down the right road. Drug Discov. Today. 2005;10:177–189. doi: 10.1016/S1359-6446(04)03321-5. [DOI] [PubMed] [Google Scholar]
- Searls D.B. Pharmacophylogenomics: genes, evolution & drug targets. Nature Rev. Drug Discov. 2003;2:613–623. doi: 10.1038/nrd1152. [DOI] [PubMed] [Google Scholar]
- Smeraldi E, Zanardi R, Benedetti F, De Bella D, Perez J, Catalano M. Polymorphism within the promoter of the serotonin transporter gene and antidepressant efficacy of fluvoxamine. Mol. Psychiatry. 1998;3:508–511. doi: 10.1038/sj.mp.4000425. [DOI] [PubMed] [Google Scholar]
- Steen V.M, Lovlie R, MacEwan T, McCreadie R.G. Dopamine D3-receptor gene variant and susceptibility to tardive dyskinesia in schizophrenic patients. Mol. Psychiatry. 1997;2:139–145. doi: 10.1038/sj.mp.4000249. [DOI] [PubMed] [Google Scholar]
- Subramanian G, Adams M.D, Ventnre J.C, Broder S. Implications of the human genome for understanding human biology and medicine. J. Am. Med. Assoc. 2001;286:2296–2307. doi: 10.1001/jama.286.18.2296. [DOI] [PubMed] [Google Scholar]
- Tantisira K.G, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 2004;13:1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- Vogel F. Moderne probleme der Humangenetik. Ergeb. Inn. Med. Kinderheilkd. 1959;12:52–125. [Google Scholar]
- Vogel C.L, Franco S.X. Clinical experience with trastuzumab (herceptin) Breast J. 2003;9:452–462. doi: 10.1046/j.1524-4741.2003.09602.x. [DOI] [PubMed] [Google Scholar]
- Weatherall D.J. Towards molecular medicine; reminiscences of the haemoglobin field, 1960–2000. Br. J. Haematol. 2001;115:729–738. doi: 10.1046/j.1365-2141.2001.03227.x. [DOI] [PubMed] [Google Scholar]
- Yamanouchi Y, Iwata N, Suzuki T, Kitajima T, Ikeda M, Ozaki N. Effect of DRD2, 5-HT2A, and COMT genes on antipsychotic response to risperidone. Pharmacogenomics J. 2003;3:356–361. doi: 10.1038/sj.tpj.6500211. [DOI] [PubMed] [Google Scholar]